Optimizing the Experimental Method for Stomata-Profiling Automation of Soybean Leaves Based on Deep Learning
Abstract
:1. Introduction
2. Results
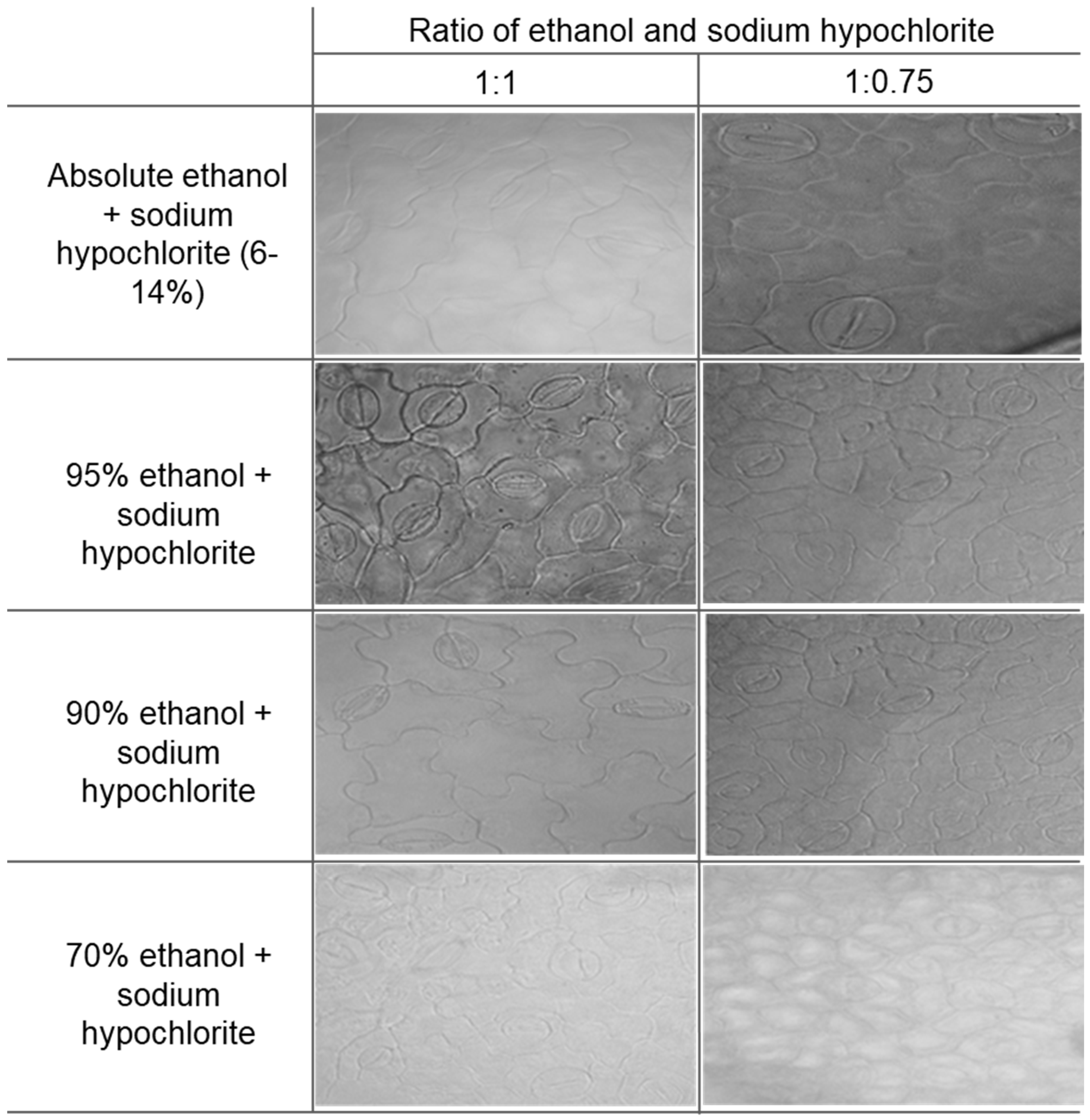
2.1. Experimental Method Optimization for Clear Stomatal Pictures
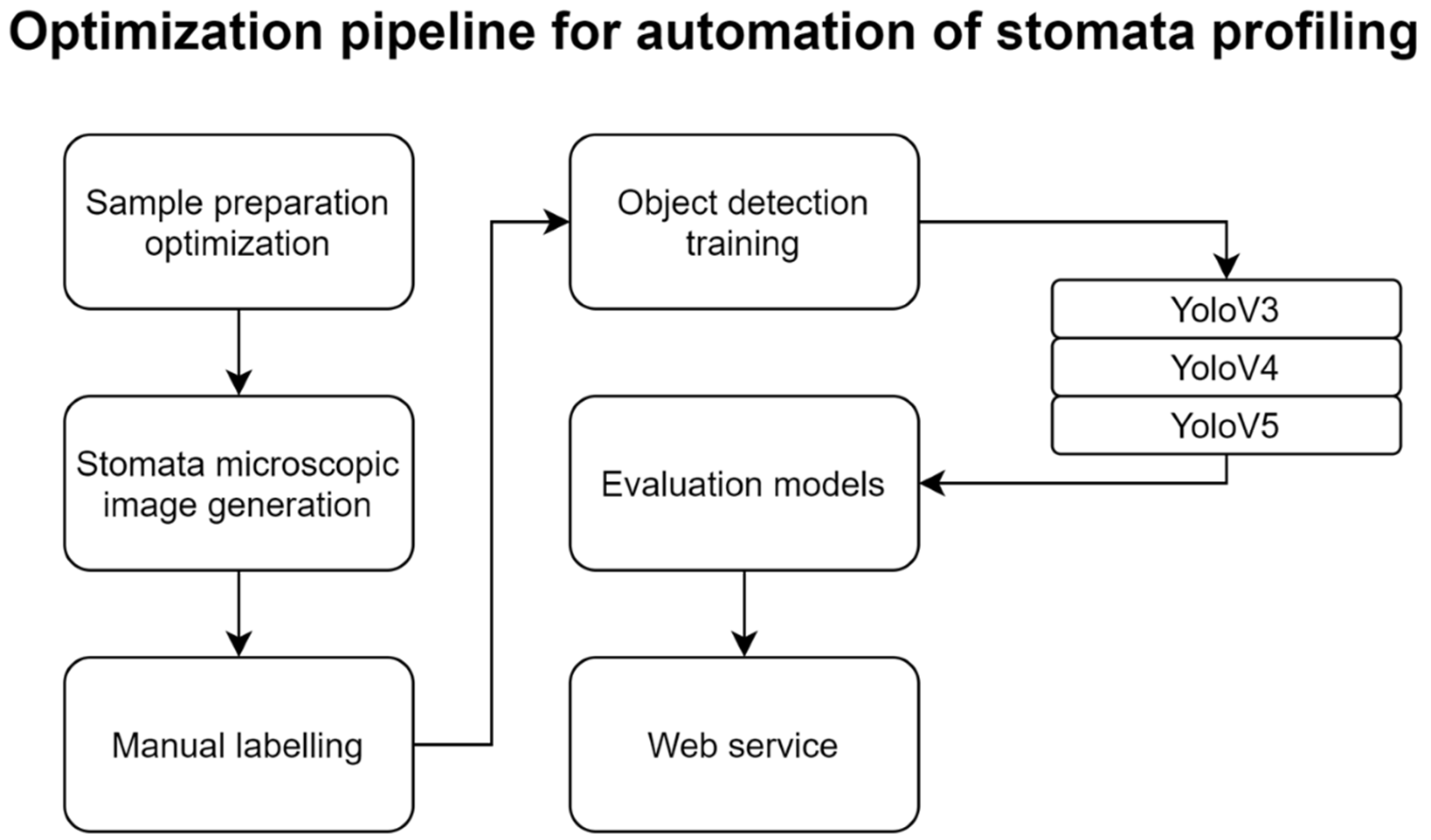
2.2. Optimization of Experimental Methods for Automating Stomata-Profiling
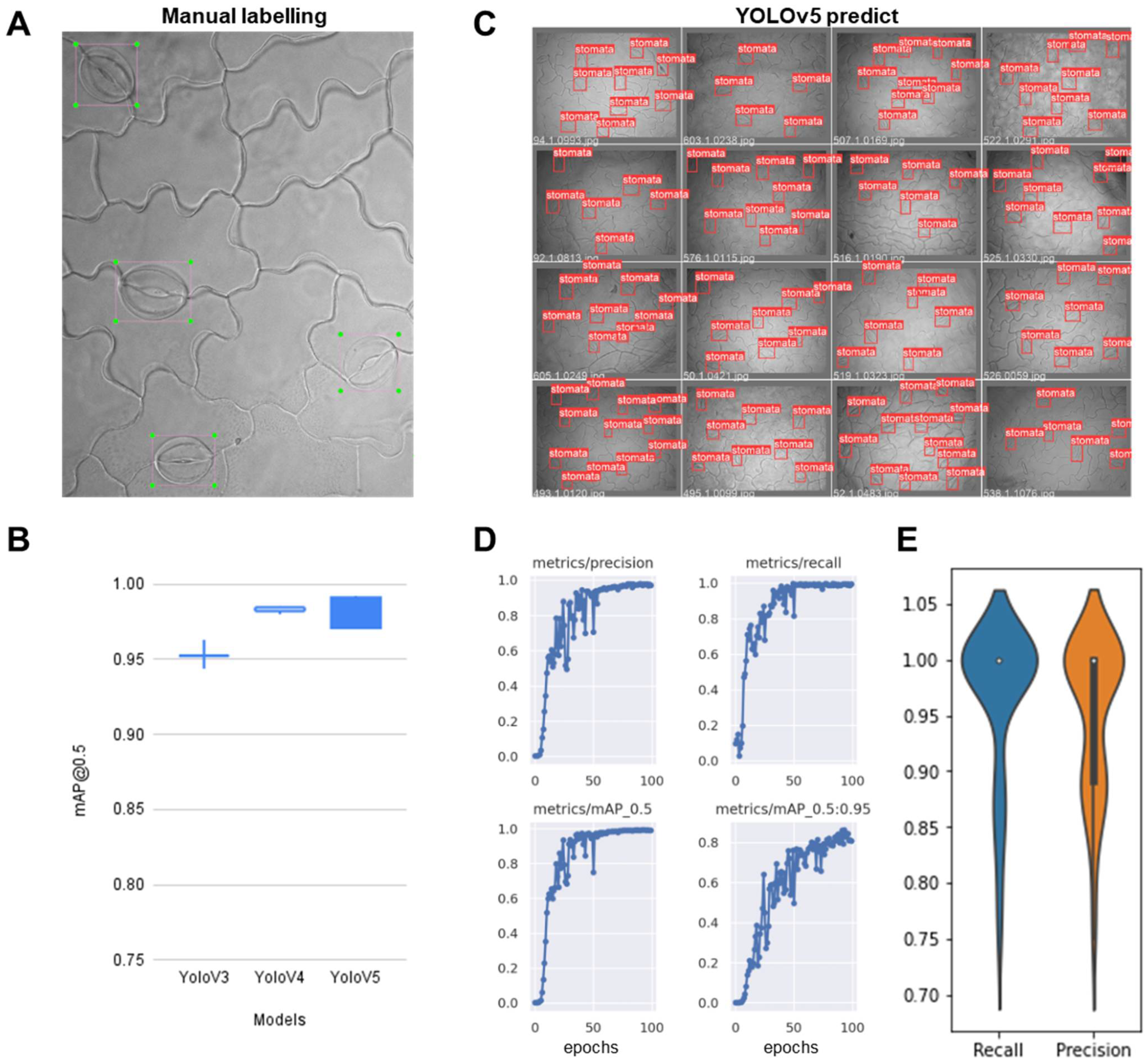
2.2.1. Training the Stomata-Detection Model
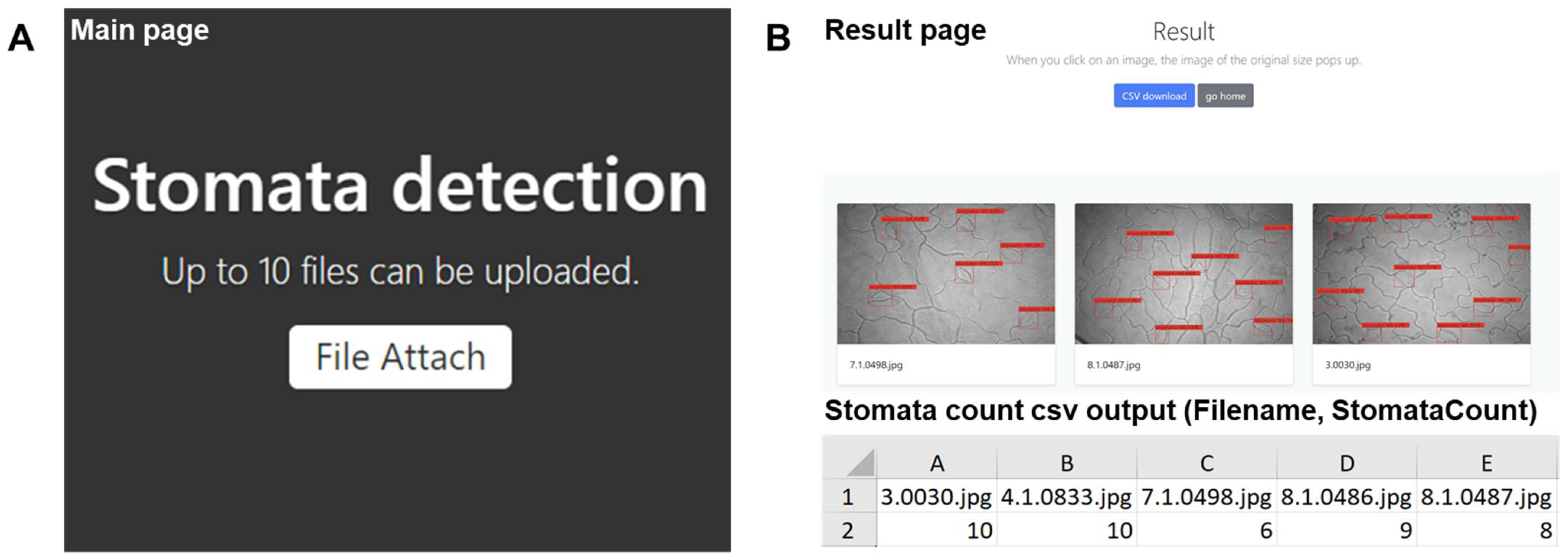
2.2.2. Development of Web Applications to Serve the Stomata-Detection Model
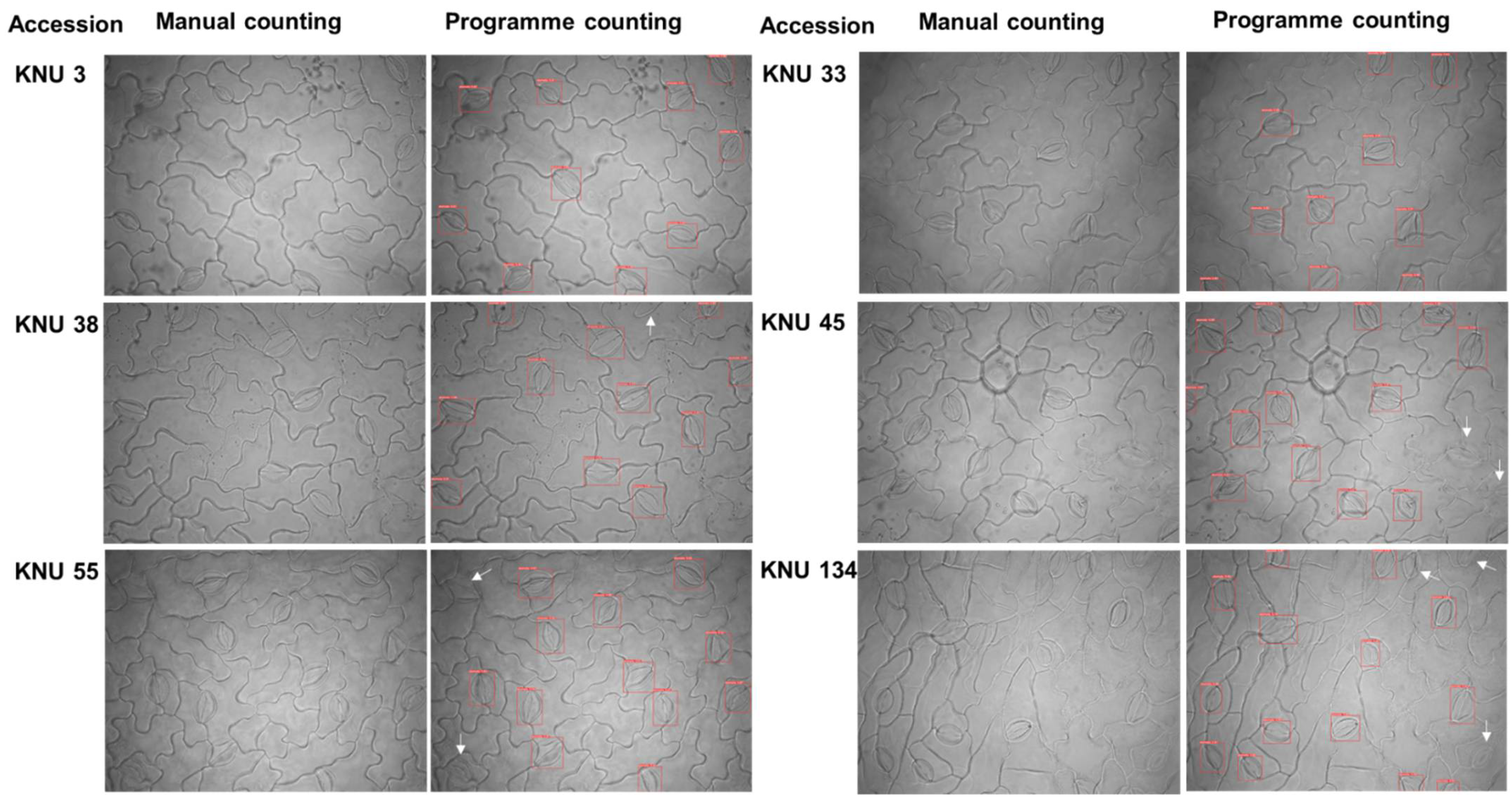
2.2.3. Validation Test Compared Manual Stomatal Counting and Web Applications of Automatic Stomata-Detection Model
3. Discussion
4. Materials and Methods
4.1. Soybean Materials
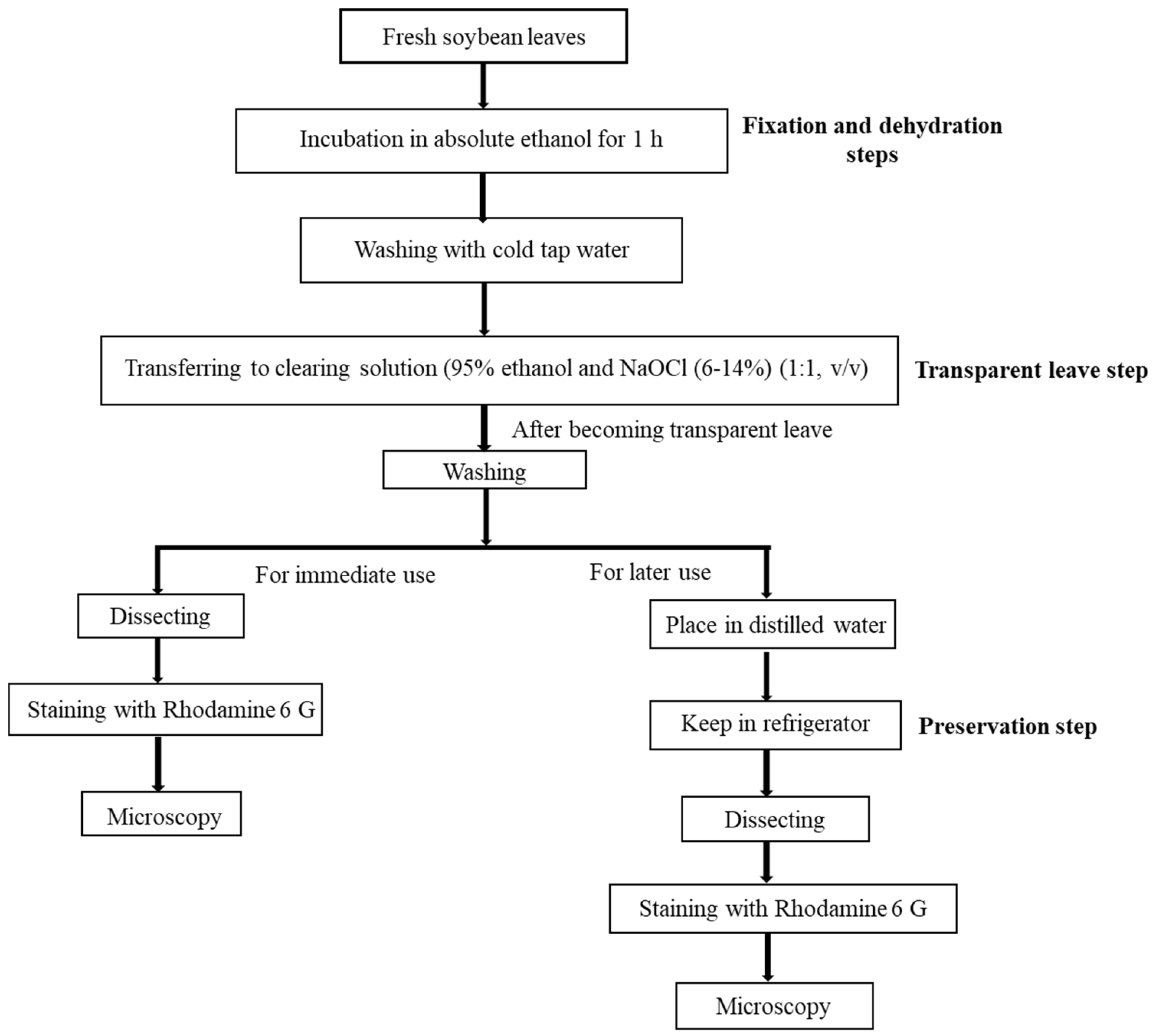
4.1.1. Experiment 1 (Development of a New Leaf-Clearing Process and Preservation Test)
4.1.2. Experiment 2 (Model Development for Automated Stomata-Profiling)
4.2. Modified Clearing Agent Method and Preservation Test to Capture Stomatal Impressions
4.3. Training Object Detection Model
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chapman, A.D. Numbers of Living Species in Australia and the World; Department of the Environment, Water Heritage and the Arts: Canberra, Australia, 2009; ISBN 9780642568601. [Google Scholar]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef] [Green Version]
