Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa
Abstract
:1. Introduction
2. Results
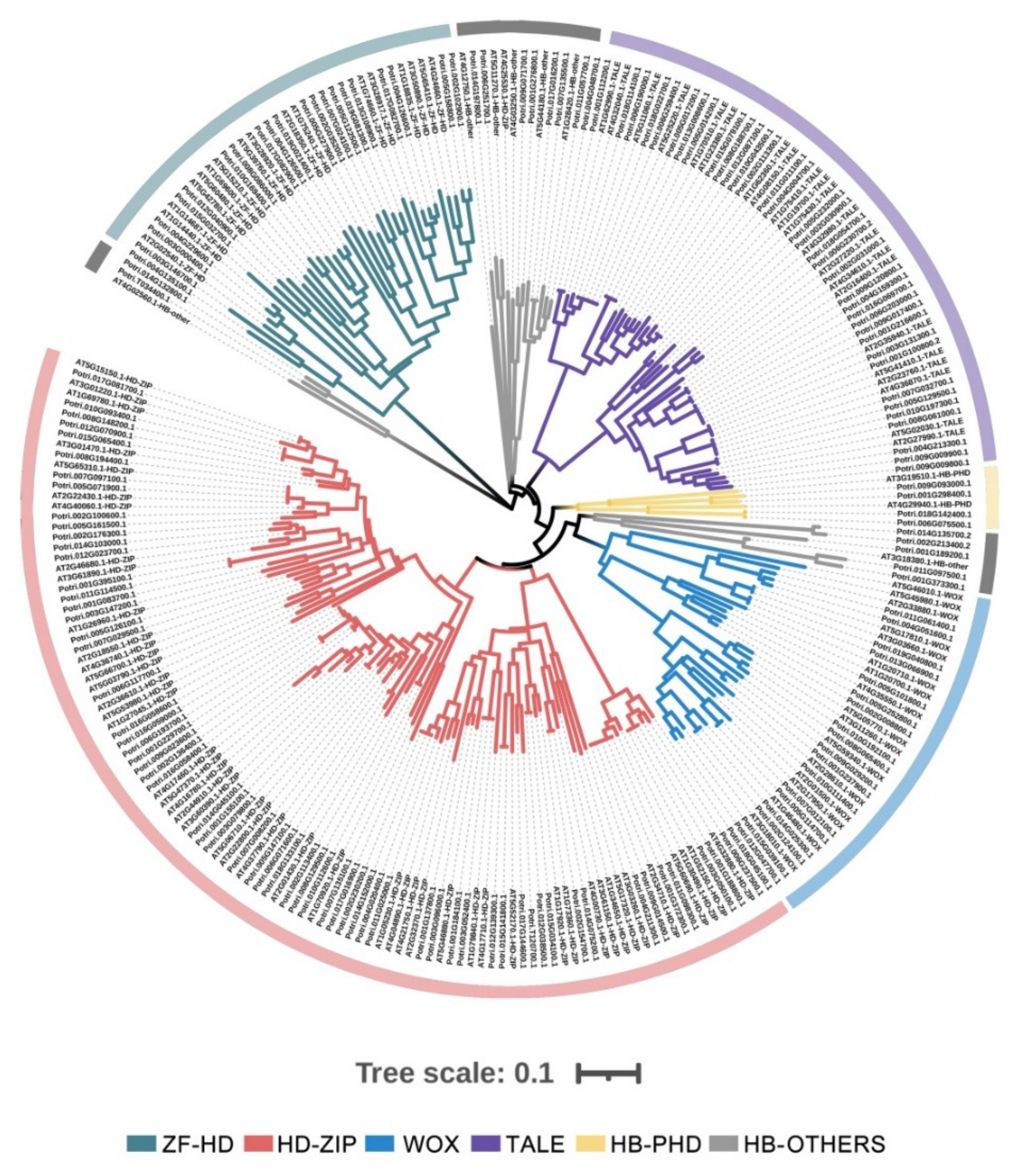
2.1. Identification and Classification of HB Genes in P. trichocarpa
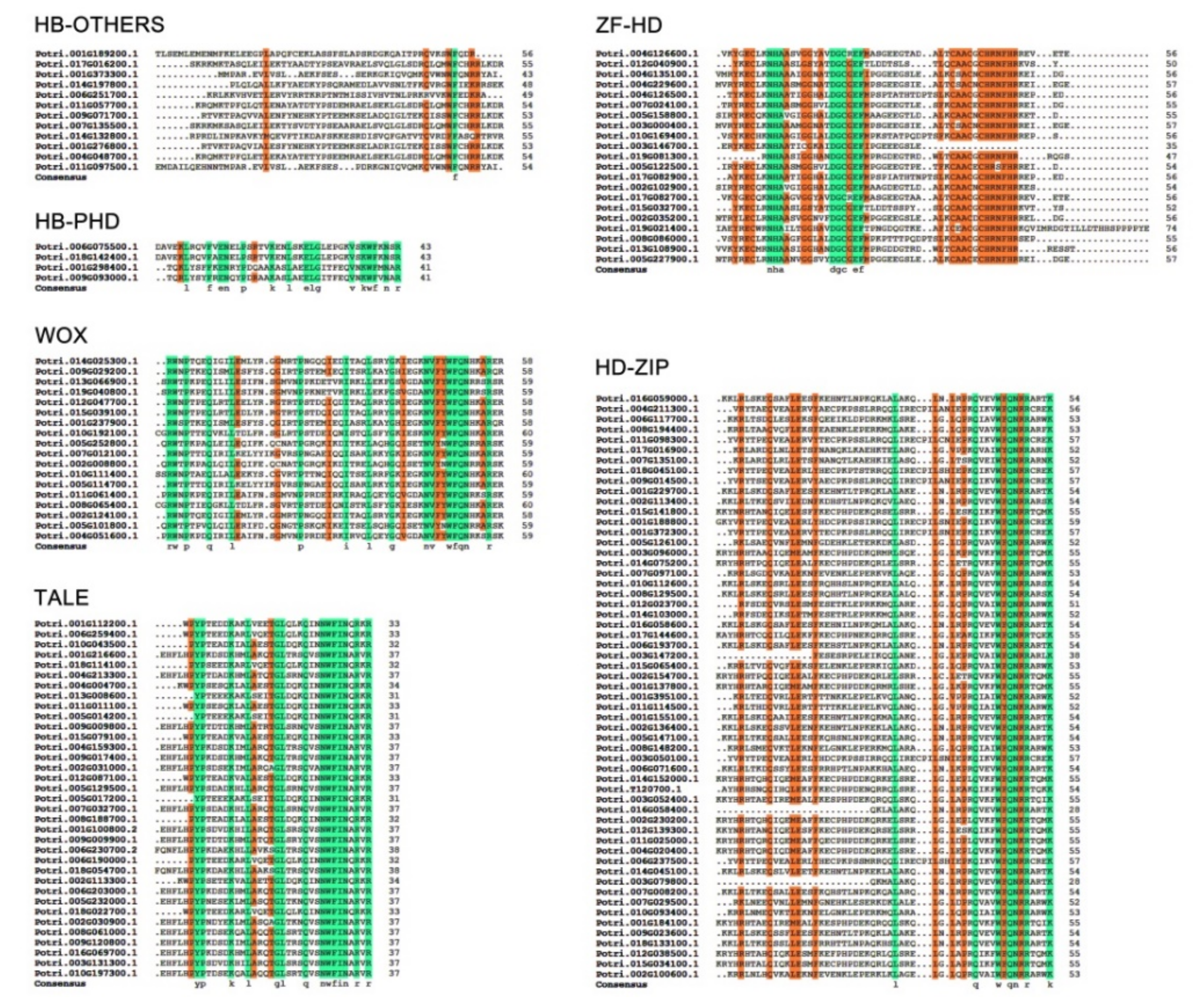
2.2. Conserved Motif Analysis
2.3. Gene Structure Analysis
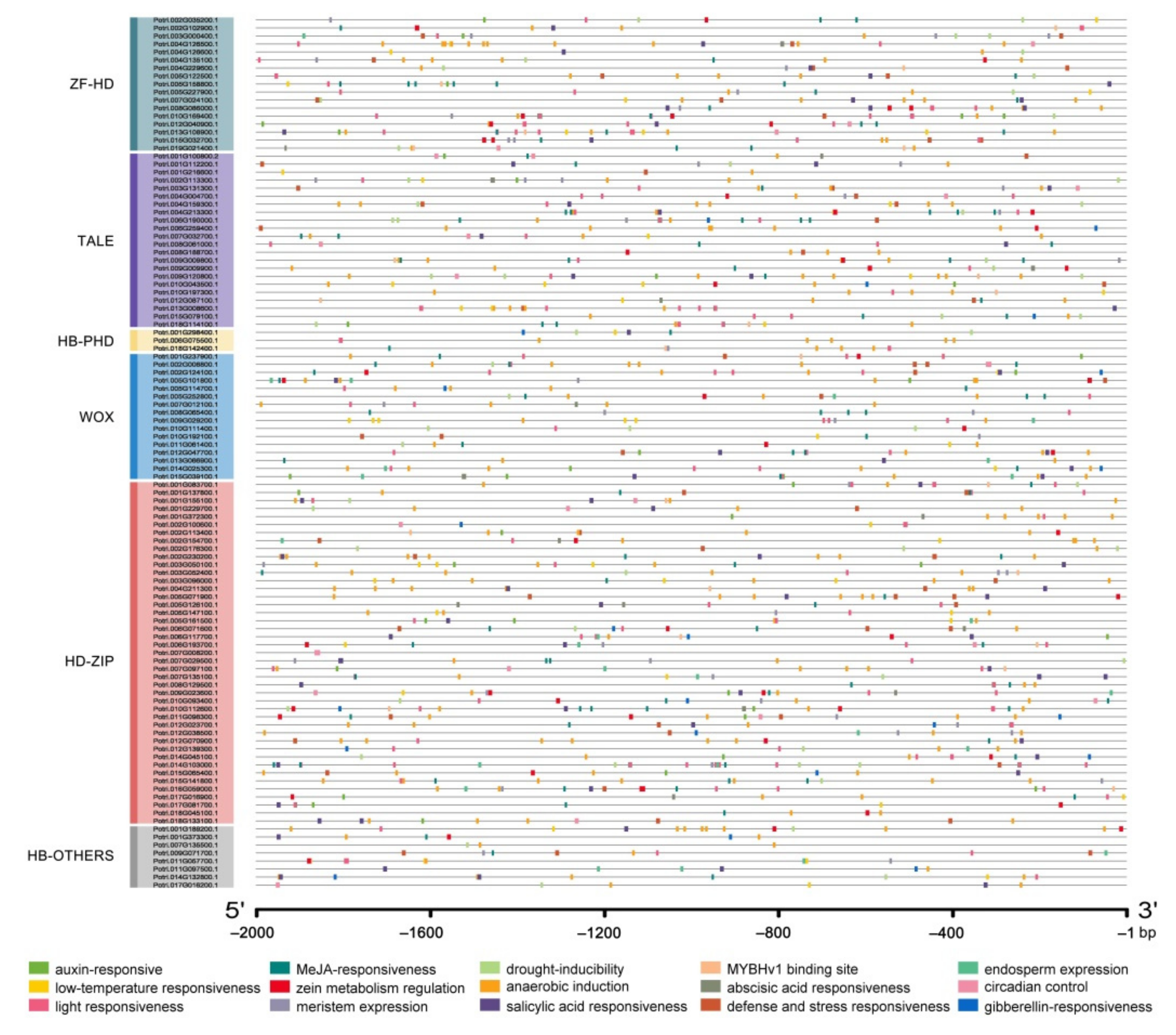
2.4. The Prediction of Cis-Acting Elements
2.5. Chromosomal Localization and Synteny Analysis
2.6. Expression Patterns of PtrHB Genes
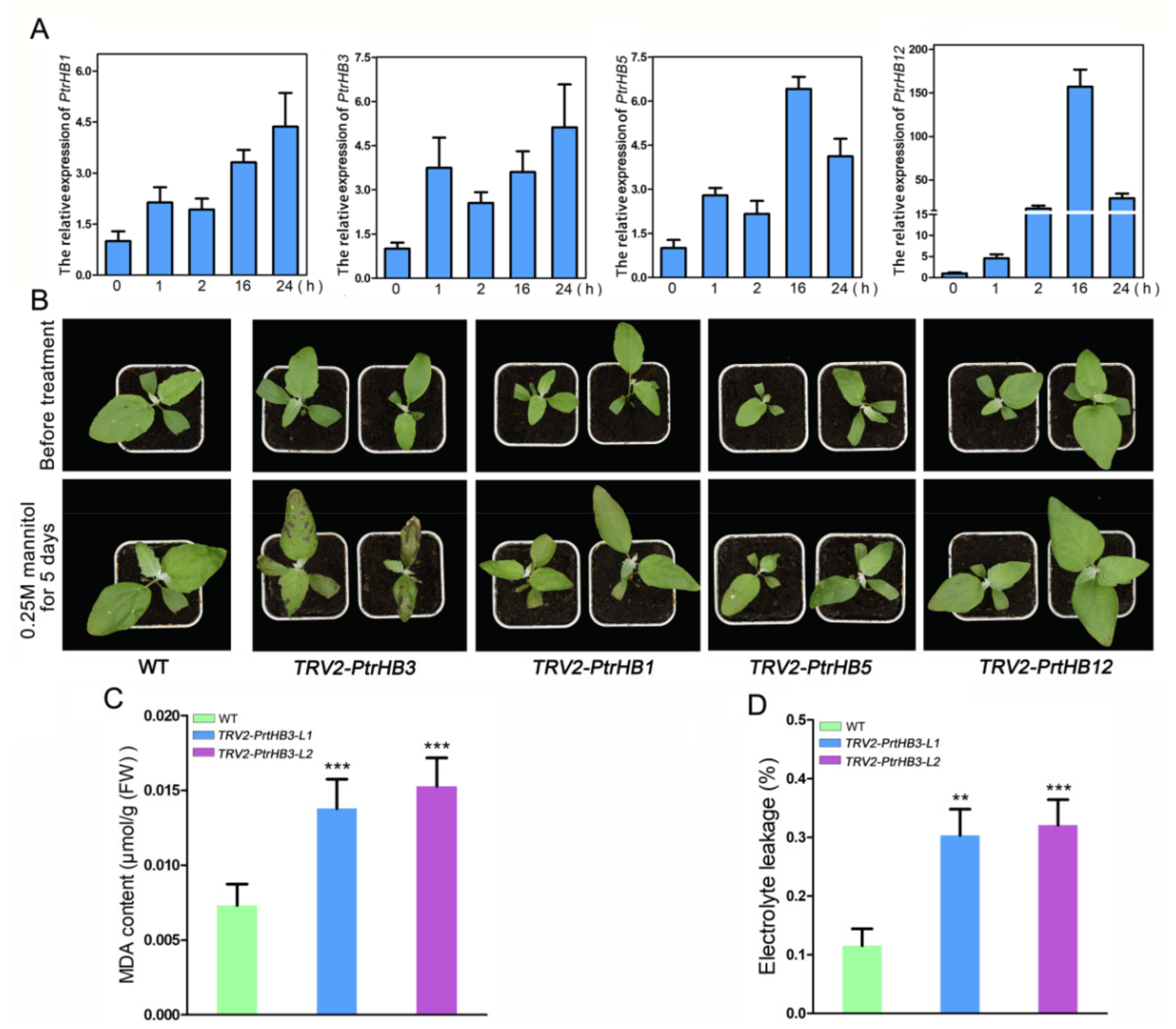
2.7. The Spatio-Temporal Expression Patterns of PtrHB1, PtrHB3, PtrHB5 and PtrHB12
2.8. Silencing PtrHB3 Results in Decreased Tolerance to Mannitol Treatment
3. Discussion
4. Materials and Methods
4.1. Genome Information
4.2. HB Gene Family Identification and Phylogenetic Tree Construct
4.3. Sequence Alignment
4.4. Analysis of Motif and Gene Structure
4.5. Synteny Analysis of PtrHB Genes
4.6. Promoter Analysis of cis-Acting Elements
4.7. Gene Expression Analysis
4.8. Plant Material and Growth Condition
4.9. The qPCR Analysis
4.10. Gene Cloning and Plasmid Construct
4.11. Agrobacterium Infiltration
4.12. Measurement of MDA and Conductivity
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mukherjee, K.; Brocchieri, L.; Burglin, T.R. A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol. Biol. Evol. 2009, 26, 2775–2794. [Google Scholar] [CrossRef]
- Garber, R.L.; Kuroiwa, A.; Gehring, W.J. Genomic and cDNA clones of the homeotic locus Antennapedia in Drosophila. EMBO J. 1983, 2, 2027–2036. [Google Scholar] [CrossRef]
- Gehring, W.J.; Affolter, M.; Burglin, T. Homeodomain proteins. Annu. Rev. Biochem. 1994, 63, 487–526. [Google Scholar] [CrossRef]
