A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant
Abstract
:1. Introduction
2. Results and Discussion
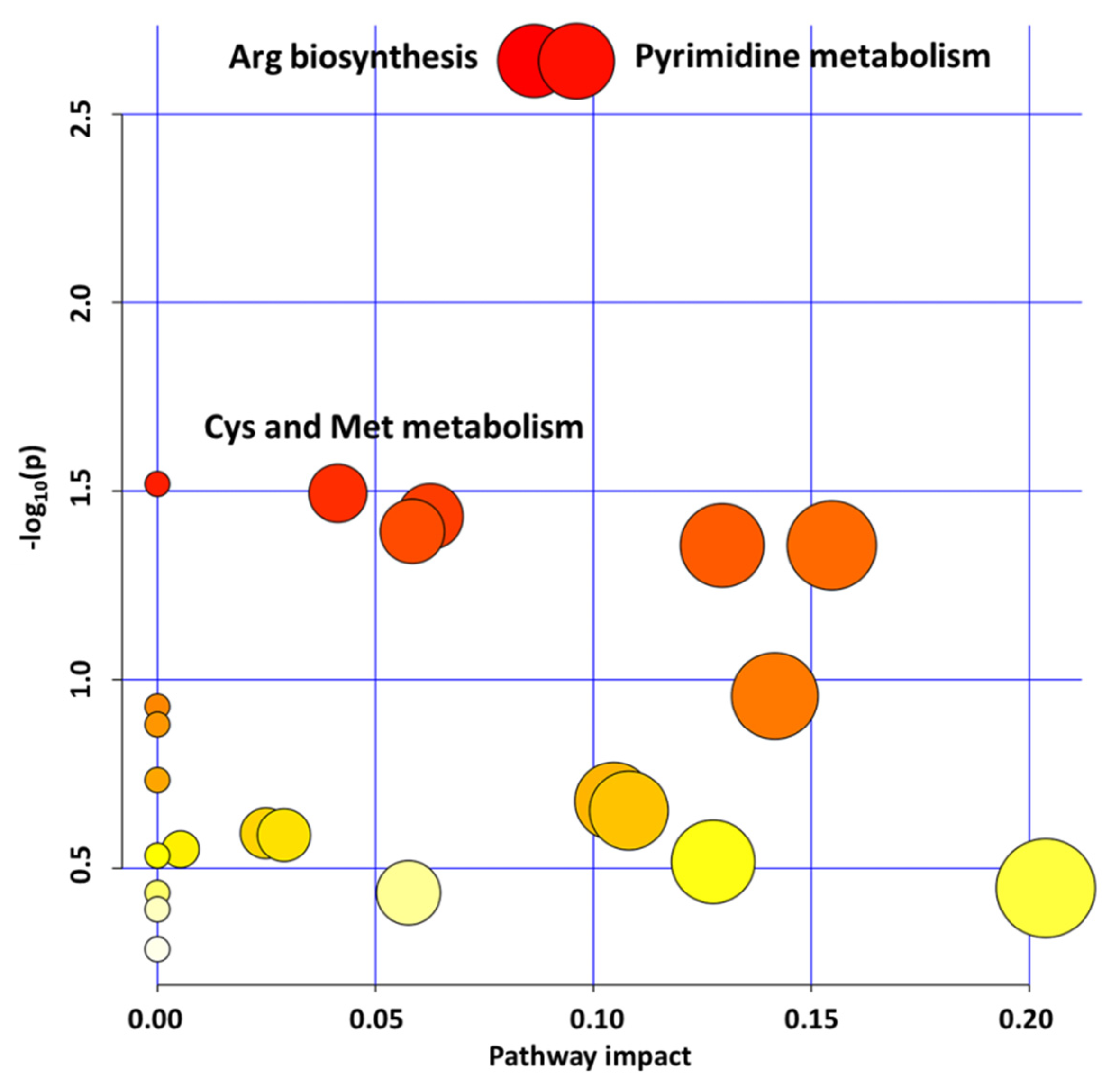
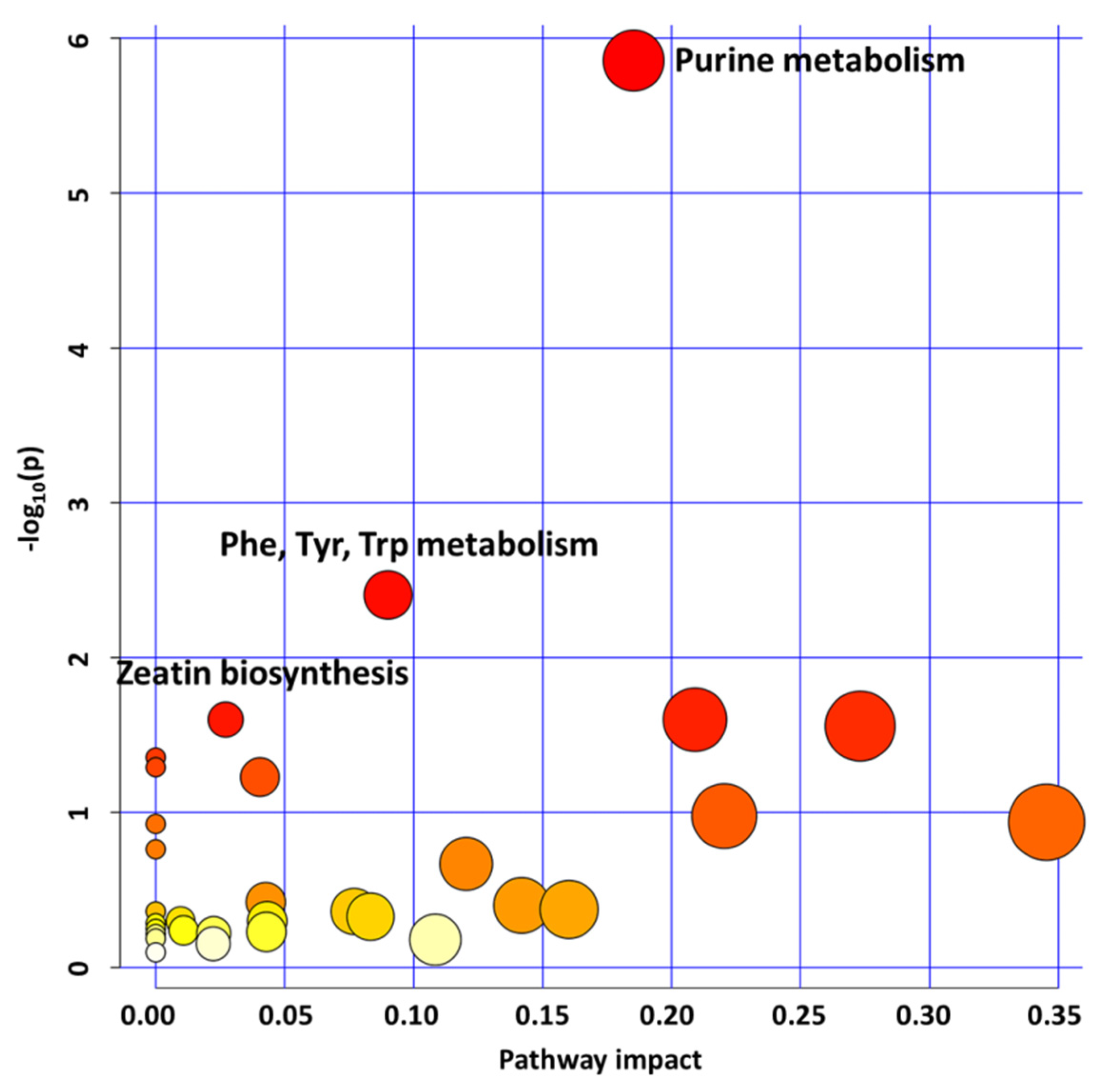
2.1. GLYI4 Knock-Down Affects Plant Metabolism
2.1.1. Pyrimidine Metabolism Upregulation Influences Redox Homeostasis
2.1.2. Aminoacid Metabolism Upregulation Promotes Cellular Energy Maintenance
2.1.3. Purine, Aromatic Amino Acid, and Zeatin Biosynthesis Downregulation Affects glyI4 Mutant Growth and Defense
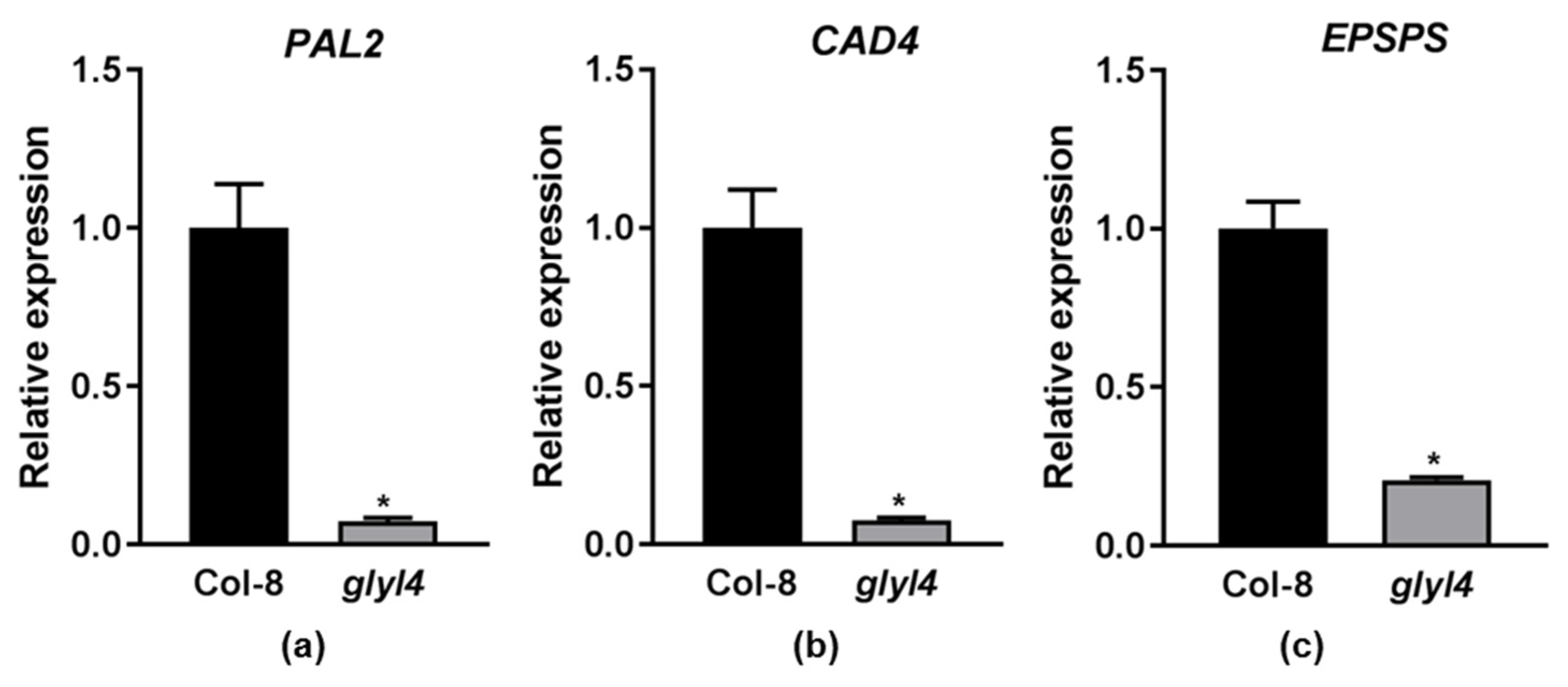
2.2. GLYI4 as a Potential Positive Regulator of Plant Growth and Defense
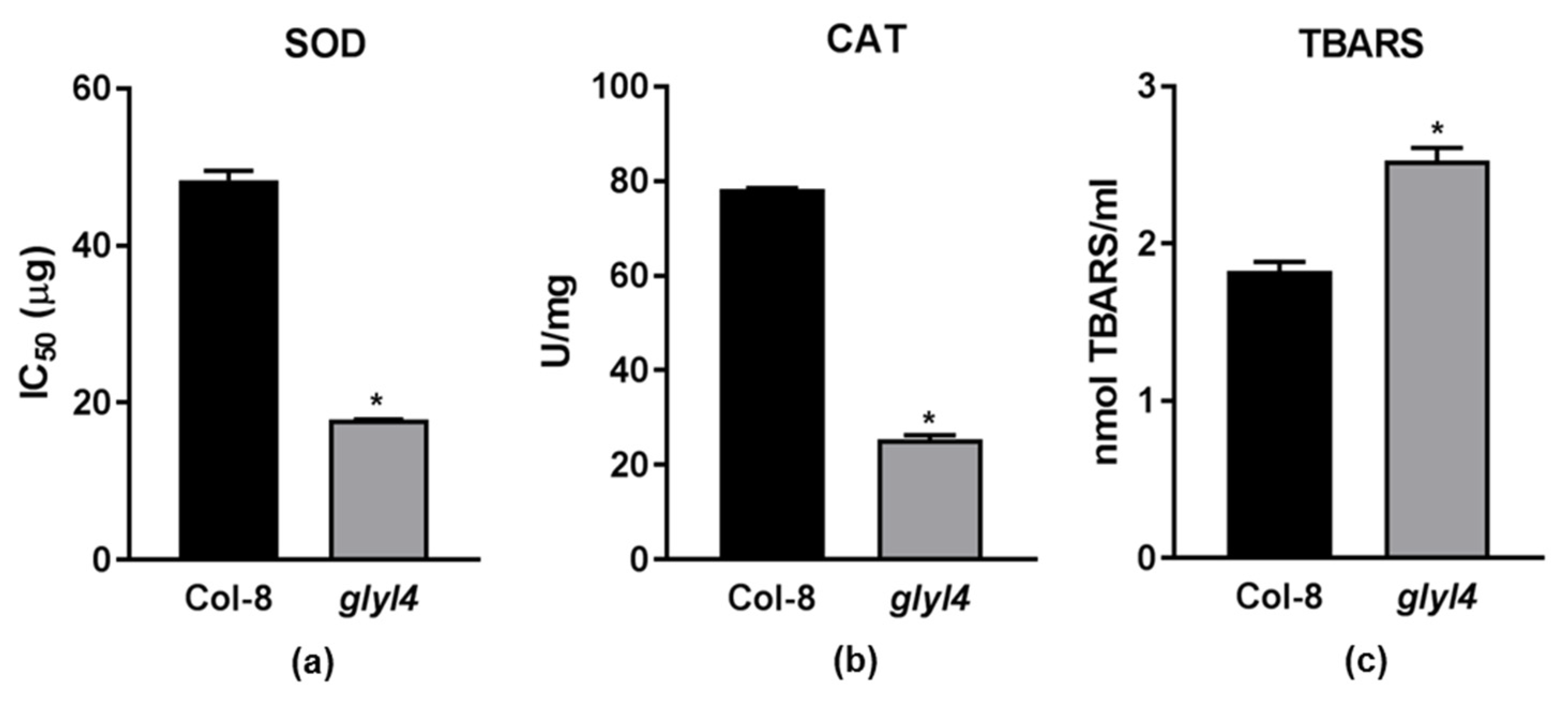
2.3. GLYI4 Affects Plant Antioxidant Responses
2.3.1. Hydrogen Peroxide Is Not Efficiently Scavenged by Antioxidant Machinery
2.3.2. glyI4 Mutant Suffers of Lipid Peroxidation
3. Materials and Methods
3.1. Plant Material and Growth Conditions
3.2. Metabolite Extraction and LC-MS Analysis
3.3. Metabolomic Data Processing
3.3.1. Data Elaboration and Statistical Analysis
3.3.2. Pathway Analysis
3.4. RNA Extraction and RT-qPCR
3.5. Protein Extraction
3.6. Superoxide Dismutase Activity Assay
3.7. Catalase Activity Assay
3.8. Thiobarbituric Acid Reactive Substance Measurement
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; VanWees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [Green Version]
- Rejeb, I.B.; Pastor, V.; Mauch-Mani, B. Plant responses to simultaneous biotic and abiotic stress: Molecular mechanisms. Plants 2014, 3, 458–475. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Wen, K.S.; Ruan, X.; Zhao, Y.X.; Wei, F.; Wang, Q. Response of plant secondary metabolites to environmental factors. Molecules 2018, 23, 762. [Google Scholar] [CrossRef] [Green Version]
- Brilli, F.; Loreto, F.; Baccelli, I. Exploiting Plant Volatile Organic Compounds (VOCs) in agriculture to improve sustainable defense strategies and productivity of crops. Front. Plant Sci. 2019, 10, 264. [Google Scholar] [CrossRef] [PubMed]
- Hoque, T.S.; Okuma, E.; Uraji, M.; Furuichi, T.; Sasaki, T.; Hoque, M.A.; Nakamura, Y.; Murata, Y. Inhibitory effects of methylglyoxal on light-induced stomatal opening and inward K+ channel activity in Arabidopsis. Biosci. Biotechnol. Biochem. 2012, 76, 617–619. [Google Scholar] [CrossRef]
- Hoque, T.S.; Uraji, M.; Tuya, A.; Nakamura, Y.; Murata, Y. Methylglyoxal inhibits seed germination and root elongation and up-regulates transcription of stress-responsive genes in ABA-dependent pathway in Arabidopsis. Plant Biol. 2012, 14, 854–858. [Google Scholar] [CrossRef]