- Lawson, T.; Kramer, D.M.; Raines, C.A. Improving yield by exploiting mechanisms underlying natural variation of photosynthesis. Curr. Opin. Biotechnol. 2012, 23, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Franks, P.J.; Adams, M.A.; Amthor, J.S.; Barbour, M.M.; Berry, J.A.; Ellsworth, D.S.; Farquhar, G.D.; Ghannoum, O.; Lloyd, J.; McDowell, N.; et al. Sensitivity of plants to changing atmospheric CO2 concentration: From the geological past to the next century. New Phytol. 2013, 197, 1077–1094. [Google Scholar] [CrossRef] [PubMed]
- Engineer, C.B.; Hashimoto-Sugimoto, M.; Negi, J.; Israelsson-Nordström, M.; Azoulay-Shemer, T.; Rappel, W.J.; Iba, K.; Schroeder, J.I. CO2 sensing and CO2 regulation of stomatal conductance: Advances and open questions. Trends Plant Sci. 2016, 21, 16–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunn, J.; Hunt, L.; Afsharinafar, M.; Meselmani, M.A.; Mitchell, A.; Howells, R.; Wallington, E.; Fleming, A.J.; Gray, J.E. Reduced stomatal density in bread wheat leads to increased water-use efficiency. J. Exp. Bot. 2019, 70, 4737–4748. [Google Scholar] [CrossRef]
- Lecoeur, J.; Wery, J.; Turc, O.; Tardieu, F. Expansion of pea leaves subjected to short water deficit: Cell number and cell size are sensitive to stress at different periods of leaf development. J. Exp. Bot. 1995, 46, 1093–1101. [Google Scholar] [CrossRef]
- Zhao, R.X.; Zhang, Q.B.; Wu, X.Y.; Wang, Y. The effects of drought on epidermal cells and stomatal density of wheat leaves. Inn. Mong. Agric. Sci. Technol. 2001, 6, 6–7. [Google Scholar]
- Galmés, J.; Flexas, J.; Savé, R.; Medrano, H. Water relations and stomatal characteristics of Mediterranean plants with different growth forms and leaf habits: Responses to water stress and recovery. Plant Soil 2007, 290, 139–155. [Google Scholar] [CrossRef]