- Derelle, R.; Lopez, P.; Le Guyader, H.; Manuel, M. Homeodomain proteins belong to the ancestral molecular toolkit of eukaryotes. Evol. Dev. 2007, 9, 212–219. [Google Scholar] [CrossRef]
- Chan, R.L.; Gago, G.M.; Palena, C.M.; Gonzalez, D.H. Homeoboxes in plant development. BBA—Gene Struct. Express 1998, 1442, 1–19. [Google Scholar] [CrossRef]
- Jin, J.; Feng, T.; De-Chang, Y.; Yu-Qi, M.; Lei, K.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88. [Google Scholar] [CrossRef]
- Windhövel, A.; Hein, I.; Dabrowa, R.; Stockhaus, J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 2001, 45, 201–214. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Ghangal, R.; Garg, R.; Jain, M. Genome-Wide Analysis of Homeobox Gene Family in Legumes: Identification, Gene Duplication and Expression Profiling. PLoS ONE 2015, 10, e0119198. [Google Scholar] [CrossRef] [PubMed]
- Haecker, A. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef]
- Chew, W.; Hrmova, M.; Lopato, S. Role of Homeodomain Leucine Zipper (HD-Zip) IV Transcription Factors in Plant Development and Plant Protection from Deleterious Environmental Factors. Int. J. Mol. Sci. 2013, 14, 8122–8147. [Google Scholar] [CrossRef]
- Keiko, S.; Tomoaki, N.; Masahiro, K.; Mitsuyasu, H. Isolation of Homeodomain–Leucine Zipper Genes from the Moss Physcomitrella patens and the Evolution of Homeodomain–Leucine Zipper Genes in Land Plants. Mol. Biol. Evol. 2001, 18, 491–502. [Google Scholar]
- Perotti, M.F.; Ribone, P.A.; Chan, R.L. Plant transcription factors from the homeodomain-leucine zipper family I. Role in development and stress responses. IUBMB Life 2017, 69, 280–289. [Google Scholar] [CrossRef] [PubMed]
