The Generic Risks and the Potential of SDN-1 Applications in Crop Plants
Abstract
:1. Introduction
2. Advantages of SDNs over Conventional Breeding
3. SDN-1 Application Types in Crop Plants
3.1. Data Collection of Market-Oriented Applications of Genome Editing Techniques in Crop Plants
- A genome editing technique (including CRISPR/Cas9, ZFN, TALENs, meganucleases, ODM or base editing) was applied in a crop plant;
- A trait was edited that may be of interest for commercialization (market-oriented trait);
- The targeted trait is expressed in the resulting genome edited plant grown.
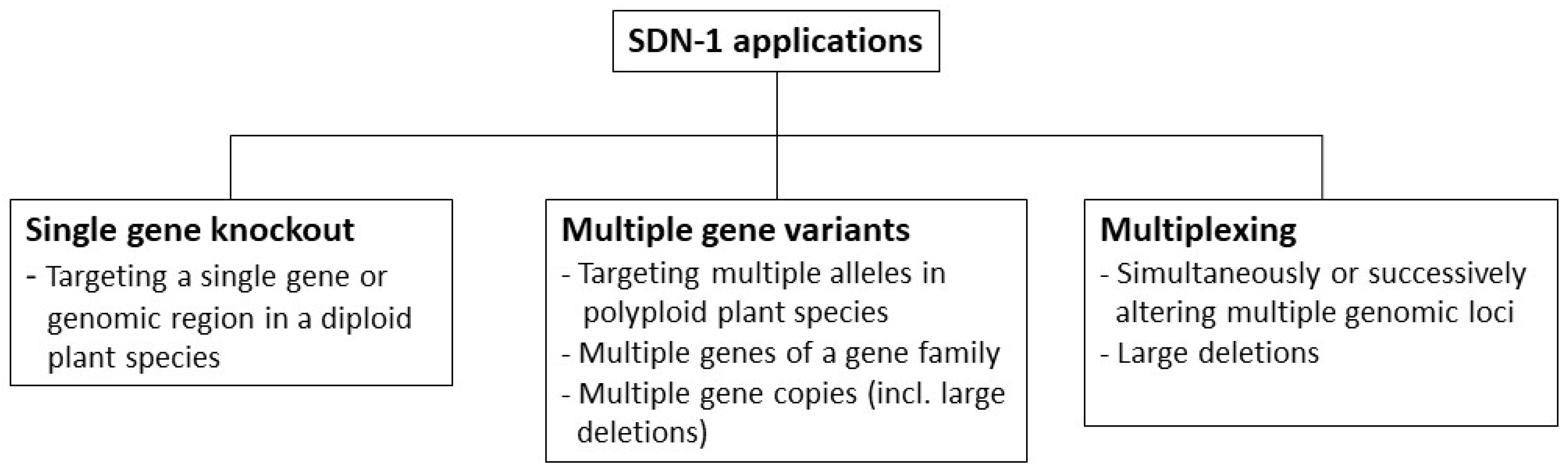
3.2. Methodology for the Categorization of SDN-1 Application Types
3.2.1. Single Gene Knockout
3.2.2. Multiple Gene Variants
3.2.3. Multiplexing
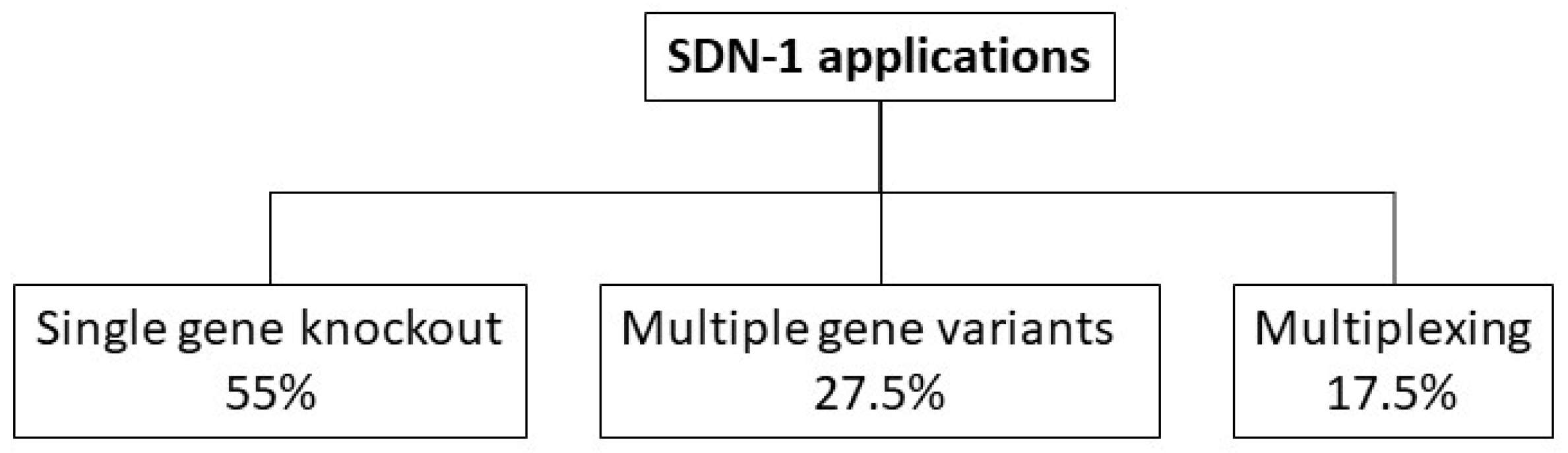
3.3. Categorization of SDN-1 Application Types
3.3.1. Results of the SDN-1 Application Type Categorization
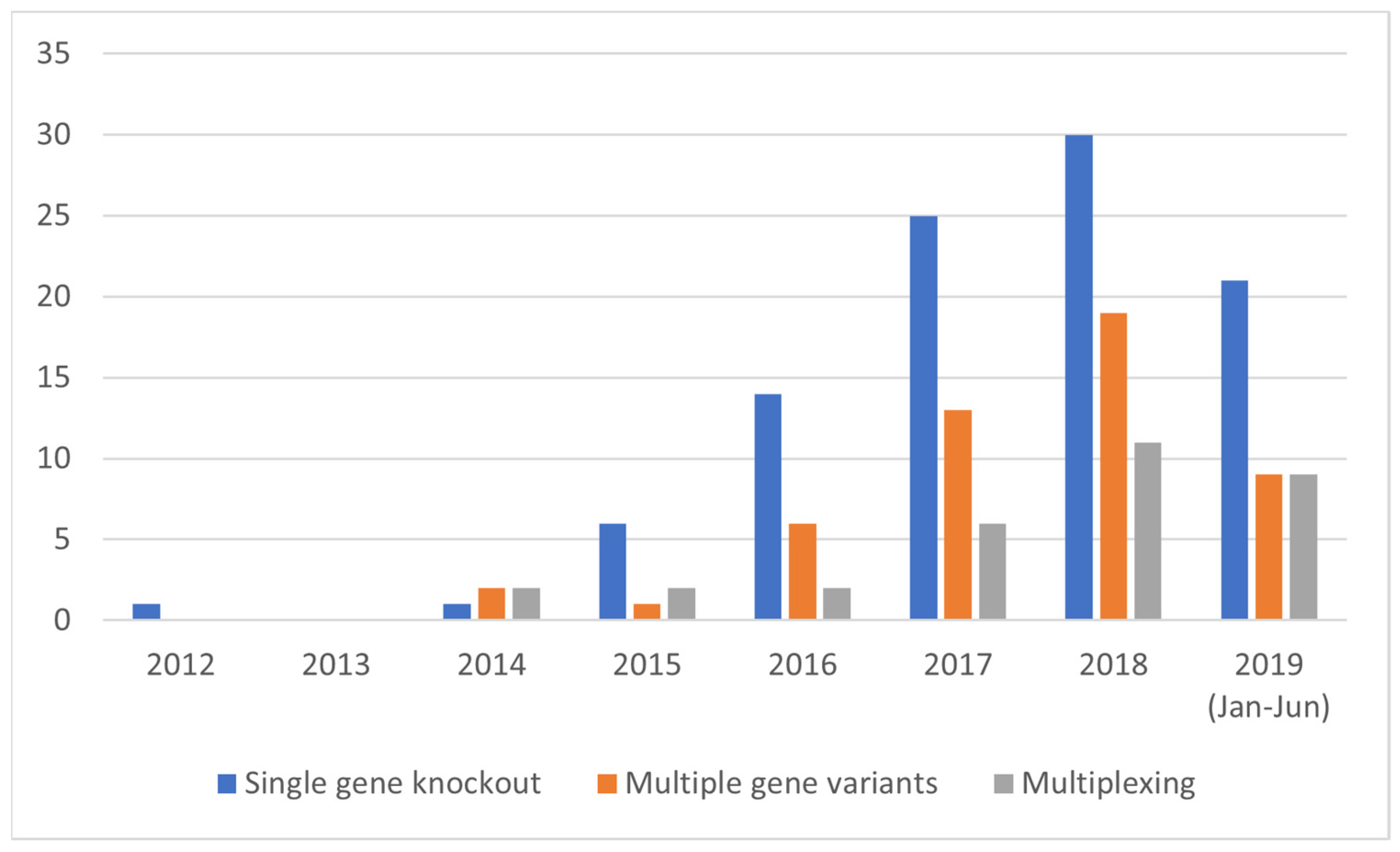
3.3.2. Development of Complex SDN-1 Applications over Time
4. Regulation and Risk Assessment of SDN-1 Applications
4.1. Generic Process-Based Risks
4.2. Product-Based Risks
4.2.1. Traits That Are Known from Previous Breeding
4.2.2. Traits Previously Unknown in Conventional Breeding
5. Discussion
6. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scientific Advisory Mechanism. New Techniques in Agricultural Biotechnology. Available online: https://ec.europa.eu/research/sam/pdf/topics/explanatory_note_new_techniques_agricultural_biotechnology.pdf (accessed on 3 March 2021).
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Crispo, M.; Mulet, A.P.; Tesson, L.; Barrera, N.; Cuadro, F.; dos Santos-Neto, P.C.; Nguyen, T.H.; Creneguy, A.; Brusselle, L.; Anegon, I.; et al. Efficient generation of myostatin knock-out sheep using CRISPR/Cas9 technology and microinjection into zygotes. PLoS ONE 2015, 10, e0136690. [Google Scholar] [CrossRef] [Green Version]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Chen, K.; Liang, Z.; Li, J.; Zhang, Y.; Zhang, K.; Liu, J.; Voytas, D.F.; Zheng, X.; et al. Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol. Plant 2013, 6, 1365–1368. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Dong, H.; Cui, Y.; Cong, L.; Zhang, D. Application of different types of CRISPR/Cas-based systems in bacteria. Microb. Cell Fact. 2020, 19, 172. [Google Scholar] [CrossRef]
- Schuster, M.; Kahmann, R. CRISPR-Cas9 genome editing approaches in filamentous fungi and oomycetes. Fungal Genet. Biol. 2019, 130, 43–53. [Google Scholar] [CrossRef]