- Shu-li, Z.; Wenfu, C.; Dianrong, M.; Fei, Z. Influence of Different Salt Level on Stomatal Character in Rice Leaves. Reclaiming Rice Cultiv. 2006, 6, 26–29. [Google Scholar]
- Beerling, D.J.; Chaloner, W.G. The impact of atmospheric CO2 and temperature changes on stomatal density: Observation from Quercus robur lammas leaves. Ann. Bot. 1993, 71, 231–235. [Google Scholar] [CrossRef]
- Limin, Y.; Mei, H.; Guangsheng, Z.; Jiandong, L. The changes in water-use efficiency and stoma density of Leymus chinensis along Northeast China Transect. Acta Ecol. Sin. 2007, 27, 16–23. [Google Scholar] [CrossRef]
- Sakoda, K.; Watanabe, T.; Sukemura, S.; Kobayashi, S.; Nagasaki, Y.; Tanaka, Y.; Shiraiwa, T. Genetic Diversity in stomatal Density among soybeans elucidated using high-throughput technique based on an algorithm for object detection. Sci. Rep. 2019, 9, 7610. [Google Scholar] [CrossRef]
- Gitz, D.C.; Baker, J.T. Methods for creating stomatal impressions directly onto archivable slides. Agron. J. 2009, 101, 232–236. [Google Scholar] [CrossRef]
- Chieco, C.; Rotondi, A.; Morrone, L.; Rapparini, F.; Baraldi, R. An ethanol-based fixation method for anatomical and micro-morphological characterization of leaves of various tree species. Biotech. Histochem. 2013, 88, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Gitz, D.C., III; Liu-Gitz, L.; Britz, S.J.; Sullivan, J.H. Ultraviolet-B effects on stomatal density, water-use efficiency, and stable carbon isotope discrimination in four glasshouse-grown soybean (Glycine max) cultivars. Environ. Exp. Bot. 2005, 53, 343–355. [Google Scholar] [CrossRef]