- Peterson, K.M.; Shyu, C.; Burr, C.A.; Horst, R.J.; Kanaoka, M.M.; Omae, M.; Sato, Y.; Torii, K.U. Arabidopsis homeodomain-leucine zipper IV proteins promote stomatal development and ectopically induce stomata beyond the epidermis. Development 2013, 140, 1924–1935. [Google Scholar] [CrossRef]
- Zhang, S.; Haider, I.; Kohlen, W.; Li, J.; Ouwerkerk, P. Function of the HD-Zip I gene OsHOX22 in ABA-mediated drought and salt tolerances in rice. Plant Mol. Biol. 2012, 80, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Annapurna, B.; Khurana, J.P.; Mukesh, J. Characterization of Rice Homeobox Genes, OsHOX22 and OsHOX24, and Over-expression of OsHOX24 in Transgenic Arabidopsis Suggest Their Role in Abiotic Stress Response. Front. Plant Sci. 2016, 7, 527. [Google Scholar]
- Himmelbach, A. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. Embo J. 2014, 21, 3029–3038. [Google Scholar] [CrossRef] [PubMed]
- Pautot, V.; Dockx, J.; Hamant, O.; Kronenberger, J.; Traas, J. KNAT2: Evidence for a Link between Knotted-like Genes and Carpel Development. Plant Cell 2001, 13, 1719–1734. [Google Scholar] [CrossRef]
- Tsuda, K.; Ito, Y.; Sato, Y.; Kurata, N. Positive Autoregulation of a KNOX Gene Is Essential for Shoot Apical Meristem Maintenance in Rice. Plant Cell 2011, 23, 4368–4381. [Google Scholar] [CrossRef]
- Sato, Y.; Sentoku, N.; Miura Hirochika, H.; Kitano Matsuoka, M. Loss-of-function mutations in the rice homeobox gene OSH15 affect the architecture of internodes resulting in dwarf plants. EMBO J. 1999, 18, 992–1002. [Google Scholar] [CrossRef]
- Shu, Y.J.; Tao, Y.; Wang, S.; Huang, L.; Yu, X. GmSBH1, a homeobox transcription factor gene, relates to growth and development and involves in response to high temperature and humidity stress in soybean. Plant Cell Rep. 2015, 34, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.M.; Zhang, S.B.; Lei, X.H.; Deng, Z.L.; Duan, E.K. Estrogen Leads to Reversible Hair Cycle Retardation through Inducing Premature Catagen and Maintaining Telogen. PLoS ONE 2012, 7, e40124. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Wang, L.; Zhang, J.; Li, J.; Zheng, H.; Chen, J.; Lu, M. WUSCHEL-related Homeobox genes in Populus tomentosa: Diversified expression patterns and a functional similarity in adventitious root formation. BMC Genom. 2014, 15, 296. [Google Scholar] [CrossRef]
- Zhang, X.; Jie, Z.; Liu, J.; Yin, J.; Zhang, D. Genome-wide analysis of WOX gene family in rice, sorghum, maize, Arabidopsis and poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Zhao, K.; Zhang, X.; Cheng, Z.; Yao, W.; Li, R.; Jiang, T.; Zhou, B. Comprehensive analysis of the three-amino-acid-loop-extension gene family and its tissue-differential expression in response to salt stress in poplar. Plant Physiol. Biochem. 2019, 136, 1–12. [Google Scholar] [CrossRef]
- Tuskan, G.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A. The Genome of Black Cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar] [PubMed]
- Leseberg, C.H.; Li, A.; Hui, K.; Duvall, M.; Long, M. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene 2006, 378, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, J.; Cai, B.; Peng, R.H.; Zhu, B.; Jin, X.F.; Xue, Y.; Gao, F.; Fu, X.Y.; Tian, Y.S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Rinaldi, C.; Duplessis, S.; Baucher, M.; Geelen, D.; Duchaussoy, F.; Meyers, B.C.; Boerjan, W.; Martin, F. Genome-wide identification of NBS resistance genes in Populus trichocarpa. Plant Mol. Biol. 2008, 66, 619–636. [Google Scholar] [CrossRef]
- He, H.; Dong, Q.; Shao, Y.; Jiang, H.; Zhu, S.; Cheng, B.; Xiang, Y. Genome-wide survey and characterization of the WRKY gene family in Populus trichocarpa. Plant Cell Rep. 2012, 31, 1199–1217. [Google Scholar] [CrossRef]