- Hoque, T.S.; Uraji, M.; Ye, W.; Hossain, M.A.; Nakamura, Y.; Murata, Y. Methylglyoxal-induced stomatal closure accompanied by peroxidase mediated ROS production in Arabidopsis. Plant Physiol. 2012, 169, 979–986. [Google Scholar] [CrossRef]
- Hoque, M.A.; Uraji, M.; Banu, M.N.A.; Mori, I.C.; Nakamura, Y.; Murata, Y. Methylglyoxal inhibition of cytosolic ascorbate peroxidase from Nicotiana tabacum. J. Biochem. Mol. Toxicol. 2012, 26, 315–321. [Google Scholar] [CrossRef]
- Mano, J. Reactive carbonyl species: Their production from lipid peroxides, action in environmental stress, and the detoxification mechanism. Plant Physiol. Biochem. 2012, 59, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Hoque, T.S.; Hossain, M.A.; Mostofa, M.G.; Burritt, D.J.; Fujita, M.; Tran, L.-S.P. Methylglyoxal: An emerging signaling molecule in plant abiotic stress responses and tolerance. Front. Plant Sci. 2016, 7, 1341. [Google Scholar] [CrossRef] [Green Version]
- Singla-Pareek, S.L.; Kaur, C.; Kumar, B.; Pareek, A.; Sopory, S.K. Reassessing plant glyoxalases: Large family and expanding functions. New Phytol. 2020, 227, 714–721. [Google Scholar] [CrossRef] [Green Version]
- Sankaranarayanan, S.; Jamshed, M.; Kumar, A.; Skori, L.; Scandola, S.; Wang, T.; Spiegel, D.; Samuel, M.A. Glyoxalase goes green: The expanding roles of glyoxalase in plants. Int. J. Mol. Sci. 2017, 18, 898. [Google Scholar] [CrossRef]
- Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. Glyoxalase and methylglyoxal as biomarkers for plant stress tolerance. Crit. Rev. Plant Sci. 2014, 33, 429–456. [Google Scholar] [CrossRef]
- Gupta, B.K.; Sahoo, K.K.; Ghosh, A.; Tripathi, A.K.; Anwar, K.; Das, P.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Manipulation of glyoxalase pathway confers tolerance to multiple stresses in rice. Plant Cell Environ. 2018, 41, 1186–1200. [Google Scholar] [CrossRef]
- Li, T.; Cheng, X.; Wang, X.; Li, G.; Wang, B.; Wang, W.; Zhang, N.; Han, Y.; Jiao, B.; Wang, Y.; et al. Glyoxalase I-4 functions downstream of NAC72 to modulate downy mildew resistance in grapevine. Plant J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Mustafiz, A.; Singh, A.K.; Pareek, A.; Sopory, S.K.; Singla-Pareek, S.L. Genome-wide analysis of rice and Arabidopsis identifies two glyoxalase genes that are highly expressed in abiotic stresses. Funct. Integr. Genom. 