- Aslam, Y.; Santhi, N. A Review of Deep Learning Approaches for Image Analysis. In Proceedings of the 2019 IEEE International Conference on Smart Systems and Inventive Technology (ICSSIT), Tirunelveli, India, 27–29 November 2019; pp. 709–714. [Google Scholar] [CrossRef]
- Zhao, Z.-Q.; Zheng, P.; Xu, S.-T.; Wu, X. Object detection with deep learning: A review. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3212–3232. [Google Scholar] [CrossRef] [Green Version]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pest’s recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Song, C.; Zhang, D. Deep learning-based object detection improvement for tomato disease. IEEE Access 2020, 8, 56607–56614. [Google Scholar] [CrossRef]
- Fetter, K.C.; Eberhardt, S.; Barclay, R.S.; Wing, S.; Keller, S.R. Stomata Counter: A neural network for automatic stomata identification and counting. New Phytol. 2019, 223, 1671–1681. [Google Scholar] [CrossRef]
- Song, W.; Li, J.; Li, K.; Chen, J.; Huang, J. An automatic method for stomatal pore detection and measurement in microscope images of plant leaf based on a convolutional neural network model. Forests 2020, 11, 954. [Google Scholar] [CrossRef]
- Metusala, D. An alternative simple method for preparing and preserving cross-section of leaves and roots in herbaceous plants: Case study in Orchidaceae. In AIP Conference Proceedings; AIP Publishing LLC: Melville, NY, USA, 2017; Volume 1862, p. 030113. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Wang, X.; Zhou, H.; Li, Y.; Zhang, J.; Yu, S.; Wang, M.; Hao, M.; Zhao, Q.; Liu, L.; et al. Comparison of sample preparation techniques for inspection of leaf epidermises using light microscopy and scanning electronic microscopy. Front. Plant Sci. 2020, 11, 133. [Google Scholar] [CrossRef]
- Sharma, N. Leaf clearing protocol to observe stomata and other cells on leaf surface. Bio-Protocol 2017, 7, e2538. [Google Scholar] [CrossRef]
- Redmon, J.; Farhadi, A. Yolov3: An incremental improvement. arXiv 2018, arXiv:1804.02767. [Google Scholar]
- Reese, W. Nginx: The high-performance web server and reverse proxy. Linux J. 2008, 173, 2. [Google Scholar]
- Cole, D.F.; Dobrenz, A.K. Stomate density of alfalfa (Medicago sativa L.) 1. Crop Sci. 1970, 10, 61–63. [Google Scholar] [CrossRef]
- Patel, I.; Gorim, L.Y.; Tanino, K.; Vandenberg, A. Diversity in surface microstructures of trichomes, epidermal cells, and stomata in lentil germplasm. Front. Plant Sci. 2021, 1385, 697692. [Google Scholar] [CrossRef] [PubMed]
- Peksen, E.; Peksen, A.; Artik, C. Comparison of leaf and stomatal characteristics in faba bean (Vicia faba L.). Res. J. Biol. Sci. 2006, 6, 360–364. [Google Scholar] [CrossRef] [Green Version]
- Ciha, A.J.; Brun, W.A. Stomatal size and frequency in soybeans 1. Crop Sci. 1975, 15, 309–313. [Google Scholar] [CrossRef]
- Tanaka, Y.; Fujii, K.; Shiraiwa, T. Variability of leaf morphology and stomatal conductance in soybean [Glycine max (L.) Merr.] cultivars. Crop Sci. 2010, 50, 2525–2532. [Google Scholar] [CrossRef]