- Wang, F.; Dong, Q.; Jiang, H.; Zhu, S.; Chen, B.; Xiang, Y. Genome-wide analysis of the heat shock transcription factors in Populus trichocarpa and Medicago truncatula. Mol. Biol. Rep. 2012, 39, 1877–1886. [Google Scholar] [CrossRef] [PubMed]
- Yer, E.; Baloglu, M.; Ayan, S. Identification and expression profiling of all Hsp family member genes under salinity stress in different poplar clones. Gene 2018, 678, 324–336. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Jin, Y.; Zhang, S.; Wang, X. Genome-wide identification and expression analyses of the homeobox transcription factor family during ovule development in seedless and seeded grapes. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Lou, Y.; Yang, K.; Shan, X.; Zhu, C.; Gao, Z. Identification of Homeobox Genes Associated with Lignification and Their Expression Patterns in Bamboo Shoots. Biomolecules 2019, 9, 862. [Google Scholar] [CrossRef] [PubMed]
- Sultan, S.E. Plant developmental responses to the environment: Eco-devo insights. Curr. Opin. Plant Biol. 2010, 13, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.; Sun, F.; Cheng, T.; Wang, Y.; Xie, K.; Zhang, C.; Xi, Y. Genome-wide identification of members of the TCP gene family in switchgrass (Panicum virgatum L.) and analysis of their expression. Gene 2019, 702, 89–98. [Google Scholar] [CrossRef]
- Seo, D.H.; Ahn, M.Y.; Park, K.Y.; Kim, E.Y.; Kim, W.T. The N-Terminal UND Motif of the Arabidopsis U-Box E3 Ligase PUB18 Is Critical for the Negative Regulation of ABA-Mediated Stomatal Movement and Determines Its Ubiquitination Specificity for Exocyst Subunit Exo70B1. Plant Cell 2016, 28, 2952–2973. [Google Scholar] [CrossRef]
