Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress
Abstract
:1. Introduction
2. Results and Discussion
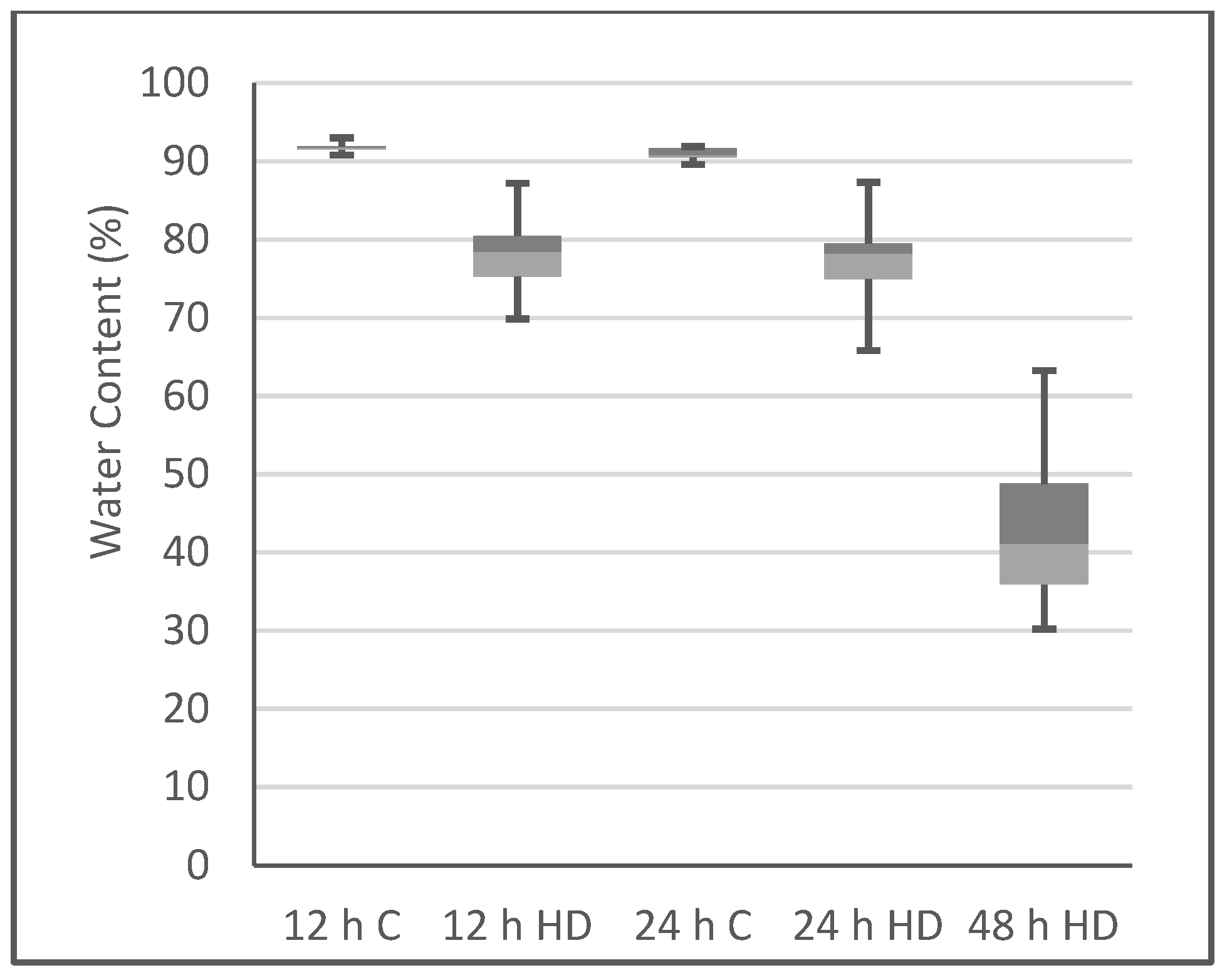
2.1. Plant Phenotype after Heat/Drought Treatments
2.2. RNA-Seq Libraries
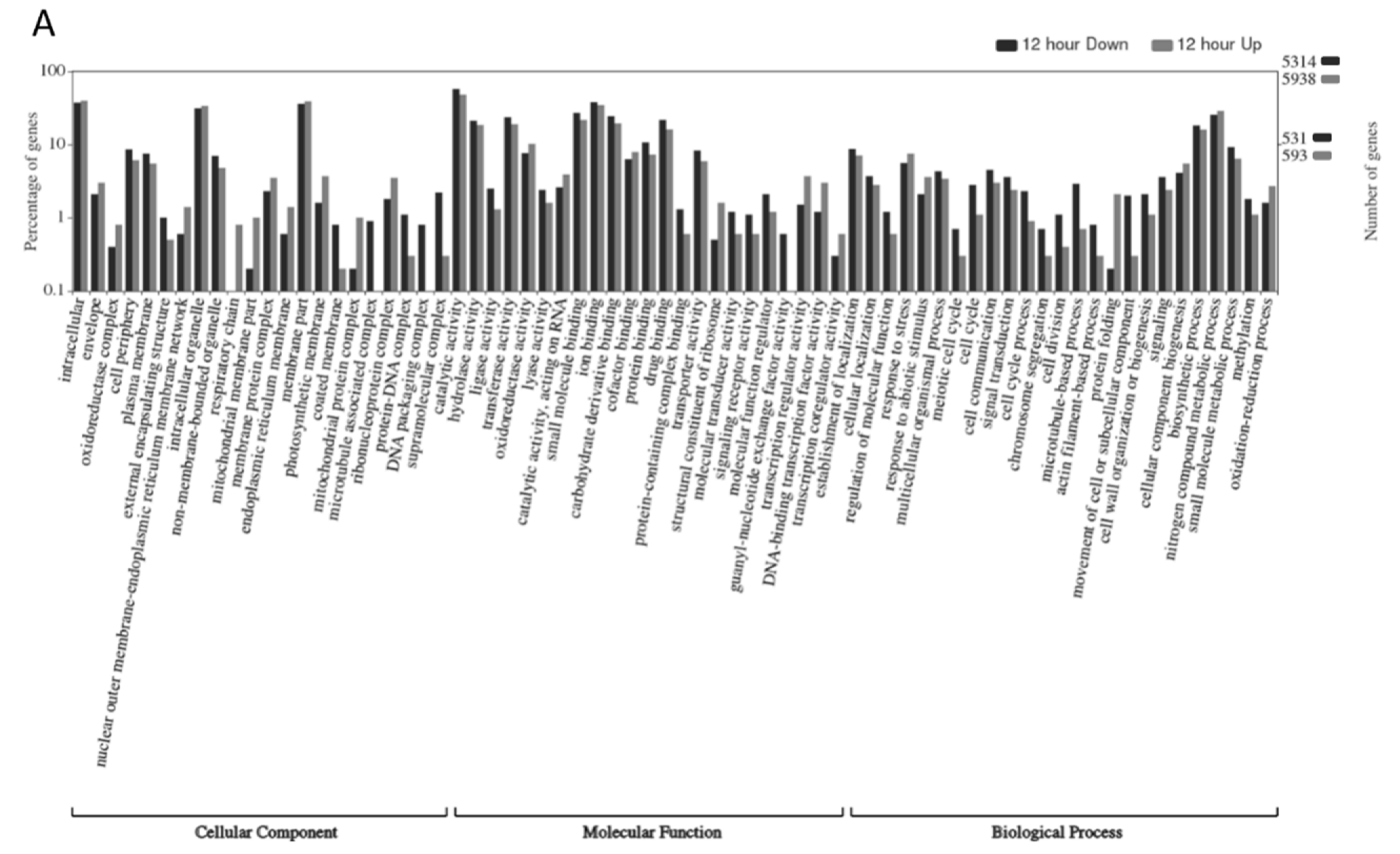