2011, 11, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Proietti, S.; Caarls, L.; Coolen, S.; Van Pelt, J.A.; Van Wees, S.C.M.; Pieterse, C.M.J. Genome-wide association study reveals novel players in defense hormone crosstalk in Arabidopsis. Plant Cell Environ. 2018, 41, 2342–2356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proietti, S.; Falconieri, G.S.; Bertini, L.; Baccelli, I.; Paccosi, E.; Belardo, A.; Timperio, A.M.; Caruso, C. GLYI4 plays a role in methylglyoxal detoxification and jasmonate-mediated stress responses in Arabidopsis thaliana. Biomolecules 2019, 9, 635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fraire-Velázquez, S.; Balderas-Hernández, V.E. Abiotic Stress in Plants and Metabolic Responses, Abiotic Stress—Plant Responses and Applications in Agriculture, Kourosh Vahdati and Charles Leslie; IntechOpen: London, UK, 2013. [Google Scholar] [CrossRef] [Green Version]
- Caldana, C.; Degenkolbe, T.; Cuadros-Inostroza, A.; Klie, S.; Sulpice, R.; Leisse, A.; Steinhauser, D.; Fernie, A.R.; Willmitzer, L.; Hannah, M.A. High-density kinetic analysis of the metabolomic and transcriptomic response of Arabidopsis to eight environmental conditions. Plant J. 2011, 67, 869–884. [Google Scholar] [CrossRef]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted metabolomics strategies—challenges and emerging directions. J. Am. Soc. Mass Spectr. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, J.D.; Pastene, D.O.; Breedijk, A.; Rodriguez, A.; Hofmann, B.B.; Sticht, C.; von Ochsenstein, E.; Allgayer, H.; van den Born, J.; Bakker, S.; et al. Methylglyoxal down-regulates the expression of cell cycle associated genes and activates the p53 pathway in human umbilical vein endothelial cells. Sci. Rep. 2019, 9, 1152. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Yamamoto, M.; Mimura, T.; Ashihara, H. Effect of short-term salt stress on the metabolic profiles of pyrimidine, purine and pyridine nucleotides in cultured cells of the mangrove tree, Bruguiera sexangula. Physiol. Plant 2006, 128, 405–414. [Google Scholar] [CrossRef]
- Bolt, R.; Zrenner, R. Purine and pyrimidine biosynthesis in higher plants. Physiol. Plant 2002, 117, 297–304. [Google Scholar] [CrossRef]
- Stasolla, C.; Katahira, R.; Thorpe, T.A.; Ashihara, H. Purine and pyrimidine nucleotide metabolism in higher plants. J. Plant Physiol. 2003, 160, 1271–1295. [Google Scholar] [CrossRef]