- Tong, S.; Chen, N.; Wang, D.; Ai, F.; Liu, B.; Ren, L.; Chen, Y.; Zhang, J.; Lou, S.; Liu, H. The U-box E3 ubiquitin ligase PalPUB79 positively regulates ABA-dependent drought tolerance via ubiquitination of PalWRKY77 in Populus. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Ramegowda, V.; Mysore, K.S.; Senthil-Kumar, M. Virus-induced gene silencing is a versatile tool for unraveling the functional relevance of multiple abiotic-stress-responsive genes in crop plants. Front. Plant Sci. 2014, 5, 323. [Google Scholar] [CrossRef]
- Senthil-Kumar, M.; Mysore, K.S. New dimensions for VIGS in plant functional genomics. Trends Plant Sci. 2011, 16, 656–665. [Google Scholar] [CrossRef]
- Shen, Z.; Jian, S.; Yao, J.; Wang, S.; Ding, M.; Zhang, H.; Qian, Z.; Nan, Z.; Gang, S.; Rui, Z. High rates of virus-induced gene silencing by tobacco rattle virus in Populus. Tree Physiol. 2015, 35, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Ye, S.; Wang, L.; Duan, Y.; Lu, W.; Liu, H.; Fan, D.; Zhang, F.; Luo, K. Heterologous gene silencing induced by tobacco rattle virus (TRV) is efficient for pursuing functional genomics studies in woody plants. Plant Cell TissueOrgan Cult. (PCTOC) 2014, 116, 163–174. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.D.; Manavella, P.A.; Dezar, C.A.; Chan, R.L. The true story of the HD-Zip family. Trends Plant Sci. 2007, 12, 419–426. [Google Scholar] [CrossRef]
- Wang, W.; Wu, P.; Li, Y.; Hou, X.L. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol. Genet. Genom. 2016, 291, 1451–1464. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Depamphilis, C.W.; Hong, M. Phylogenetic Analysis of the Plant-specific Zinc Finger-Homeobox and Mini Zinc Finger Gene Families. J. Integr. Plant Biol. 2010, 50, 1031–1045. [Google Scholar]