2.3. Gene Ontology Analyses
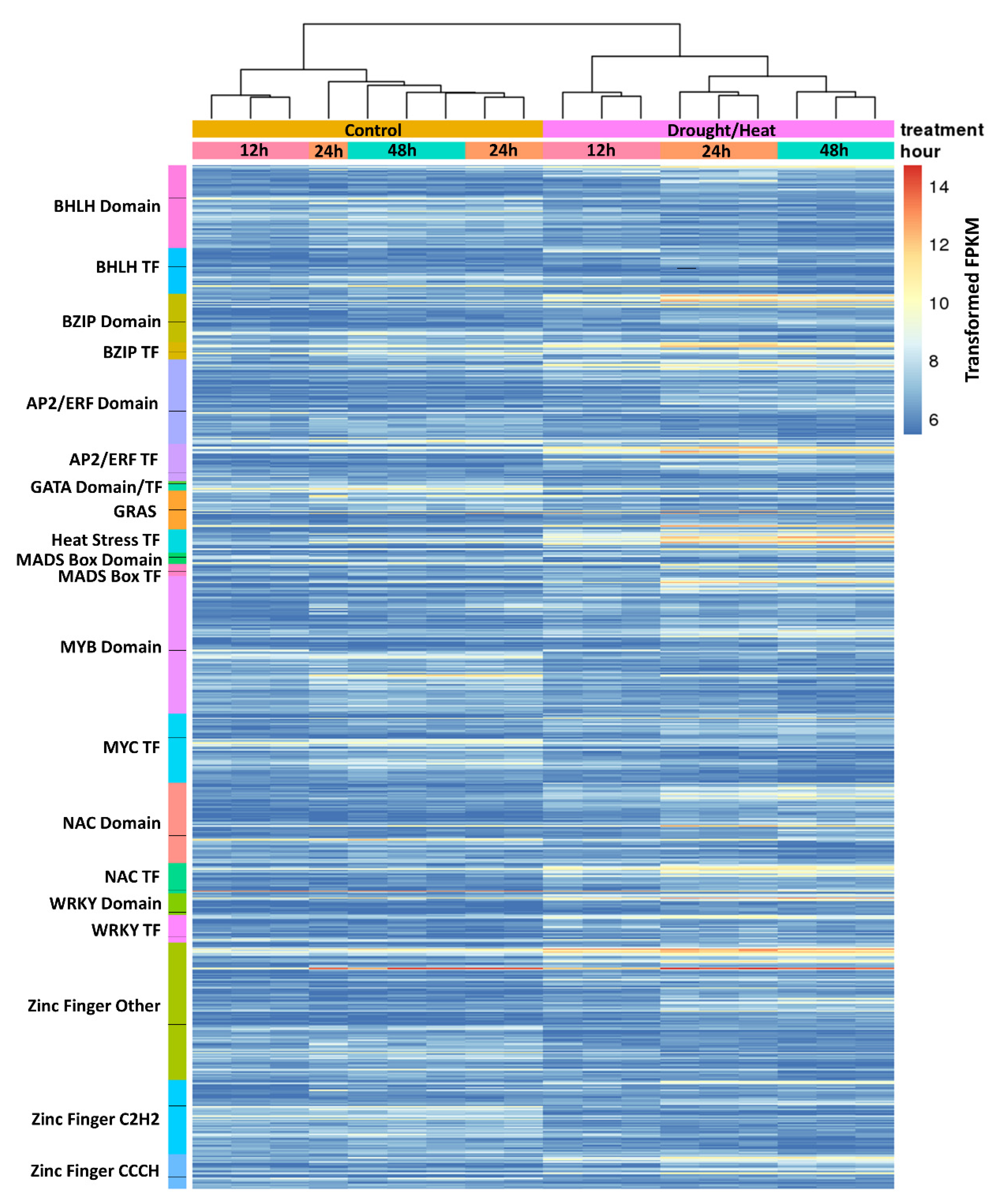
2.4. Analysis of Transcription Factors