- Kirkham, M.B. Principles of Soil and Plant Water Relations; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar] [CrossRef]
- Bozoglu, H.; Karayel, R. Investigation of stomata densities in Pea (Pisum sativum L.) lines/cultivars. Res. J. Biol. Sci. 2006, 6, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Pallas, J.; James, E. Photosynthetic traits of selected peanut genotypes. Peanut Sci. 1982, 9, 14–17. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, M.A.; Lin, X.; Chan, T.F.; Ham, J.; Shin, T.S.; Ercisli, S.; Golokhvast, K.S.; Lam, H.M.; Chung, G. Korean wild soybeans (Glycine soja Sieb & Zucc.): Geographic distribution and germplasm conservation. Agronomy 2020, 10, 214. [Google Scholar] [CrossRef] [Green Version]
- Hong, T.; Lin, H.; He, D. Characteristics and correlations of leaf stomata in different Aleurites montana provenances. PLoS ONE 2018, 13, e0208899. [Google Scholar] [CrossRef]
- Harrison, E.L.; Arce Cubas, L.; Gray, J.E.; Hepworth, C. The influence of stomatal morphology and distribution on photosynthetic gas exchange. Plant J. 2020, 101, 768–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papanatsiou, M.; Amtmann, A.; Blatt, M.R. Stomatal spacing safeguards stomatal dynamics by facilitating guard cell ion transport independent of the epidermal solute reservoir. Plant Physiol. 2016, 172, 254–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasco, A.; Thadeo, M.; Conover, M.; Daly, D.C. Preparation of samples for leaf architecture studies, a method for mounting cleared leaves. Appl. Plant Sci. 2014, 2, 1400038. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.M. The Handbook of Histopathological Practices in Aquatic Environments: Guide to Histology for Environmental Toxicology; Academic Press: Cambridge, MA, USA, 2017. [Google Scholar]