- Fang, J.; Uchiumi, T.; Yagi, M.; Matsumoto, S.; Amamoto, R.; Takazaki, S.; Yamaza, H.; Nonaka, K.; Kang, D. Dihydro-orotate dehydrogenase is physically associated with the respiratory complex and its loss leads to mitochondrial dysfunction. Biosci. Rep. 2013, 33, e00021. [Google Scholar] [CrossRef]
- Handy, D.E.; Loscalzo, J. Redox Regulation of Mitochondrial Function. Antioxid. Redox Signal. 2012, 16, 1323–1367. [Google Scholar] [CrossRef]
- Schmidt, B.; de Assis, A.M.; Battu, C.E.; Rieger, D.K.; Hansen, F.; Sordi, F.; Longoni, A.; Hoefel, A.L.; Farina, M.; Gonçalves, C.A.; et al. Effects of glyoxal or methylglyoxal on the metabolism of amino acids, lactate, glucose and acetate in the cerebral cortex of young and adult rats. Brain Res. 2010, 1315, 19–24. [Google Scholar] [CrossRef]
- Slocum, R.D. Genes, enzymes and regulation of arginine biosynthesis in plants. Plant Physiol. Biochem. 2005, 43, 729–745. [Google Scholar] [CrossRef]
- Winter, G.; Todd, C.D.; Trovato, M.; Forlani, G.; Funck, D. Physiological implications of arginine metabolism in plants. Front. Plant Sci. 2015, 6, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikiforova, V.; Kempa, S.; Zeh, M.; Maimann, S.; Kreft, O.; Casazza, A.P.; Riedel, K.; Tauberger, E.; Hoefgen, R.; Hesse, H. Engineering of cysteine and methionine biosynthesis in potato. Amino Acids 2002, 22, 259–278. [Google Scholar] [CrossRef] [PubMed]
- Romero, L.C.; Aroca, M.Á.; Laureano-Marín, A.M.; Moreno, I.; García, I.; Gotor, C. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol. Plant 2014, 7, 264–276. [Google Scholar] [CrossRef] [Green Version]
- Hell, R.; Wirtz, M. Molecular biology, biochemistry and cellular physiology of cysteine metabolism in Arabidopsis thaliana. In The Arabidopsis Book; American Society of Plant Biologists: Rockville, MD, USA, 2011; Volume 9, p. e0154. [Google Scholar] [CrossRef] [Green Version]
- Yadav, S.K.; Singla-Pareek, S.L.; Ray, M.; Reddy, M.K.; Sopory, S.K. Transgenic tobacco plants overexpressing glyoxalase enzymes resist an increase in methylglyoxal and maintain higher reduced glutathione levels under salinity stress. FEBS Lett. 2005, 579, 6265–6271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusano, M.; Fukushima, A.; Redestig, H.; Kobayashi, M.; Otsuki, H.; Onouchi, H.; Naito, S.; Hirai, M.Y.; Saito, K. Comparative metabolomics charts the impact of genotype-dependent methionine accumulation in Arabidopsis thaliana. Amino Acids 2010, 39, 1013–1021. [Google Scholar] [CrossRef]