- Silva, G.; Poirot, L.; Galetto, R.; Smith, J.; Montoya, G.; Duchateau, P.; Pâques, F. Meganucleases and other tools for targeted genome engineering: Perspectives and challenges for gene therapy. Curr. Gene Ther. 2011, 11, 11–27. [Google Scholar] [CrossRef] [Green Version]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef] [Green Version]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Eckerstorfer, M.F.; Dolezel, M.; Heissenberger, A.; Miklau, M.; Reichenbecher, W.; Steinbrecher, R.A.; Wassmann, F. An EU perspective on biosafety considerations for plants developed by genome editing and other new genetic modification techniques (nGMs). Front. Bioeng. Biotechnol. 2019, 7, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Wilhelm, R. What is the available evidence for the range of applications of genome-editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map. Environ. Evid. 2019, 8, 27. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Peng, N. Endogenous CRISPR-Cas System-Based Genome Editing and Antimicrobials: Review and Prospects. Front. Microbiol. 2019, 10, 2471. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Ran, F.A.; Hsu, P.D.; Konermann, S.; Shehata, S.I.; Dohmae, N.; Ishitani, R.; Zhang, F.; Nureki, O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 2014, 156, 935–949. [Google Scholar] [CrossRef] [Green Version]
- Gorbunova, V.V.; Levy, A.A. How plants make ends meet: DNA double-strand break repair. Trends Plant Sci. 1999, 4, 263–269. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.X.; Mao, L.; Chen, B.; Xu, Y.; et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 23890. [Google Scholar] [CrossRef] [PubMed]
- Petolino, J.F.; Kumar, S. Transgenic trait deployment using designed nucleases. Plant Biotechnol. J. 2016, 14, 503–509. [Google Scholar] [CrossRef]
- Puchta, H. The repair of double-strand breaks in plants: Mechanisms and consequences for genome evolution. J. Exp. Bot. 2004, 56, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef]
- Sauer, N.J.; Narvaez-Vasquez, J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Woodward, M.J.; Mihiret, Y.A.; Lincoln, T.A.; Segami, R.E.; Sanders, S.L.; et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 2016, 170, 1917–1928. [Google Scholar] [CrossRef] [Green Version]
- Judgement of the Court (Grand Chamber), 25 July 2018 in Case C-528/16. Available online: http://curia.europa.eu/juris/document/document.jsf?text=&docid=204387&pageIndex=0&doclang=en&mode=req&dir=&occ=first&part=1&cid=133112 (accessed on 10 February 2021).
