Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline
Abstract
:1. Introduction
2. Results
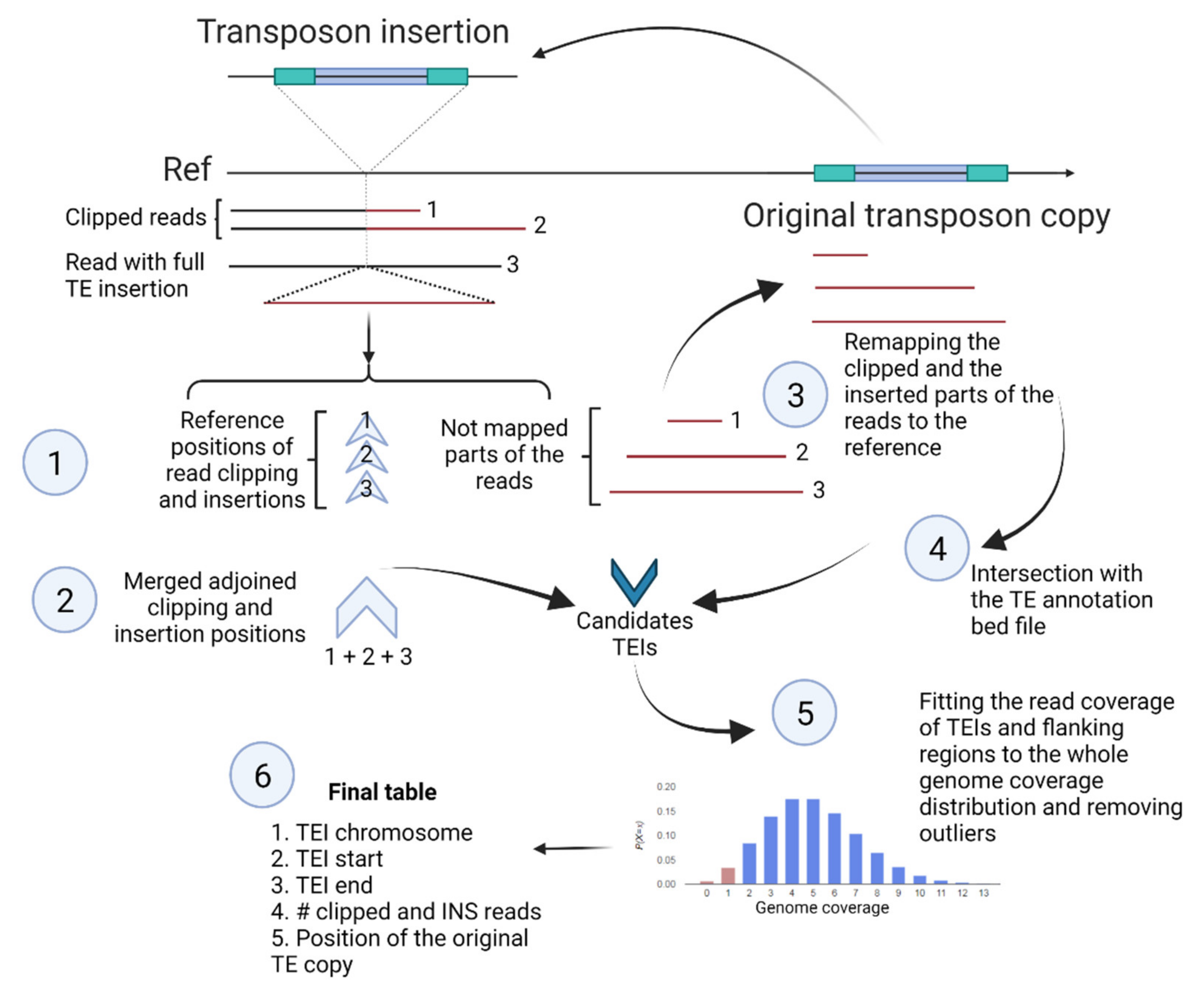
2.1. Nanotei—A New Pipeline for Genome-Wide Transposon Insertion Detection from Nanopore Data
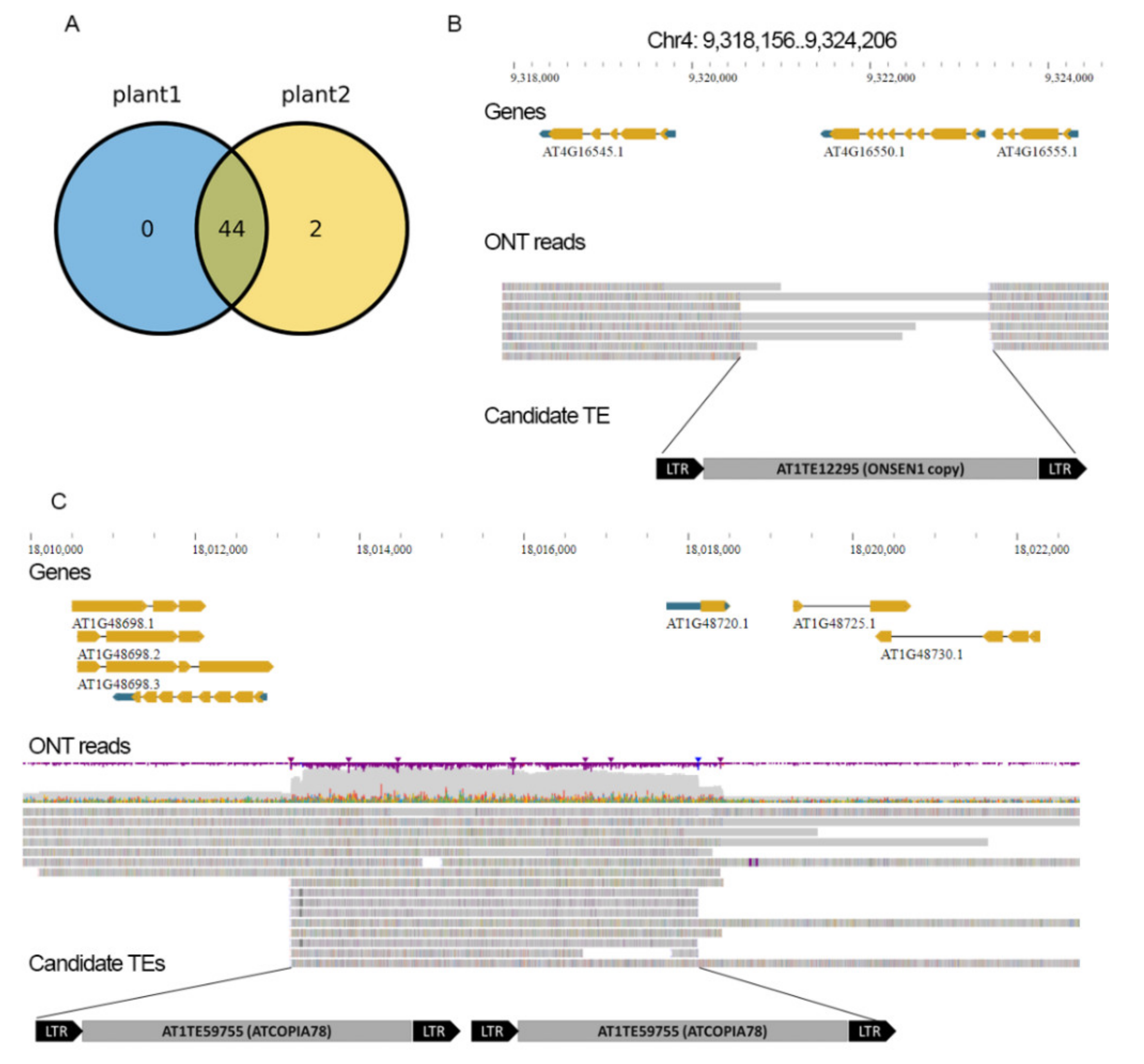
2.2. Numerous TE Copies Are Hidden in TAIR10 Genome Assembly Gaps
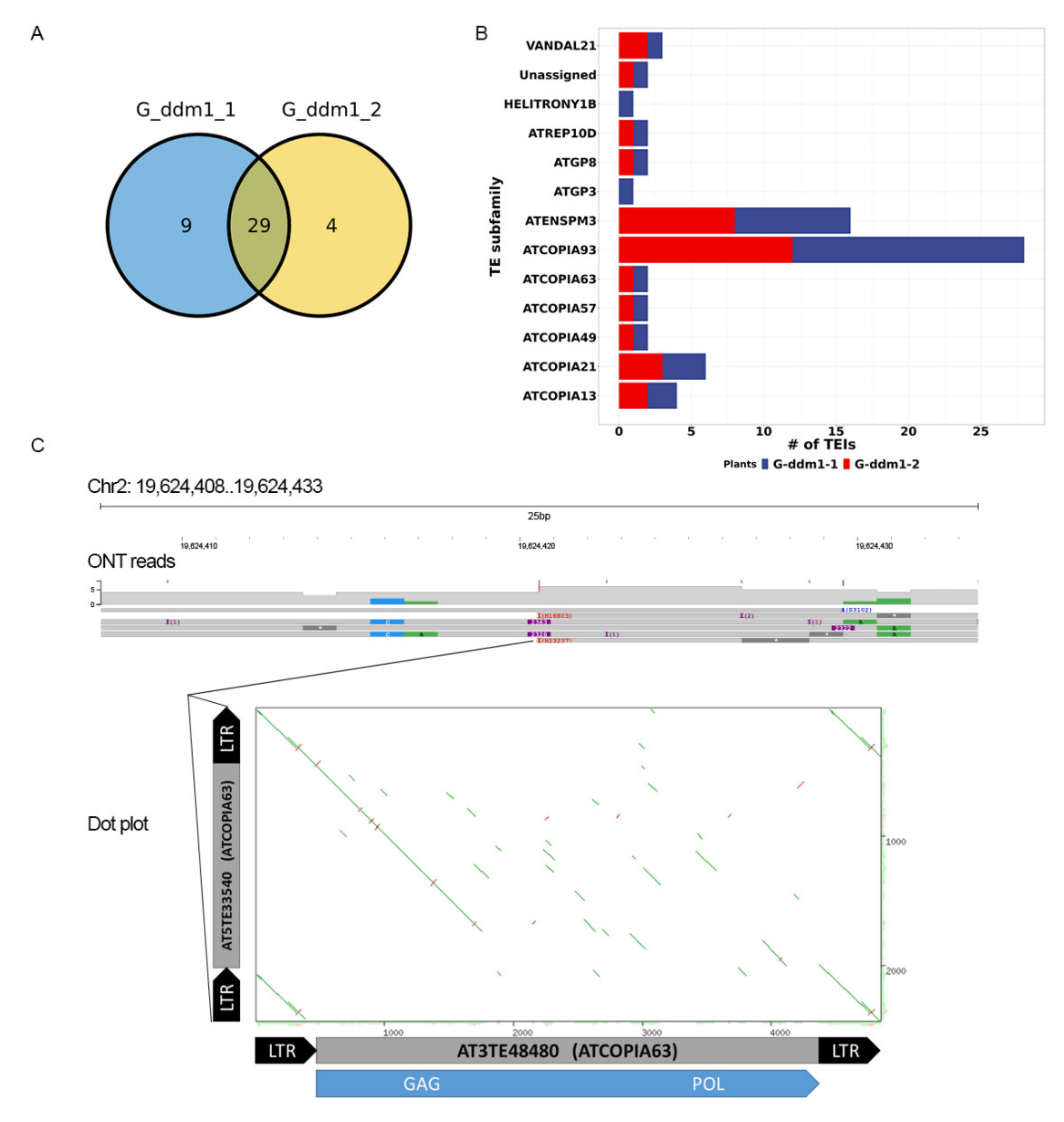
2.3. Known and New Active TEs of the ddm1 Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. HMW DNA Isolation and Size Selection
4.3. Nanopore Sequencing and Basecalling
4.4. Nanotei Pipeline
4.5. Manual Curation of TEIs
4.6. Statistics and Data Visualization
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rabanus-Wallace, M.T.; Hackauf, B.; Mascher, M.; Lux, T.; Wicker, T.; Gundlach, H.; Baez, M.; Houben, A.; Mayer, K.F.X.; Guo, L.; et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021, 53, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Bortolini Silveira, A.; Mayhew, G.F.; LeBlanc, C.; Martienssen, R.A.; Jeddeloh, J.A.; Colot, V. The Arabidopsis thaliana mobilome and its impact at the species level. eLife 2016, 5, e15716. [Google Scholar] [CrossRef]
- Song, X.; Cao, X. Transposon-mediated epigenetic regulation contributes to phenotypic diversity and environmental adaptation in rice. Curr. Opin. Plant Biol. 2017, 36, 111–118. [Google Scholar] [CrossRef]
- Baduel, P.; Quadrana, L.; Hunter, B.; Bomblies, K.; Colot, V. Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation. Nat. Commun. 2019, 10, 5818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baduel, P.; Quadrana, L. Jumpstarting evolution: How transposition can facilitate adaptation to rapid environmental changes. Curr. Opin. Plant Biol. 2021, 61, 102043. [Google Scholar] [CrossRef] [PubMed]
- Vitte, C.; Fustier, M.-A.; Alix, K.; Tenaillon, M.I. The bright side of transposons in crop evolution. Brief. Funct. Genom. 2014, 13, 276–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgante, M.; Brunner, S.; Pea, G.; Fengler, K.; Zuccolo, A.; Rafalski, A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005, 37, 997–1002. [Google Scholar] [CrossRef]