- Lai, M.; Lü, B. Tissue preparation for microscopy and histology. In Comprehensive Sampling and Sample Preparation; Academic Press: Cambridge, MA, USA, 2012. [Google Scholar] [CrossRef]
- Panzacchi, S.; Gnudi, F.; Mandrioli, D.; Montella, R.; Strollo, V.; Merrick, B.A.; Belpoggi, F.; Tibaldi, E. Effects of short and long-term alcohol-based fixation on Sprague-Dawley rat tissue morphology, protein and nucleic acid preservation. Acta Histochem. 2019, 121, 750–760. [Google Scholar] [CrossRef]
- Soukup, A.; Tylová, E. Essential methods of plant sample preparation for light microscopy. In Plant Cell Morphogenesis; Humana Press: Totowa, NJ, USA, 2014; pp. 1–23. [Google Scholar] [CrossRef]
- Gardner, R.O. An overview of botanical clearing technique. Stain. Technol. 1975, 50, 99–105. [Google Scholar] [CrossRef]
- García-Gutiérrez, E.; Ortega-Escalona, F.; Angeles, G. A novel, rapid technique for clearing leaf tissues. Appl. Plant Sci. 2020, 8, e11391. [Google Scholar] [CrossRef]
- Lee, C.; Choi, M.-S.; Kim, H.-T.; Yun, H.-T.; Lee, B.; Chung, Y.-S.; Kim, R.W.; Choi, H.K. Soybean [Glycine max (L.) Merrill]: Importance as a crop and pedigree reconstruction of Korean varieties. Plant Breed. Biotechnol. 2015, 3, 179–196. [Google Scholar] [CrossRef] [Green Version]