- Kirma, M.; Araújo, W.L.; Fernie, A.R.; Galili, G. The multifaceted role of aspartate-family amino acids in plant metabolism. J. Exp. Bot. 2012, 63, 4995–5001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, A.K.; Witte, C.-P. The biochemistry of nitrogen mobilization: Purine ring catabolism. Trends Plant Sci. 2011, 16, 381–387. [Google Scholar] [CrossRef]
- Yobi, A.; Wone, B.W.M.; Xu, W.; Alexander, D.C.; Guo, L.; Ryals, J.A.; Olivier, M.J.; Cushman, J.C. Metabolomic profiling in Selaginella lepidophylla at various hydration states provides new insights into the mechanistic basis of desiccation tolerance. Mol. Plant 2013, 6, 369–385. [Google Scholar] [CrossRef] [Green Version]
- Rose, M.T.; Rose, T.J.; Pariasca-Tanaka, J.; Yoshihashi, T.; Neuweger, H.; Goesmann, A.; Frei, M.; Wissuwa, M. Root metabolic response of rice (Oryza sativa L.) genotypes with contrasting tolerance to zinc deficiency and bicarbonate excess. Planta 2012, 236, 959–973. [Google Scholar] [CrossRef]
- Kanani, H.; Dutta, B.; Klapa, M.I. Individual vs. combinatorial effect of elevated CO2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: Comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Syst. Biol. 2010, 4, 177. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, S.; Kounosu, Y.; Shimada, H.; Sakamoto, A. Arabidopsis xanthine dehydrogenase mutants defective in purine degradation show a compromised protective response to drought and oxidative stress. Plant Biotechnol. 2014, 31, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Misra, B.B.; de Armas, E.; Tong, Z.; Chen, S. Metabolomic responses of guard cells and mesophyll cells to bicarbonate. PLoS ONE 2015, 10, e0144206. [Google Scholar] [CrossRef]
- Vivancos, P.D.; Driscoll, S.P.; Bulman, C.A.; Ying, L.; Emami, K.; Treumann, A.; Mauve, C.; Noctor, G.; Foyer, C.H. Perturbations of amino acid metabolism associated with glyphosate-dependent inhibition of shikimic acid metabolism affect cellular redox homeostasis and alter the abundance of proteins involved in photosynthesis and photorespiration. Plant Physiol. 2011, 157, 256–268. [Google Scholar] [CrossRef] [Green Version]
- Tzin, V.; Galili, G. The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. Arab. Book 2010, 8, e0132. [Google Scholar] [CrossRef] [Green Version]
- Fraser, C.M.; Chapple, C. The Phenylpropanoid Pathway in Arabidopsis. Arab. Book 2011, 9, e0152. [Google Scholar] [CrossRef] [Green Version]