- Domínguez, M.; Dugas, E.; Benchouaia, M.; Leduque, B.; Jiménez-Gómez, J.M.; Colot, V.; Quadrana, L. The impact of transposable elements on tomato diversity. Nat. Commun. 2020, 11, 4058. [Google Scholar] [CrossRef]
- Carpentier, M.C.; Manfroi, E.; Wei, F.J.; Wu, H.P.; Lasserre, E.; Llauro, C.; Debladis, E.; Akakpo, R.; Hsing, Y.I.; Panaud, O. Retrotranspositional landscape of Asian rice revealed by 3000 genomes. Nat. Commun. 2019, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Lye, Z.N.; Purugganan, M.D. Copy Number Variation in Domestication. Trends Plant Sci. 2019, 24, 352–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonge, M.; Wang, X.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell 2020, 182, 145–161.e23. [Google Scholar] [CrossRef]
- Akakpo, R.; Carpentier, M.-C.; Ie Hsing, Y.; Panaud, O. The impact of transposable elements on the structure, evolution and function of the rice genome. New Phytol. 2020, 226, 44–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makałowski, W.; Pande, A.; Gotea, V.; Makałowska, I. Transposable Elements and Their Identification. In Evolutionary Genomics: Statistical and Computational Methods; Anisimova, M., Ed.; Humana Press: Totowa, NJ, USA, 2012; Volume 1, pp. 337–359. [Google Scholar] [CrossRef]
- Ewing, A.D. Transposable element detection from whole genome sequence data. Mob. DNA 2015, 6, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabot, F.; Picault, N.; El-Baidouri, M.; Llauro, C.; Chaparro, C.; Piegu, B.; Roulin, A.; Guiderdoni, E.; Delabastide, M.; McCombie, R.; et al. Transpositional landscape of the rice genome revealed by paired-end mapping of high-throughput re-sequencing data. Plant J. 2011, 66, 241–246. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Slotkin, R.K. The current revolution in transposable element biology enabled by long reads. Curr. Opin. Plant Biol. 2020, 54, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Kirov, I.; Dudnikov, M.; Merkulov, P.; Shingaliev, A.; Omarov, M.; Kolganova, E.; Sigaeva, A.; Karlov, G.; Soloviev, A. Nanopore RNA Sequencing Revealed Long Non-Coding and LTR Retrotransposon-Related RNAs Expressed at Early Stages of Triticale SEED Development. Plants 2020, 9, 1794. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, M.; VanKuren, N.W.; Zhao, R.; Zhang, X.; Kalsow, S.; Emerson, J.J. Hidden genetic variation shapes the structure of functional elements in Drosophila. Nat. Genet. 2018, 50, 20–25. [Google Scholar] [CrossRef] [Green Version]
- Rech, G.E.; Radío, S.; Guirao-Rico, S.; Aguilera, L.; Horvath, V.; Green, L.; Lindstadt, H.; Jamilloux, V.; Quesneville, H.; González, J. Population-scale long-read sequencing uncovers transposable elements contributing to gene expression variation and associated with adaptive signatures in Drosophila melanogaster. bioRxiv 2021. [Google Scholar] [CrossRef]
- Debladis, E.; Llauro, C.; Carpentier, M.-C.; Mirouze, M.; Panaud, O. Detection of active transposable elements in Arabidopsis thaliana using Oxford Nanopore Sequencing technology. BMC Genom. 2017, 18, 537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ni, P.; Huang, N.; Nie, F.; Zhang, J.; Zhang, Z.; Wu, B.; Bai, L.; Liu, W.; Xiao, C.-L.; Luo, F.; et al. Genome-wide detection of cytosine methylations in plant from Nanopore data using deep learning. Nat. Commun. 2021, 12, 5976. [Google Scholar] [CrossRef] [PubMed]
- Belser, C.; Istace, B.; Denis, E.; Dubarry, M.; Baurens, F.-C.; Falentin, C.; Genete, M.; Berrabah, W.; Chèvre, A.-M.; Delourme, R.; et al. Chromosome-scale assemblies of plant genomes using nanopore long reads and optical maps. Nat. Plants 2018, 4, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Borges-Monroy, R.; Viswanadham, V.V.; Lee, S.; Li, H.; Lee, E.A.; Park, P.J. Comprehensive identification of transposable element insertions using multiple sequencing technologies. Nat. Commun. 2021, 12, 3836. [Google Scholar] [CrossRef] [PubMed]