- Khatun, K.; Nath, U.K.; Robin, A.H.K.; Park, J.I.; Lee, D.J.; Kim, M.B.; Kim, C.K.; Lim, K.B.; Nou, I.S.; Chung, M.Y. Genome-wide analysis and expression profiling of zinc finger homeodomain ( ZHD ) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genom. 2017, 18, 1–16. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008, 275, 2845–2861. [Google Scholar] [CrossRef]
- Khan, N.; Hu, C.M.; Khan, W.A.; Wang, W.; Ke, H.; Huijie, D.; Zhishuo, Z.; Hou, X. Genome-wide Identification, Classification, and Expression Pattern of Homeobox Gene Family in Brassica rapa under Various Stresses. Sci. Rep. 2018, 8, 1–17. [Google Scholar]
- Lai, W.; Zhu, C.; Hu, Z.; Liu, S.; Zhou, Y. Identification and Transcriptional Analysis of Zinc Finger-Homeodomain (ZF-HD) Family Genes in Cucumber. Biochem. Genet. 2021, 59, 884–901. [Google Scholar] [CrossRef]
- Sardiello, M.; Cairo, S.; Fontanella, B.; Ballabio, A.; Meroni, G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol. Biol. 2008, 8, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Affolter, M.; Schier, A.; Gehring, W.J. Homeodomain proteins and the regulation of gene expression. Curr. Opin. Cell Biol. 1990, 2, 485–495. [Google Scholar] [CrossRef]
- Bürglin, T.R.; Affolter, M. Homeodomain proteins: An update. Chromosoma 2016, 125, 497–521. [Google Scholar] [CrossRef]
- Holland, P. Evolution of homeobox genes. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 31–45. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bai, B.; Wen, F.; Zhao, M.; Xia, Q.; Yang, D.H.; Wang, G. Genome-Wide Identification and Expression Analysis of HD-ZIP I Gene Subfamily in Nicotiana tabacum. Genes 2019, 10, 575. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Liu, A.; Zhang, Y.; Zhou, Y.; Li, D.; Dossa, K.; Zhou, R.; Zhang, X.; You, J. Genome-wide characterization and expression analysis of the HD-Zip gene family in response to drought and salinity stresses in sesame. BMC Genom. 2019, 20, 748. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Wang, H.L.; Chen, J.; Tian, Q.; Wang, S.; Xia, X.; Yin, W. Identification and validation of reference genes for Populus euphratica gene expression analysis during abiotic stresses by quantitative real-time PCR. Physiol. Plant. 2014, 152, 529–545. [Google Scholar] [CrossRef]
- Wang, J.; Sun, P.P.; Chen, C.L.; Yin, W.; Fu, X.Z.; Liu, J.H. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 2011, 62, 2899. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Tong, S.; Chen, N.; Liu, B.; Bai, Q.; Chen, Y.; Bi, H.; Zhang, Z.; Lou, S.; Tang, H. The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus. Plant J. 2021, 105, 1258–1273. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, J.; Sun, Y.; Wang, L.; Jiang, Y.; Chen, N.; Tong, S. Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa. Plants 2021, 10, 2284. https://doi.org/10.3390/plants10112284
Hou J, Sun Y, Wang L, Jiang Y, Chen N, Tong S. Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa. Plants. 2021; 10(11):2284. https://doi.org/10.3390/plants10112284
Chicago/Turabian StyleHou, Jing, Yan Sun, Lei Wang, Yuanzhong Jiang, Ningning Chen, and Shaofei Tong. 2021. "Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa" Plants 10, no. 11: 2284. https://doi.org/10.3390/plants10112284
APA StyleHou, J., Sun, Y., Wang, L., Jiang, Y., Chen, N., & Tong, S. (2021). Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa. Plants, 10(11), 2284. https://doi.org/10.3390/plants10112284