- Study on the Status of New Genomic Techniques under Union Law and in Light of the Court of Justice Ruling in Case C-528/16. Available online: https://ec.europa.eu/food/system/files/2021-04/gmo_mod-bio_ngt_eu-study.pdf (accessed on 4 June 2021).
- Towards a Scientifically Justified, Differentiated Regulation of Genome Edited Plants in the EU. Available online: https://www.leopoldina.org/uploads/tx_leopublication/2019_Stellungnahme_Genomeditierte_Pflanzen_web.pdf (accessed on 5 January 2021).
- The Regulation of Genome-Edited Plants in the European Union. Available online: https://www.leopoldina.org/uploads/tx_leopublication/2020_EASAC_Genome-Edited_Plants_Web_01.pdf (accessed on 4 June 2021).
- Kawall, K.; Cotter, J.; Then, C. Broadening the GMO Risk Assessment in the EU for Genome Editing Technologies in Agriculture. Environ. Sci. Eur. 2020, 32, 106. [Google Scholar] [CrossRef]
- Scientific Critique of Leopoldina and EASAC Statements on Genome Edited Plants in the EU. Available online: https://ensser.org/wp-content/uploads/2021/04/Greens-EFA-GMO-Study-1.pdf (accessed on 4 June 2021).
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.D.; Asyraf Md Hatta, M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, K.; Wang, Y.; Zhang, R.; Zhang, H.; Gao, C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019, 70, 667–697. [Google Scholar] [CrossRef] [PubMed]
- Eckerstorfer, M.F.; Grabowski, M.; Lener, M.; Engelhard, M.; Simon, S.; Dolezel, M.; Heissenberger, A.; Lüthi, C. Biosafety of genome editing applications in plant breeding: Considerations for a focused case-specific risk assessment in the EU. BioTech 2021, 10, 10. [Google Scholar] [CrossRef]
- Monroe, J.G.; Srikant, T.; Carbonell-Bejerano, P.; Exposito-Alonso, M.; Weng, M.-L.; Rutter, M.T.; Fenster, C.B.; Weigel, D. Mutation bias shapes gene evolution in Arabidopsis thaliana. bioRxiv 2020. [Google Scholar] [CrossRef]
- Belfield, E.J.; Ding, Z.J.; Jamieson, F.J.C.; Visscher, A.M.; Zheng, S.J.; Mithani, A.; Harberd, N.P. DNA mismatch repair preferentially protects genes from mutation. Genome Res. 2018, 28, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Kawall, K. New possibilities on the horizon: Genome editing makes the whole genome accessible for changes. Front. Plant Sci. 2019, 10, 525. [Google Scholar] [CrossRef]
- Pele, A.; Rousseau-Gueutin, M.; Chevre, A.M. Speciation success of polyploid plants closely relates to the regulation of meiotic recombination. Front. Plant Sci. 2018, 9, 907. [Google Scholar] [CrossRef] [PubMed]
- Glover, N.M.; Redestig, H.; Dessimoz, C. Homoeologs: What Are They and How Do We Infer Them? Trends Plant Sci. 2016, 21, 609–621. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, C.; Fransz, P.; Rönspies, M.; Dreissig, S.; Fuchs, J.; Heckmann, S.; Houben, A.; Puchta, H. Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat. Commun. 2020, 11, 4418. [Google Scholar] [CrossRef] [PubMed]
- Loewe, L. Genetic muation. Nat. Educ. 2008, 1, 113. [Google Scholar]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSalpha. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yazdi, P.G.; Pedersen, B.A.; Taylor, J.F.; Khattab, O.S.; Chen, Y.H.; Chen, Y.; Jacobsen, S.E.; Wang, P.H. Increasing nucleosome occupancy is correlated with an increasing mutation rate so long as DNA repair machinery is intact. PLoS ONE 2015, 10, e0136574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schuster-Bockler, B.; Lehner, B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 2012, 488, 504–507. [Google Scholar] [CrossRef]
- Martincorena, I.; Seshasayee, A.S.; Luscombe, N.M. Evidence of non-random mutation rates suggests an evolutionary risk management strategy. Nature 2012, 485, 95–98. [Google Scholar] [CrossRef] [PubMed]
- Frigola, J.; Sabarinathan, R.; Mularoni, L.; Muinos, F.; Gonzalez-Perez, A.; Lopez-Bigas, N. Reduced mutation rate in exons due to differential mismatch repair. Nat. Genet. 2017, 49, 1684–1692. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Popodi, E.; Tang, H.; Foster, P.L. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 2774–2783. [Google Scholar] [CrossRef] [Green Version]
- Foster, P.L.; Niccum, B.A.; Popodi, E.; Townes, J.P.; Lee, H.; MohammedIsmail, W.; Tang, H. Determinants of Base-Pair Substitution Patterns Revealed by Whole-Genome Sequencing of DNA Mismatch Repair Defective Escherichia coli. Genetics 2018, 209, 1029–1042. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Lehner, B. Clustered mutation signatures reveal that error-prone DNA repair targets mutations to active genes. Cell 2017, 170, 534–547.e23. [Google Scholar] [CrossRef] [Green Version]
- Niccum, B.A.; Lee, H.; MohammedIsmail, W.; Tang, H.; Foster, P.L. The spectrum of replication errors in the absence of error correction assayed across the whole genome of Escherichia coli. Genetics 2018, 209, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhang, Y.; Zhang, Z.; Zheng, Y.; Du, L.; Zhu, B. Preferential protection of genetic fidelity within open chromatin by the mismatch repair machinery. J. Biol. Chem. 2016, 291, 17692–17705. [Google Scholar] [CrossRef] [Green Version]
- Fang, J.; Huang, Y.; Mao, G.; Yang, S.; Rennert, G.; Gu, L.; Li, H.; Li, G.M. Cancer-driving H3G34V/R/D mutations block H3K36 methylation and H3K36me3-MutSalpha interaction. Proc. Natl. Acad. Sci. USA 2018, 115, 9598–9603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Gu, L.; Li, G.M. H3K36me3-mediated mismatch repair preferentially protects actively transcribed genes from mutation. J. Biol. Chem. 2018, 293, 7811–7823. [Google Scholar] [CrossRef] [Green Version]
- Naegeli, H.; Bresson, J.-L.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; Nogue, F.; et al. Evaluation of existing guidelines for their adequacy for the molecular characterisation and environmental risk assessment of genetically modified plants obtained through synthetic biology. EFSA J. 2021, 19, e06301. [Google Scholar] [CrossRef] [PubMed]
- Aktualisierung der Übersicht über Nutz- und Zierpflanzen, die Mittels neuer Molekularbiologischer Techniken für die Bereiche Ernährung, Landwirtschaft und Gartenbau Erzeugt Wurden—Marktorientierte Anwendungen. Available online: https://www.bmel.de/SharedDocs/Downloads/DE/_Landwirtschaft/Gruene-Gentechnik/NMT_Uebersicht-Zier-Nutzpflanzen.pdf?__blob=publicationFile&v=3 (accessed on 16 August 2021).