- Jeong, N.; Kim, K.S.; Jeong, S.; Kim, J.Y.; Park, S.K.; Lee, J.S.; Jeong, S.C.; Kang, S.T.; Ha, B.K.; Kim, D.Y.; et al. Korean soybean core collection: Genotypic and phenotypic diversity population structure and genome-wide association study. PLoS ONE 2019, 14, e0224074. [Google Scholar] [CrossRef] [PubMed]
- Eisele, J.F.; Fäßler, F.; Bürgel, P.F.; Chaban, C. A rapid and simple method for microscopy-based stomata analyses. PLoS ONE 2016, 11, e0164576. [Google Scholar] [CrossRef] [PubMed]
- Chollet, F. Others Keras. Available online: https://keras.io (accessed on 30 September 2021).


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sultana, S.N.; Park, H.; Choi, S.H.; Jo, H.; Song, J.T.; Lee, J.-D.; Kang, Y.J. Optimizing the Experimental Method for Stomata-Profiling Automation of Soybean Leaves Based on Deep Learning. Plants 2021, 10, 2714. https://doi.org/10.3390/plants10122714
Sultana SN, Park H, Choi SH, Jo H, Song JT, Lee J-D, Kang YJ. Optimizing the Experimental Method for Stomata-Profiling Automation of Soybean Leaves Based on Deep Learning. Plants. 2021; 10(12):2714. https://doi.org/10.3390/plants10122714
Chicago/Turabian StyleSultana, Syada Nizer, Halim Park, Sung Hoon Choi, Hyun Jo, Jong Tae Song, Jeong-Dong Lee, and Yang Jae Kang. 2021. "Optimizing the Experimental Method for Stomata-Profiling Automation of Soybean Leaves Based on Deep Learning" Plants 10, no. 12: 2714. https://doi.org/10.3390/plants10122714
APA StyleSultana, S. N., Park, H., Choi, S. H., Jo, H., Song, J. T., Lee, J.-D., & Kang, Y. J. (2021). Optimizing the Experimental Method for Stomata-Profiling Automation of Soybean Leaves Based on Deep Learning. Plants, 10(12), 2714. https://doi.org/10.3390/plants10122714