- Garai, S.; Bhowal, B.; Kaur, C.; Singla-Pareek, S.L.; Sopory, S.K. What signals the glyoxalase pathway in plants? Physiol. Mol. Biol. Plants 2021. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishiyama, R.; Le, D.T.; Watanabe, Y.; Matsui, A.; Tanaka, M.; Seki, M.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Phan Tran, L.-S. Transcriptome analyses of a salt-tolerant cytokinin-deficient mutant reveal differential regulation of salt stress response by cytokinin deficiency. PLoS ONE 2012, 7, e32124. [Google Scholar] [CrossRef] [Green Version]
- Schäfer, M.; Brütting, C.; Canales, I.M.; Großkinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis-zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Zhang, J.; Tschaplinski, T.J.; Tuskan, G.A.; Chen, J.-G.; Muchero, W. Regulation of lignin biosynthesis and its role in growth-defense tradeoffs. Front. Plant Sci. 2018, 9, 1427. [Google Scholar] [CrossRef] [Green Version]
- Govender, N.T.; Mahmood, M.; Seman, I.A.; Wong, M.-Y. The phenylpropanoid pathway and lignin in defense against Ganoderma boninense colonized root tissues in Oil Palm (Elaeis guineensis Jacq.). Front. Plant Sci. 2017, 8, 1395. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and biological functions in plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boudet, A. Lignins and lignification: Selected issues. Plant Physiol. Biochem. 2000, 38, 81–96. [Google Scholar] [CrossRef]
- Donaldson, L.A. Lignification and lignin topochemistry—An ultrastructural view. Phytochemistry 2001, 57, 859–873. [Google Scholar] [CrossRef]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef] [PubMed]
- Vanholme, R.; Morreel, K.; Ralph, J.; Boerjan, W. Lignin engineering. Curr. Opin. Plant Biol. 2008, 11, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Ana, R.D.; Ian, A.D. Panama disease: Cell wall reinforcement in banana roots in response to elicitors from Fusarium oxysporum f. sp. cubense race four. Am. Phytopathol. Soc. 2000, 90, 1173–1180. [Google Scholar] [CrossRef] [Green Version]
- Tiburzy, R.; Reisener, H.J. Resistance of wheat to Puccinia graminis f.sp. tritici: Association of the hypersensitive reaction with the cellular accumulation of lignin-like material and callose. Physiol. Mol. Plant Pathol. 1990, 36, 109–120. [Google Scholar] [CrossRef]