Differentially Regulated during Heat/Drought Stress
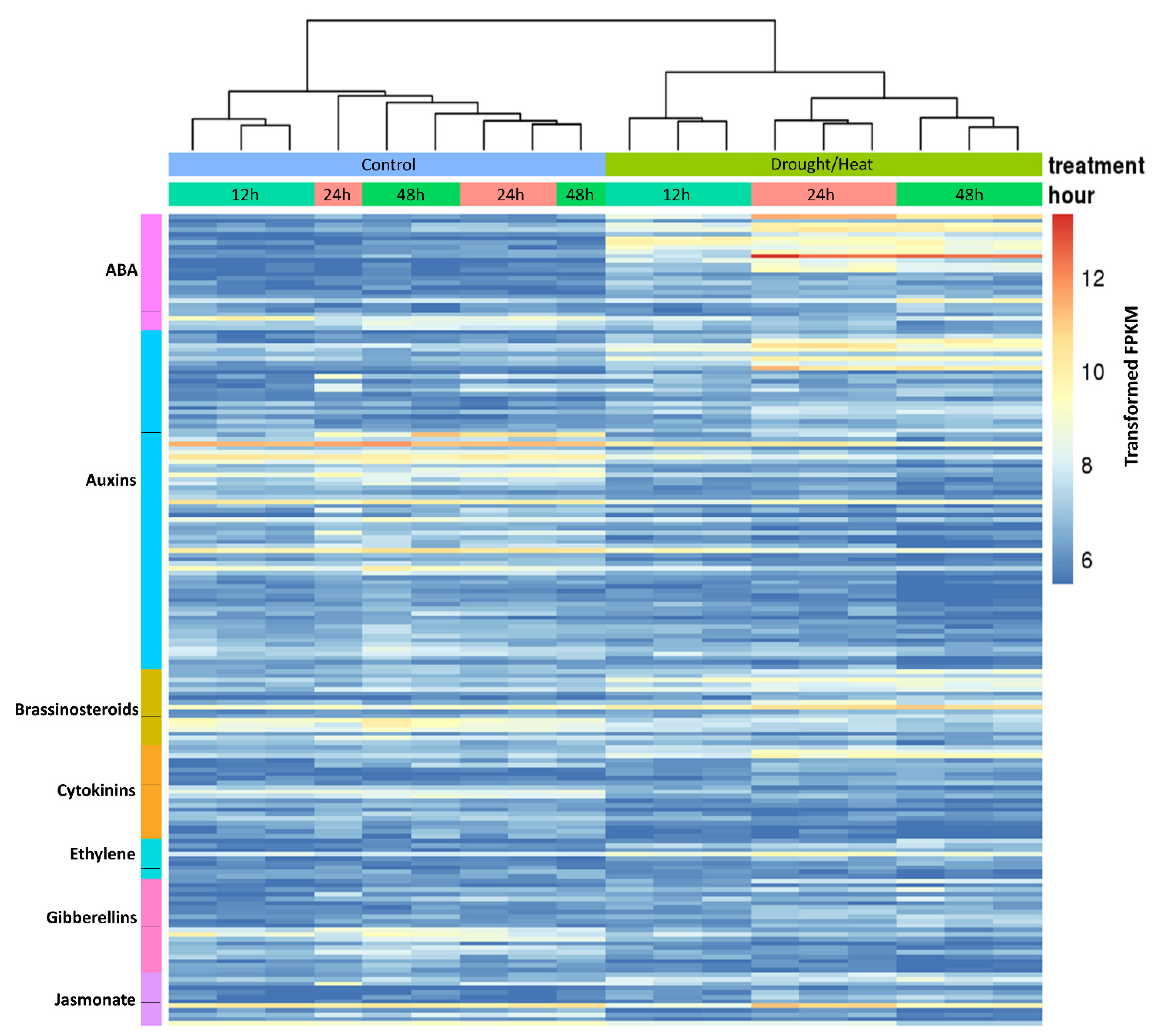
2.5. Analysis of Hormone-Related Genes