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome edited crops touch the market: A view on the global development and regulatory environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef]
- First Commercial Sale of Calyxt High Oleic Soybean Oil on the US Market. Available online: https://calyxt.com/wp-content/uploads/2019/02/20190226_PR-Calyno-Commercialization.pdf (accessed on 21 January 2021).
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient increase of ɣ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017, 7, 7057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Launch of the Genome-Edited Tomato with Increased GABA in Japan. Available online: https://sanatech-seed.com/en/20201211-2-2/ (accessed on 2 August 2021).
- Wang, Y.; Meng, Z.; Liang, C.; Meng, Z.; Wang, Y.; Sun, G.; Zhu, T.; Cai, Y.; Guo, S.; Zhang, R.; et al. Increased lateral root formation by CRISPR/Cas9-mediated editing of arginase genes in cotton. Sci. China Life Sci. 2017, 60, 524–527. [Google Scholar] [CrossRef]
- Wang, R.; Tavano, E.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Demorest, Z.L.; Coffman, A.; Baltes, N.J.; Stoddard, T.J.; Clasen, B.M.; Luo, S.; Retterath, A.; Yabandith, A.; Gamo, M.E.; Bissen, J.; et al. Direct stacking of sequence-specific nuclease-induced mutations to produce high oleic and low linolenic soybean oil. BMC Plant Biol. 2016, 16, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Wu, B.; Wang, J.; Zhu, W.; Nie, H.; Qian, J.; Huang, W.; Fang, Z. Blocking amino acid transporter OsAAP3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol. J. 2018, 16, 1710–1722. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wang, G.; Liu, Z. Efficient genome editing of wild strawberry genes, vector development and validation. Plant Biotechnol. J. 2018, 16, 1868–1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, Y.B.; Li, J.; Qin, R.Y.; Xu, R.F.; Li, H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016, 90, 49–62. [Google Scholar] [CrossRef]
- Am I Regulated under 7 CFR Part 340. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/18-051-01_air_response_signed.pdf (accessed on 11 February 2021).
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell 2017, 169, 1142–1155. [Google Scholar] [CrossRef] [Green Version]
- Roldan, M.V.G.; Perilleux, C.; Morin, H.; Huerga-Fernandez, S.; Latrasse, D.; Benhamed, M.; Bendahmane, A. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 2017, 7, 4402. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef]
- Filler Hayut, S.; Melamed Bessudo, C.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 15605. [Google Scholar] [CrossRef]
- Abe, F.; Haque, E.; Hisano, H.; Tanaka, T.; Kamiya, Y.; Mikami, M.; Kawaura, K.; Endo, M.; Onishi, K.; Hayashi, T.; et al. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 2019, 28, 1362–1369. [Google Scholar] [CrossRef] [Green Version]
- Sawai, S.; Ohyama, K.; Yasumoto, S.; Seki, H.; Sakuma, T.; Yamamoto, T.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; Aoki, T.; et al. Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 2014, 26, 3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.-J.; Yoo, C.G.; Flanagan, A.; Pu, Y.; Debnath, S.; Ge, Y.; Ragauskas, A.J.; Wang, Z.-Y. Defined tetra-allelic gene disruption of the 4-coumarate:coenzyme A ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol. Biofuels 2017, 10, 284. [Google Scholar] [CrossRef] [Green Version]
- Wen, S.; Liu, H.; Li, X.; Chen, X.; Hong, Y.; Li, H.; Lu, Q.; Liang, X. TALEN-mediated targeted mutagenesis of fatty acid desaturase 2 (FAD2) in peanut (Arachis hypogaea L.) promotes the accumulation of oleic acid. Plant Mol. Biol. 2018, 97, 177–185. [Google Scholar] [CrossRef]
- Jiang, W.Z.; Henry, I.M.; Lynagh, P.G.; Comai, L.; Cahoon, E.B.; Weeks, D.P. Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol. J. 2017, 15, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Jia, S.; Yobi, A.; Ge, Z.; Sato, S.J.; Zhang, C.; Angelovici, R.; Clemente, T.E.; Holding, D.R. Editing of an alpha-Kafirin gene family increases, digestibility and protein quality in sorghum. Plant Physiol. 2018, 177, 1425–1438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schachtsiek, J.; Stehle, F. Nicotine-free, nontransgenic tobacco (Nicotiana tabacum L.) edited by CRISPR-Cas9. Plant Biotechnol. J. 2019, 17, 2228–2230. [Google Scholar] [CrossRef] [Green Version]
- Sanchez-Leon, S.; Gil-Humanes, J.; Ozuna, C.V.; Gimenez, M.J.; Sousa, C.; Voytas, D.F.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Kannan, B.; Jung, J.H.; Moxley, G.W.; Lee, S.M.; Altpeter, F. TALEN-mediated targeted mutagenesis of more than 100 COMT copies/alleles in highly polyploid sugarcane improves saccharification efficiency without compromising biomass yield. Plant Biotechnol. J. 2018, 16, 856–866. [Google Scholar] [CrossRef] [PubMed]
- Fister, A.S.; Landherr, L.; Maximova, S.N.; Guiltinan, M.J. Transient expression of CRISPR/Cas9 machinery targeting TcNPR3 enhances defense response in Theobroma cacao. Front. Plant Sci. 2018, 9, 268. [Google Scholar] [CrossRef] [Green Version]