- Eynck, C.; Seguin-swartz, G.; Clarke, W.E.; Parkin, I.A.P. Monolignol biosynthesis is associated with resistance to Sclerotinia sclerotiorum in Camelina sativa. Mol. Plant Pathol. 2012, 13, 887–899. [Google Scholar] [CrossRef]
- Tronchet, M.; Balagué, C.; Kroj, T.; Jouanin, L.; Roby, D. Cinnamyl alcohol dehydrogenases-C and D, key enzymes in lignin biosynthesis, play an essential role in disease resistance in Arabidopsis. Mol. Plant Pathol. 2010, 11, 83–92. [Google Scholar] [CrossRef]
- Wu, Z.; Han, S.; Zhou, H.; Tuang, Z.K.; Wang, Y.; Jin, Y.; Shi, H.; Yang, W. Cold stress activates disease resistance in Arabidopsis thaliana through a salicylic acid dependent pathway. Plant Cell Environ. 2019, 42, 2645–2663. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.J.; Hong, Y.S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef] [PubMed]
- Molin, W. Glyphosate, a Unique Global Herbicide. J. E. Franz, M.K. Mao, and J. A. Sikorski, ACS Monograph 189, 1997. 653 pp. Weed Technol. 1997, 12, 564–565. [Google Scholar] [CrossRef]
- Yang, X.; Beres, Z.T.; Jin, L.; Parrish, J.T.; Zhao, W.; Mackey, D.; Snow, A.A. Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana. PLoS ONE 2017, 12, e0175820. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Nan, P.; Gu, Z.; Ge, X.; Feng, Y.-Q.; Lu, B.-R. Overexpressing exogenous 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) genes increases fecundity and auxin content of transgenic Arabidopsis plants. Front. Plant Sci. 2018, 9, 233. [Google Scholar] [CrossRef] [Green Version]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Elstner, E.F. Mechanisms of oxygen activation in different compartments of plant cells. In Active Oxygen/Oxidative Stress and Plant Metabolism; Pell, E.J., Steffen, K.L., Eds.; American Society of Plant Physiologists Press: Rockville, MD, USA, 1991; Volume 1, pp. 13–25. [Google Scholar]
- Cai, Y.T.; Zhang, H.; Qi, Y.P.; Ye, X.; Huang, Z.-R.; Guo, J.-X.; Chen, L.-S.; Yang, L.-T. Responses of reactive oxygen species and methylglyoxal metabolisms to magnesium-deficiency differ greatly among the roots, upper and lower leaves of Citrus sinensis. BMC Plant Biol. 2019, 19, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Van Breusegem, F.; Noctor, G. Catalase function in plants: A focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [Green Version]
- Escobar, J.A.; Rubio, M.A.; Lissi, E.A. SOD and catalase inactivation by singlet oxygen and peroxyl radicals. Free Radic. Biol. Med. 1996, 20, 285–290. [Google Scholar] [CrossRef]