- Ewing, A.D.; Smits, N.; Sanchez-Luque, F.J.; Faivre, J.; Brennan, P.M.; Richardson, S.R.; Cheetham, S.W.; Faulkner, G.J. Nanopore Sequencing Enables Comprehensive Transposable Element Epigenomic Profiling. Mol. Cell 2020, 80, 915–928.e915. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Emery, S.B.; Flasch, D.A.; Wang, Y.; Kwan, K.Y.; Kidd, J.M.; Moran, J.V.; Mills, R.E. Identification and characterization of occult human-specific LINE-1 insertions using long-read sequencing technology. Nucleic Acids Res. 2019, 48, 1146–1163. [Google Scholar] [CrossRef]
- Jeddeloh, J.A.; Bender, J.; Richards, E.J. The DNA methylation locus DDM1 is required for maintenance of gene silencing in Arabidopsis. Genes Dev. 1998, 12, 1714–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, A.; Yonebayashi, S.; Watanabe, K.; Toyama, T.; Shimada, H.; Kakutani, T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature 2001, 411, 212–214. [Google Scholar] [CrossRef] [PubMed]
- Saze, H.; Kakutani, T. Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 2007, 26, 3641–3652. [Google Scholar] [CrossRef] [PubMed]
- Vongs, A.; Kakutani, T.; Martienssen, R.A.; Richards, E.J. Arabidopsis thaliana DNA methylation mutants. Science 1993, 260, 1926–1928. [Google Scholar] [CrossRef]
- Tsukahara, S.; Kobayashi, A.; Kawabe, A.; Mathieu, O.; Miura, A.; Kakutani, T. Bursts of retrotransposition reproduced in Arabidopsis. Nature 2009, 461, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Kawabe, A.; Etcheverry, M.; Ito, T.; Toyoda, A.; Fujiyama, A.; Colot, V.; Tarutani, Y.; Kakutani, T. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J. 2013, 32, 2407–2417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panda, K.; Slotkin, R.K. Long-Read cDNA Sequencing Enables a “Gene-Like” Transcript Annotation of Transposable Elements. Plant Cell 2020, 32, 2687–2698. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Etcheverry, M.; Gilly, A.; Caillieux, E.; Madoui, M.-A.; Guy, J.; Bortolini Silveira, A.; Engelen, S.; Baillet, V.; Wincker, P.; et al. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat. Commun. 2019, 10, 3421. [Google Scholar] [CrossRef]
- Chaparro, C.; Gayraud, T.; de Souza, R.F.; Domingues, D.S.; Akaffou, S.; Laforga Vanzela, A.L.; Kochko, A.d.; Rigoreau, M.; Crouzillat, D.; Hamon, S.; et al. Terminal-Repeat Retrotransposons with GAG Domain in Plant Genomes: A New Testimony on the Complex World of Transposable Elements. Genome Biol. Evol. 2015, 7, 493–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirov, I.; Omarov, M.; Merkulov, P.; Dudnikov, M.; Gvaramiya, S.; Kolganova, E.; Komakhin, R.; Karlov, G.; Soloviev, A. Genomic and Transcriptomic Survey Provides New Insight into the Organization and Transposition Activity of Highly Expressed LTR Retrotransposons of Sunflower (Helianthus annuus L.). Int. J. Mol. Sci. 2020, 21, 9331. [Google Scholar] [CrossRef] [PubMed]
- Mirouze, M.; Reinders, J.; Bucher, E.; Nishimura, T.; Schneeberger, K.; Ossowski, S.; Cao, J.; Weigel, D.; Paszkowski, J.; Mathieu, O. Selective epigenetic control of retrotransposition in Arabidopsis. Nature 2009, 461, 427–430. [Google Scholar] [CrossRef] [PubMed]
- Lanciano, S.; Mirouze, M. Transposable elements: All mobile, all different, some stress responsive, some adaptive? Curr. Opin. Genet. Dev. 2018, 49, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Pucker, B.; Kleinbölting, N.; Weisshaar, B. Large scale genomic rearrangements in selected Arabidopsis thaliana T-DNA lines are caused by T-DNA insertion mutagenesis. BMC Genom. 2021, 22, 599. [Google Scholar] [CrossRef] [PubMed]