- Jung, J.H.; Altpeter, F. TALEN mediated targeted mutagenesis of the caffeic acid O-methyltransferase in highly polyploid sugarcane improves cell wall composition for production of bioethanol. Plant Mol. Biol. 2016, 92, 131–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morineau, C.; Bellec, Y.; Tellier, F.; Gissot, L.; Kelemen, Z.; Nogue, F.; Faure, J.D. Selective gene dosage by CRISPR-Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017, 15, 729–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Simmonds, J.; Pan, Q.; Davidson, D.; He, F.; Battal, A.; Akhunova, A.; Trick, H.N.; Uauy, C.; Akhunov, E. Gene editing and mutagenesis reveal inter-cultivar differences and additivity in the contribution of TaGW2 homoeologues to grain size and weight in wheat. Theor. Appl. Genet. 2018, 131, 2463–2475. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, D.; Zhang, D.; Zhao, X.; Cao, X.; Dong, L.; Liu, J.; Chen, K.; Zhang, H.; Gao, C.; et al. Analysis of the functions of TaGW2 homoeologs in wheat grain weight and protein content traits. Plant J. 2018, 94, 857–866. [Google Scholar] [CrossRef] [Green Version]
- Okada, A.; Arndell, T.; Borisjuk, N.; Sharma, N.; Watson-Haigh, N.S.; Tucker, E.J.; Baumann, U.; Langridge, P.; Whitford, R. CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol. J. 2019, 17, 1905–1913. [Google Scholar] [CrossRef] [Green Version]
- Shen, L.; Hua, Y.; Fu, Y.; Li, J.; Liu, Q.; Jiao, X.; Xin, G.; Wang, J.; Wang, X.; Yan, C.; et al. Rapid generation of genetic diversity by multiplex CRISPR/Cas9 genome editing in rice. Sci. China Life Sci. 2017, 60, 506–515. [Google Scholar] [CrossRef]
- Li, X.; Zhou, W.; Ren, Y.; Tian, X.; Lv, T.; Wang, Z.; Fang, J.; Chu, C.; Yang, J.; Bu, Q. High-efficiency breeding of early-maturing rice cultivars via CRISPR/Cas9-mediated genome editing. J. Genet. Genom. 2017, 44, 175–178. [Google Scholar] [CrossRef]
- Li, S.; Shen, L.; Hu, P.; Liu, Q.; Zhu, X.; Qian, Q.; Wang, K.; Wang, Y. Developing disease-resistant thermosensitive male sterile rice by multiplex gene editing. J. Integr. Plant Biol. 2019, 61, 1201–1205. [Google Scholar] [CrossRef]
- Zsogon, A.; Cermak, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Zhu, M.; Xu, Z.; Xu, Q. Assessment of the effect of ten heading time genes on reproductive transition and yield components in rice using a CRISPR/Cas9 system. Theor. Appl. Genet. 2019, 132, 1887–1896. [Google Scholar] [CrossRef] [PubMed]
- Am I Regulated under 7 CFR Part 340. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/air_isu_ting_rice.pdf (accessed on 12 February 2021).
- Shibuya, K.; Watanabe, K.; Ono, M. CRISPR/Cas9-mediated mutagenesis of the EPHEMERAL1 locus that regulates petal senescence in Japanese morning glory. Plant Physiol. Biochem. 2018, 131, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Kui, L.; Chen, H.; Zhang, W.; He, S.; Xiong, Z.; Zhang, Y.; Yan, L.; Zhong, C.; He, F.; Chen, J.; et al. Building a genetic manipulation tool box for orchid biology: Identification of constitutive promoters and application of CRISPR/Cas9 in the orchid, Dendrobium officinale. Front. Plant Sci. 2017, 7, 2036. [Google Scholar] [CrossRef]
- Xu, J.; Kang, B.C.; Naing, A.H.; Bae, S.J.; Kim, J.S.; Kim, H.; Kim, C.K. CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol. J. 2020, 18, 287–297. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Chen, L.; Sun, S.; Wu, C.; Yao, W.; Jiang, B.; Han, T.; Hou, W. CRISPR/Cas9-mediated deletion of large genomic fragments in soybean. Int. J. Mol. Sci. 2018, 19, 3835. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Jiang, G.; Yang, L.; Qiu, L.; He, P.; Nong, C.; Wang, Y.; He, Y.; Xing, Y. Gene diagnosis and targeted breeding for blast-resistant Kongyu 131 without changing regional adaptability. J. Genet. Genom. 2018, 45, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Long, L.; Tian, X.; Xu, F.; Liu, J.; Singh, P.K.; Botella, J.R.; Song, C. Genome editing in cotton with the CRISPR/Cas9 system. Front. Plant Sci. 2017, 8, 1364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massel, K.; Lam, Y.; Wong, A.C.S.; Hickey, L.T.; Borrell, A.K.; Godwin, I.D. Hotter, drier, CRISPR: The latest edit on climate change. Theor. Appl. Genet. 2021, 134, 1691–1709. [Google Scholar] [CrossRef] [PubMed]
- Curtin, S.J.; Xiong, Y.; Michno, J.M.; Campbell, B.W.; Stec, A.O.; Cermak, T.; Starker, C.; Voytas, D.F.; Eamens, A.L.; Stupar, R.M. CRISPR/Cas9 and TALENs generate heritable mutations for genes involved in small RNA processing of Glycine max and Medicago truncatula. Plant Biotechnol. J. 2018, 16, 1125–1137. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Njuguna, E.; Griet, C.; Aesaert, S.; Neyt, P.; Anami, S.; van Lijsebettens, M. Modulation of energy homeostasis in maize and Arabidopsis to develop lines tolerant to drought, genotoxic and oxidative stresses. Afrika Focus 2017, 30, 66–76. [Google Scholar] [CrossRef] [Green Version]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [CrossRef] [PubMed]
- Blanvillain-Baufumé, S.; Reschke, M.; Solé, M.; Auguy, F.; Doucoure, H.; Szurek, B.; Meynard, D.; Portefaix, M.; Cunnac, S.; Guiderdoni, E.; et al. Targeted promoter editing for rice resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14-inducing TAL effectors. Plant Biotechnol. J. 2017, 15, 306–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390–392. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Ge, X.; Luo, X.; Wang, P.; Fan, Q.; Hu, G.; Xiao, J.; Li, F.; Wu, J. Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against Verticillium dahliae in allotetraploid upland cotton. Front. Plant Sci. 2018, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-mediated multiplex genome editing of the BnWRKY11 and BnWRKY70 genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2716. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Wang, C.; Liu, P.; Lei, C.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tu, M.; Wang, D.; Liu, J.; Li, Y.; Li, Z.; Wang, Y.; Wang, X. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol. J. 2018, 16, 844–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Bai, Q.; Xu, P.; Wu, T.; Guo, D.; Peng, Y.; Zhang, H.; Deng, X.; Chen, X.; Luo, M.; et al. Mutation in rice Abscisic Acid2 results in cell death, enhanced disease-resistance, altered seed dormancy and development. Front. Plant Sci. 2018, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Clasen, B.M.; Stoddard, T.J.; Luo, S.; Demorest, Z.L.; Li, J.; Cedrone, F.; Tibebu, R.; Davison, S.; Ray, E.E.; Daulhac, A.; et al. Improving cold storage and processing traits in potato through targeted gene knockout. Plant Biotechnol. J. 2016, 14, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, D.; Jin, L.; Ruan, Y.; Shen, W.H.; Liu, C. Histone lysine methyltransferases BnaSDG8.A and BnaSDG8.C are involved in the floral transition in Brassica napus. Plant J. 2018, 95, 672–685. [Google Scholar] [CrossRef]
- Huang, C.; Sun, H.; Xu, D.; Chen, Q.; Liang, Y.; Wang, X.; Xu, G.; Tian, J.; Wang, C.; Li, D.; et al. ZmCCT9 enhances maize adaptation to higher latitudes. Proc. Natl. Acad. Sci. USA 2018, 115, E334–E341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wu, B.; Lu, K.; Wei, Q.; Qian, J.; Chen, Y.; Fang, Z. The Amino Acid Permease 5 (OsAAP5) regulates tiller number and grain yield in rice. Plant Physiol. 2019, 180, 1031–1045. [Google Scholar] [CrossRef]
- Ozseyhan, M.E.; Kang, J.; Mu, X.; Lu, C. Mutagenesis of the FAE1 genes significantly changes fatty acid composition in seeds of Camelina sativa. Plant Physiol. Biochem. 2018, 123, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Nakayasu, M.; Akiyama, R.; Lee, H.J.; Osakabe, K.; Osakabe, Y.; Watanabe, B.; Sugimoto, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; et al. Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 2018, 131, 70–77. [Google Scholar] [CrossRef] [PubMed]
- McGinn, M.; Phippen, W.B.; Chopra, R.; Bansal, S.; Jarvis, B.A.; Phippen, M.E.; Dorn, K.M.; Esfahanian, M.; Nazarenus, T.J.; Cahoon, E.B.; et al. Molecular tools enabling pennycress (Thlaspi arvense) as a model plant and oilseed cash cover crop. Plant Biotechnol. J. 2019, 17, 776–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Qi, X.; Zhu, J.; Liu, C.; Zhang, X.; Cheng, B.; Mao, L.; Xie, C. Supersweet and waxy: Meeting the diverse demands for specialty maize by genome editing. Plant Biotechnol. J. 2019, 17, 1853–1855. [Google Scholar] [CrossRef]
- Kelliher, T.; Starr, D.; Su, X.; Tang, G.; Chen, Z.; Carter, J.; Wittich, P.E.; Dong, S.; Green, J.; Burch, E.; et al. One-step genome editing of elite crop germplasm during haploid induction. Nat. Biotechnol. 2019, 37, 287–292. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, Y.; Liu, C.; Qi, X.; Jiao, Y.; Wang, D.; Wang, Y.; Liu, Z.; Chen, C.; Chen, B.; Tian, X.; et al. Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants 2019, 5, 575–580. [Google Scholar] [CrossRef]
- Shi, Q.S.; Wang, K.Q.; Li, Y.L.; Zhou, L.; Xiong, S.X.; Han, Y.; Zhang, Y.F.; Yang, N.Y.; Yang, Z.N.; Zhu, J. OsPKS1 is required for sexine layer formation, which shows functional conservation between rice and Arabidopsis. Plant Sci. 2018, 277, 145–154. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Niu, B.; Long, Y.; Li, G.; Tang, J.; Zhang, Y.; Ren, D.; Liu, Y.G.; Chen, L. Suppression or knockout of SaF/SaM overcomes the Sa-mediated hybrid male sterility in rice. J. Integr. Plant Biol. 2017, 59, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhang, D.; Chen, M.; Liang, W.; Wei, J.; Qi, Y.; Yuan, Z. Development of japonica photo-sensitive genic male sterile rice lines by editing carbon starved anther using CRISPR/Cas9. J. Genet. Genom. 2016, 43, 415–419. [Google Scholar] [CrossRef]
- Andersson, M.; Turesson, H.; Nicolia, A.; Falt, A.S.; Samuelsson, M.; Hofvander, P. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017, 36, 117–128. [Google Scholar] [CrossRef] [Green Version]
- Veillet, F.; Chauvin, L.; Kermarrec, M.P.; Sevestre, F.; Merrer, M.; Terret, Z.; Szydlowski, N.; Devaux, P.; Gallois, J.L.; Chauvin, J.E. The Solanum tuberosum GBSSI gene: A target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 2019, 38, 1065–1080. [Google Scholar] [CrossRef]
- Kusano, H.; Ohnuma, M.; Mutsuro-Aoki, H.; Asahi, T.; Ichinosawa, D.; Onodera, H.; Asano, K.; Noda, T.; Horie, T.; Fukumoto, K.; et al. Establishment of a modified CRISPR/Cas9 system with increased mutagenesis frequency using the translational enhancer dMac3 and multiple guide RNAs in potato. Sci. Rep. 2018, 8, 13753. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Zhu, C.; Zheng, M.; Liu, M.; Zhang, D.; Liu, B.; Li, Q.; Si, J.; Ren, X.; Song, H. CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic. Res. 2019, 6, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Wang, C.; Liu, Q.; Shen, Y.; Hua, Y.; Wang, J.; Lin, J.; Wu, M.; Sun, T.; Cheng, Z.; Mercier, R.; et al. Clonal seeds from hybrid rice by simultaneous genome engineering of meiosis and fertilization genes. Nat. Biotechnol. 2019, 37, 283–286. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garland, S. EU policy must change to reflect the potential of gene editing for addressing climate change. Glob. Food Secur. 2021, 28, 100496. [Google Scholar] [CrossRef]
- Directive 2001/18/EC of the European Parliament and of the Council of 12 March 2001 on the Deliberate Release into the Environment of Genetically Modified Organisms and Repealing Council Directive 90/220/EEC. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/HTML/?uri=CELEX:32001L0018&from=EN (accessed on 28 April 2021).