- Orendi, G.; Zimmermann, P.; Baar, C.; Zentgraf, U. Loss of stress-induced expression of catalase3 during leaf senescence in Arabidopsis thaliana is restricted to oxidative stress. Plant Sci. 2001, 161, 301–314. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Kobayashi, Y.; Matsumoto, H. Lipid Peroxidation Is an Early Symptom Triggered by Aluminum, But Not the Primary Cause of Elongation Inhibition in Pea Roots. Plant Phys. 2001, 125, 199–208. [Google Scholar] [CrossRef] [Green Version]
- Hasanuzzaman, M.; Nahar, K.; Hossain, M.S.; Mahmud, J.A.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated actions of glyoxalase and antioxidant defense systems in conferring abiotic stress tolerance in plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, C.A.; O’Maille, G.; Want, E.J.; Qin, C.; Trauger, S.A.; Brandon, T.R.; Custodio, D.E.; Abagyan, R.; Siuzdaket, G. METLIN: A metabolite mass spectral database. Ther. Drug Monit. 2005, 27, 747–751. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Mandal, R.; Sinelnikov, I.; Broadhurst, D.; Wishart, D.S. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 2012, 40, W127–W133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertini, L.; Cozzolino, F.; Proietti, S.; Falconieri, G.S.; Iacobucci, I.; Salvia, R.; Falabella, P.; Monti, M.; Caruso, C. What antarctic plants can tell us about climate changes: Temperature as a driver for metabolic reprogramming. Biomolecules 2021, 11, 1094. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proietti, S.; Bertini, L.; Falconieri, G.S.; Baccelli, I.; Timperio, A.M.; Caruso, C. A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant. Plants 2021, 10, 2464. https://doi.org/10.3390/plants10112464
Proietti S, Bertini L, Falconieri GS, Baccelli I, Timperio AM, Caruso C. A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant. Plants. 2021; 10(11):2464. https://doi.org/10.3390/plants10112464
Chicago/Turabian StyleProietti, Silvia, Laura Bertini, Gaia Salvatore Falconieri, Ivan Baccelli, Anna Maria Timperio, and Carla Caruso. 2021. "A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant" Plants 10, no. 11: 2464. https://doi.org/10.3390/plants10112464
APA StyleProietti, S., Bertini, L., Falconieri, G. S., Baccelli, I., Timperio, A. M., & Caruso, C. (2021). A Metabolic Profiling Analysis Revealed a Primary Metabolism Reprogramming in Arabidopsis glyI4 Loss-of-Function Mutant. Plants, 10(11), 2464. https://doi.org/10.3390/plants10112464