- Pucker, B.; Rückert, C.; Stracke, R.; Viehöver, P.; Kalinowski, J.; Weisshaar, B. Twenty-Five Years of Propagation in Suspension Cell Culture Results in Substantial Alterations of the Arabidopsis Thaliana Genome. Genes 2019, 10, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavrak, V.V.; Lettner, N.; Jamge, S.; Kosarewicz, A.; Bayer, L.M.; Mittelsten Scheid, O. How a Retrotransposon Exploits the Plant’s Heat Stress Response for Its Activation. PLoS Genet. 2014, 10, e1004115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thieme, M.; Lanciano, S.; Balzergue, S.; Daccord, N.; Mirouze, M.; Bucher, E. Inhibition of RNA polymerase II allows controlled mobilisation of retrotransposons for plant breeding. Genome Biol. 2017, 18, 134. [Google Scholar] [CrossRef]
- Pecinka, A.; Dinh, H.Q.; Baubec, T.; Rosa, M.; Lettner, N.; Scheid, O.M. Epigenetic Regulation of Repetitive Elements Is Attenuated by Prolonged Heat Stress in Arabidopsis. Plant Cell 2010, 22, 3118–3129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tittel-Elmer, M.; Bucher, E.; Broger, L.; Mathieu, O.; Paszkowski, J.; Vaillant, I. Stress-Induced Activation of Heterochromatic Transcription. PLoS Genet. 2010, 6, e1001175. [Google Scholar] [CrossRef] [PubMed]
- Roquis, D.; Robertson, M.; Yu, L.; Thieme, M.; Julkowska, M.; Bucher, E. Genomic impact of stress-induced transposable element mobility in Arabidopsis. Nucleic Acids Res. 2021, 49, 10431–10447. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wang, B.; Yang, X.; Jia, Y.; Xu, Y.; Jia, P.; Dang, N.; Wang, S.; Xu, T.; Zhao, X.; Gao, S.; et al. High-quality Arabidopsis thaliana Genome Assembly with Nanopore and HiFi Long Reads. Genom. Proteom. Bioinform. 2021. [Google Scholar] [CrossRef] [PubMed]
- Tanskanen, J.A.; Sabot, F.; Vicient, C.; Schulman, A.H. Life without GAG: The BARE-2 retrotransposon as a parasite’s parasite. Gene 2007, 390, 166–174. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Cock, P.J.A.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [Green Version]