- EFSA. Guidance on the Environmental Risk Assessment of Genetically Modified Plants. EFSA J. 2010, 8, 111. [Google Scholar] [CrossRef]
- EFSA. Guidance for Risk Assessment of Food and Feed from Genetically Modified Plants. EFSA J. 2011, 9, 2150. [Google Scholar] [CrossRef]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Kosicki, M.; Allen, F.; Bradley, A. Cas9-induced large deletions and small indels are controlled in a convergent fashion. bioRxiv 2020. [Google Scholar] [CrossRef]
- Lalonde, S.; Stone, O.A.; Lessard, S.; Lavertu, A.; Desjardins, J.; Beaudoin, M.; Rivas, M.; Stainier, D.Y.R.; Lettre, G. Frameshift indels introduced by genome editing can lead to in-frame exon skipping. PLoS ONE 2017, 12, e0178700. [Google Scholar] [CrossRef] [PubMed]
- Kapahnke, M.; Banning, A.; Tikkanen, R. Random splicing of several exons caused by a single base change in the target exon of CRISPR/Cas9 mediated gene knockout. Cells 2016, 5, 45. [Google Scholar] [CrossRef]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar] [CrossRef] [Green Version]
- Wolt, J.D.; Wang, K.; Sashital, D.; Lawrence-Dill, C.J. Achieving plant CRISPR targeting that limits off-target effects. Plant Genome 2016, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Sharpe, J.J.; Cooper, T.A. Unexpected consequences: Exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017, 18, 109. [Google Scholar] [CrossRef] [Green Version]
- Adikusuma, F.; Piltz, S.; Corbett, M.A.; Turvey, M.; McColl, S.R.; Helbig, K.J.; Beard, M.R.; Hughes, J.; Pomerantz, R.T.; Thomas, P.Q. Large deletions induced by Cas9 cleavage. Nature 2018, 560, E8–E9. [Google Scholar] [CrossRef] [Green Version]
- Tuladhar, R.; Yeu, Y.; Tyler Piazza, J.; Tan, Z.; Rene Clemenceau, J.; Wu, X.; Barrett, Q.; Herbert, J.; Mathews, D.H.; Kim, J.; et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019, 10, 4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, R.; Yasuhiko, Y.; Aisaki, K.I.; Kitajima, S.; Kanno, J.; Hirabayashi, Y. Exosome-mediated horizontal gene transfer occurs in double-strand break repair during genome editing. Commun. Biol. 2019, 2, 57. [Google Scholar] [CrossRef] [PubMed]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.J.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef]
- Skryabin, B.V.; Kummerfeld, D.-M.; Gubar, L.; Seeger, B.; Kaiser, H.; Stegemann, A.; Roth, J.; Meuth, S.G.; Pavenstädt, H.; Sherwood, J.; et al. Pervasive head-to-tail insertions of DNA templates mask desired CRISPR-Cas9–mediated genome editing events. Sci. Adv. 2020, 6, eaax2941. [Google Scholar] [CrossRef] [Green Version]
- Weisheit, I.; Kroeger, J.A.; Malik, R.; Klimmt, J.; Crusius, D.; Dannert, A.; Dichgans, M.; Paquet, D. Detection of deleterious on-target effects after HDR-mediated CRISPR editing. Cell Rep. 2020, 31, 107689. [Google Scholar] [CrossRef] [PubMed]
- Michno, J.M.; Virdi, K.; Stec, A.O.; Liu, J.; Wang, X.; Xiong, Y.; Stupar, R.M. Integration, abundance, and transmission of mutations and transgenes in a series of CRISPR/Cas9 soybean lines. BMC Biotechnol. 2020, 20, 10. [Google Scholar] [CrossRef] [Green Version]
- Norris, A.L.; Lee, S.S.; Greenlees, K.J.; Tadesse, D.A.; Miller, M.F.; Lombardi, H.A. Template plasmid integration in germline genome-edited cattle. Nat. Biotechnol. 2020, 38, 163–164. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, J.; Zhou, R.; Garcia, S.P.; Iyer, S.; Lareau, C.A.; Aryee, M.J.; Joung, J.K. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 2019, 569, 433–437. [Google Scholar] [CrossRef]
- Liu, M.; Zhang, W.; Xin, C.; Yin, J.; Shang, Y.; Ai, C.; Li, J.; Meng, F.-L.; Hu, J. Global detection of DNA repair outcomes induced by CRISPR–Cas9. Nucleic Acids Res. 2021, 49, 8732–8742. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.; Tian, J.; Li, R.; Chen, X.; Luo, Z.; Chen, M.; Zhao, X.; Zhang, D.; Persson, S.; Yuan, Z.; et al. Investigation of CRISPR/Cas9-induced SD1 rice mutants highlights the importance of molecular characterization in plant molecular breeding. J. Genet. Genom. 2020, 47, 273–280. [Google Scholar] [CrossRef]
- Burgio, G.; Teboul, L. Anticipating and identifying collateral damage in genome editing. Trends Genet. 2020, 36, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Commission Implementing Regulation (EU) No 503/2013 of 3 April 2013 on Applications for Authorisation of Genetically Modified Food and Feed in Accordance with Regulation (EC) No 1829/2003 of the European Parliament and of the Council and Amending Commission Regulations (EC) No 641/2004 and (EC) No 1981/2006. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?qid=1565205598558&uri=CELEX:32013R0503 (accessed on 7 February 2021).
- Brinkman, E.K.; Chen, T.; de Haas, M.; Holland, H.A.; Akhtar, W.; van Steensel, B. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Mol. Cell 2018, 70, 801–813.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taagen, E.; Bogdanove, A.J.; Sorrells, M.E. Counting on crossovers: Controlled recombination for plant breeding. Trends Plant Sci. 2020, 25, 455–465. [Google Scholar] [CrossRef] [Green Version]
- Mou, H.; Smith, J.L.; Peng, L.; Yin, H.; Moore, J.; Zhang, X.O.; Song, C.Q.; Sheel, A.; Wu, Q.; Ozata, D.M.; et al. CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol. 2017, 18, 108. [Google Scholar] [CrossRef]
- Gelvin, S.B. Integration of Agrobacterium T-DNA into the plant genome. Annu. Rev. Genet. 2017, 51, 195–217. [Google Scholar] [CrossRef] [PubMed]
- Forsbach, A.; Schubert, D.; Lechtenberg, B.; Gils, M.; Schmidt, R. A comprehensive characterization of single-copy T-DNA insertions in the Arabidopsis thaliana genome. Plant Mol. Biol. 2003, 52, 161–176. [Google Scholar] [CrossRef]
- Jupe, F.; Rivkin, A.C.; Michael, T.P.; Zander, M.; Motley, S.T.; Sandoval, J.P.; Slotkin, R.K.; Chen, H.; Castanon, R.; Nery, J.R.; et al. The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet. 2019, 15, e1007819. [Google Scholar] [CrossRef] [Green Version]
- Makarevitch, I.; Svitashev, S.K.; Somers, D.A. Complete sequence analysis of transgene loci from plants transformed via microprojectile bombardment. Plant Mol. Biol. 2003, 52, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Windels, P.; De Buck, S.; Van Bockstaele, E.; De Loose, M.; Depicker, A. T-DNA integration in Arabidopsis chromosomes. Presence and origin of filler DNA sequences. Plant Physiol. 2003, 133, 2061–2068. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Nannas, N.J.; Fu, F.-F.; Shi, J.; Aspinwall, B.; Parrott, W.A.; Dawe, R.K. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 2019, 31, 368–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilson, A.K.; Latham, J.R.; Steinbrecher, R.A. Transformation-induced mutations in transgenic plants: Analysis and biosafety implications. Biotechnol. Genet. Eng. Rev. 2006, 23, 209–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liu, Z.B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Wu, J.; He, C. Induction of chromosomal inversion by integration of T-DNA in the rice genome. J. Genet. Genom. 2010, 37, 189–196. [Google Scholar] [CrossRef]
- Clark, K.A.; Krysan, P.J. Chromosomal translocations are a common phenomenon in Arabidopsis thaliana T-DNA insertion lines. Plant J. 2010, 64, 990–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nacry, P.; Camilleri, C.; Courtial, B.; Caboche, M.; Bouchez, D. Major chromosomal rearrangements induced by T-DNA transformation in Arabidopsis. Genetics 1998, 149, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.K.; Pawlowski, W.P.; Makarevitch, I.; Plank, D.W.; Somers, D.A. Complex transgene locus structures implicate multiple mechanisms for plant transgene rearrangement. Plant J. 2002, 32, 433–445. [Google Scholar] [CrossRef] [Green Version]
- Pawlowski, W.P.; Somers, D.A. Transgene inheritance in plants genetically engineered by microprojectile bombardment. Mol. Biotechnol. 1996, 6, 17–30. [Google Scholar] [CrossRef]
- Overview of EFSA and European National Authorities’ Scientic Opinions on the Risk Assessment of Plants Developed through New Genomic Techniques. Available online: https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/j.efsa.2021.6314 (accessed on 10 June 2021).
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstadler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Yu, H.; Lin, T.; Meng, X.; Du, H.; Zhang, J.; Liu, G.; Chen, M.; Jing, Y.; Kou, L.; Li, X.; et al. A route to de novo domestication of wild allotetraploid rice. Cell 2021, 184, 1156–1170.e14. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.R. Road map for domesticating multi-genome rice using gene editing. Nature 2021, 591, 537–538. [Google Scholar] [CrossRef]
- Zhou, J.; Xin, X.; He, Y.; Chen, H.; Li, Q.; Tang, X.; Zhong, Z.; Deng, K.; Zheng, X.; Akher, S.A.; et al. Multiplex QTL editing of grain-related genes improves yield in elite rice varieties. Plant Cell Rep. 2019, 38, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Rostoks, N. Implications of the EFSA scientific opinion on Site Directed Nucleases 1 and 2 for risk assessment of genome-edited plants in the EU. Agronomy 2021, 11, 572. [Google Scholar] [CrossRef]
- Current and Future Market Applications of New Genomic Techniques. Available online: https://publications.jrc.ec.europa.eu/repository/handle/JRC123830 (accessed on 11 June 2021).
- Walkowiak, S.; Gao, L.; Monat, C.; Haberer, G.; Kassa, M.T.; Brinton, J.; Ramirez-Gonzalez, R.H.; Kolodziej, M.C.; Delorean, E.; Thambugala, D.; et al. Multiple wheat genomes reveal global variation in modern breeding. Nature 2020, 588, 277–283. [Google Scholar] [CrossRef]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Gonda, I.; Sun, H.; Ma, Q.; Bao, K.; Tieman, D.M.; Burzynski-Chang, E.A.; Fish, T.L.; Stromberg, K.A.; Sacks, G.L.; et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat. Genet. 2019, 51, 1044–1051. [Google Scholar] [CrossRef]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef] [PubMed]
- Applicability of the EFSA Opinion on Site-Directed Nucleases Type 3 for the Safety Assessment of Plants Developed Using Site-Directed Nucleases Type 1 and 2 and Oligonucleotide-Directed Mutagenesis. Available online: https://efsa.onlinelibrary.wiley.com/doi/epdf/10.2903/j.efsa.2020.6299 (accessed on 11 June 2021).
| Category of Market-Orientated Applications | Single Gene Knockouts | Multiple Gene Variants | Multiplexing | Total |
|---|---|---|---|---|
| Abiotic stress resistance | 3 | 2 | 1 | 6 |
| Biotic stress resistance | 15 | 9 | 4 | 28 |
| Agronomic value | 35 | 11 | 15 | 61 |
| Food and feed quality | 24 | 18 | 5 | 47 |
| Herbicide tolerant plants | 2 | - | 1 | 3 |
| Enhanced breeding | 17 | 2 | 1 | 20 |
| Industrial utilization | 1 | 7 | - | 8 |
| Multiple traits | 1 | - | 4 | 5 |
| In total | 98 | 49 | 31 | 178 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kawall, K. The Generic Risks and the Potential of SDN-1 Applications in Crop Plants. Plants 2021, 10, 2259. https://doi.org/10.3390/plants10112259
Kawall K. The Generic Risks and the Potential of SDN-1 Applications in Crop Plants. Plants. 2021; 10(11):2259. https://doi.org/10.3390/plants10112259
Chicago/Turabian StyleKawall, Katharina. 2021. "The Generic Risks and the Potential of SDN-1 Applications in Crop Plants" Plants 10, no. 11: 2259. https://doi.org/10.3390/plants10112259
APA StyleKawall, K. (2021). The Generic Risks and the Potential of SDN-1 Applications in Crop Plants. Plants, 10(11), 2259. https://doi.org/10.3390/plants10112259