- Priyam, A.; Woodcroft, B.J.; Rai, V.; Moghul, I.; Munagala, A.; Ter, F.; Chowdhary, H.; Pieniak, I.; Maynard, L.J.; Gibbins, M.A.; et al. Sequenceserver: A Modern Graphical User Interface for Custom BLAST Databases. Mol. Biol. Evol. 2019, 36, 2922–2924. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. ggplot2. WIREs Comp. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Buels, R.; Yao, E.; Diesh, C.M.; Hayes, R.D.; Munoz-Torres, M.; Helt, G.; Goodstein, D.M.; Elsik, C.G.; Lewis, S.E.; Stein, L.; et al. JBrowse: A dynamic web platform for genome visualization and analysis. Genome Biol. 2016, 17, 66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kirov, I.; Merkulov, P.; Dudnikov, M.; Polkhovskaya, E.; Komakhin, R.A.; Konstantinov, Z.; Gvaramiya, S.; Ermolaev, A.; Kudryavtseva, N.; Gilyok, M.; et al. Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline. Plants 2021, 10, 2681. https://doi.org/10.3390/plants10122681
Kirov I, Merkulov P, Dudnikov M, Polkhovskaya E, Komakhin RA, Konstantinov Z, Gvaramiya S, Ermolaev A, Kudryavtseva N, Gilyok M, et al. Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline. Plants. 2021; 10(12):2681. https://doi.org/10.3390/plants10122681
Chicago/Turabian StyleKirov, Ilya, Pavel Merkulov, Maxim Dudnikov, Ekaterina Polkhovskaya, Roman A. Komakhin, Zakhar Konstantinov, Sofya Gvaramiya, Aleksey Ermolaev, Natalya Kudryavtseva, Marina Gilyok, and et al. 2021. "Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline" Plants 10, no. 12: 2681. https://doi.org/10.3390/plants10122681
APA StyleKirov, I., Merkulov, P., Dudnikov, M., Polkhovskaya, E., Komakhin, R. A., Konstantinov, Z., Gvaramiya, S., Ermolaev, A., Kudryavtseva, N., Gilyok, M., Divashuk, M. G., Karlov, G. I., & Soloviev, A. (2021). Transposons Hidden in Arabidopsis thaliana Genome Assembly Gaps and Mobilization of Non-Autonomous LTR Retrotransposons Unravelled by Nanotei Pipeline. Plants, 10(12), 2681. https://doi.org/10.3390/plants10122681