Differentially Regulated during Heat/Drought Stress
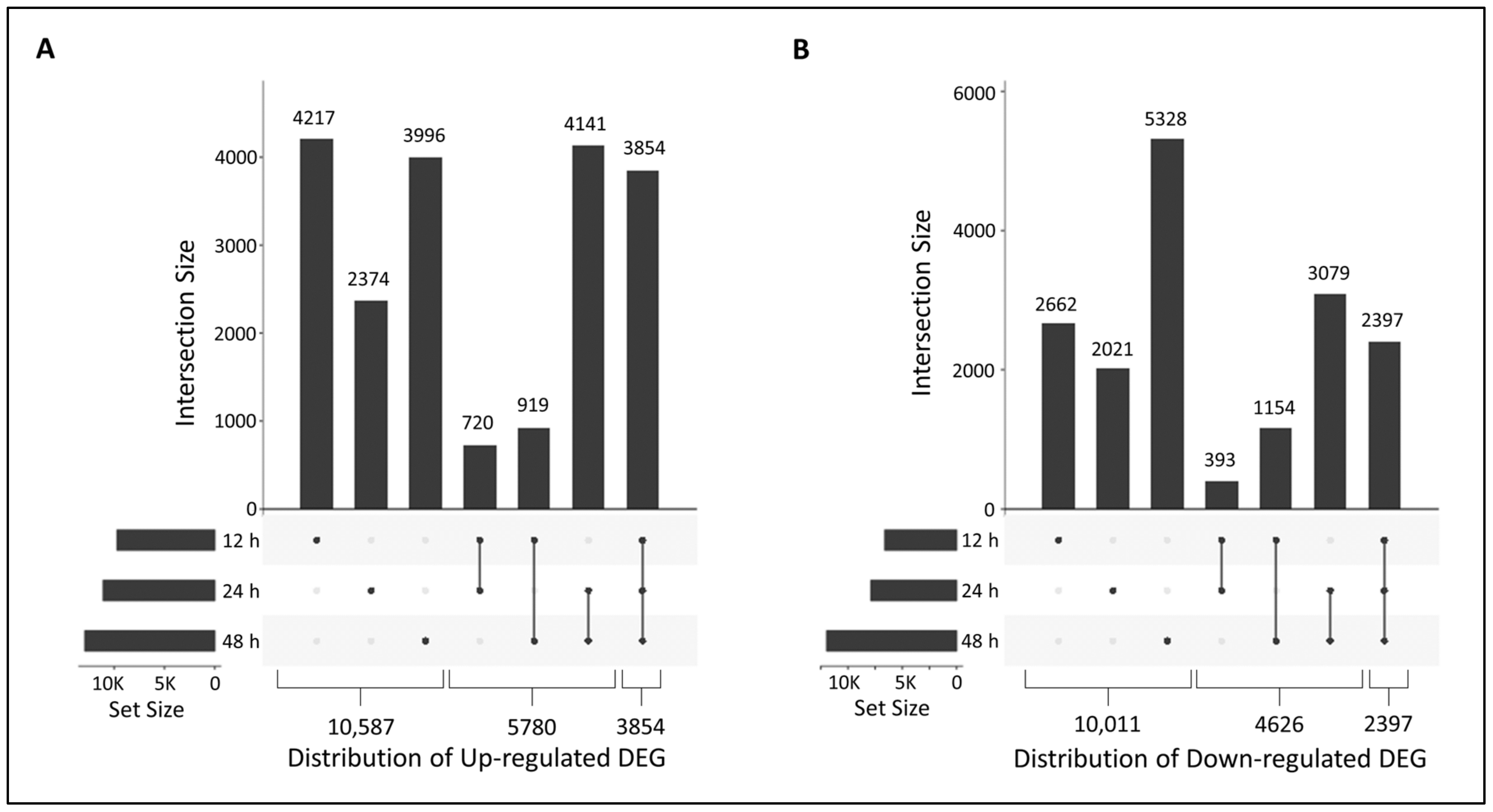
2.6. Functional Analysis of DEGs
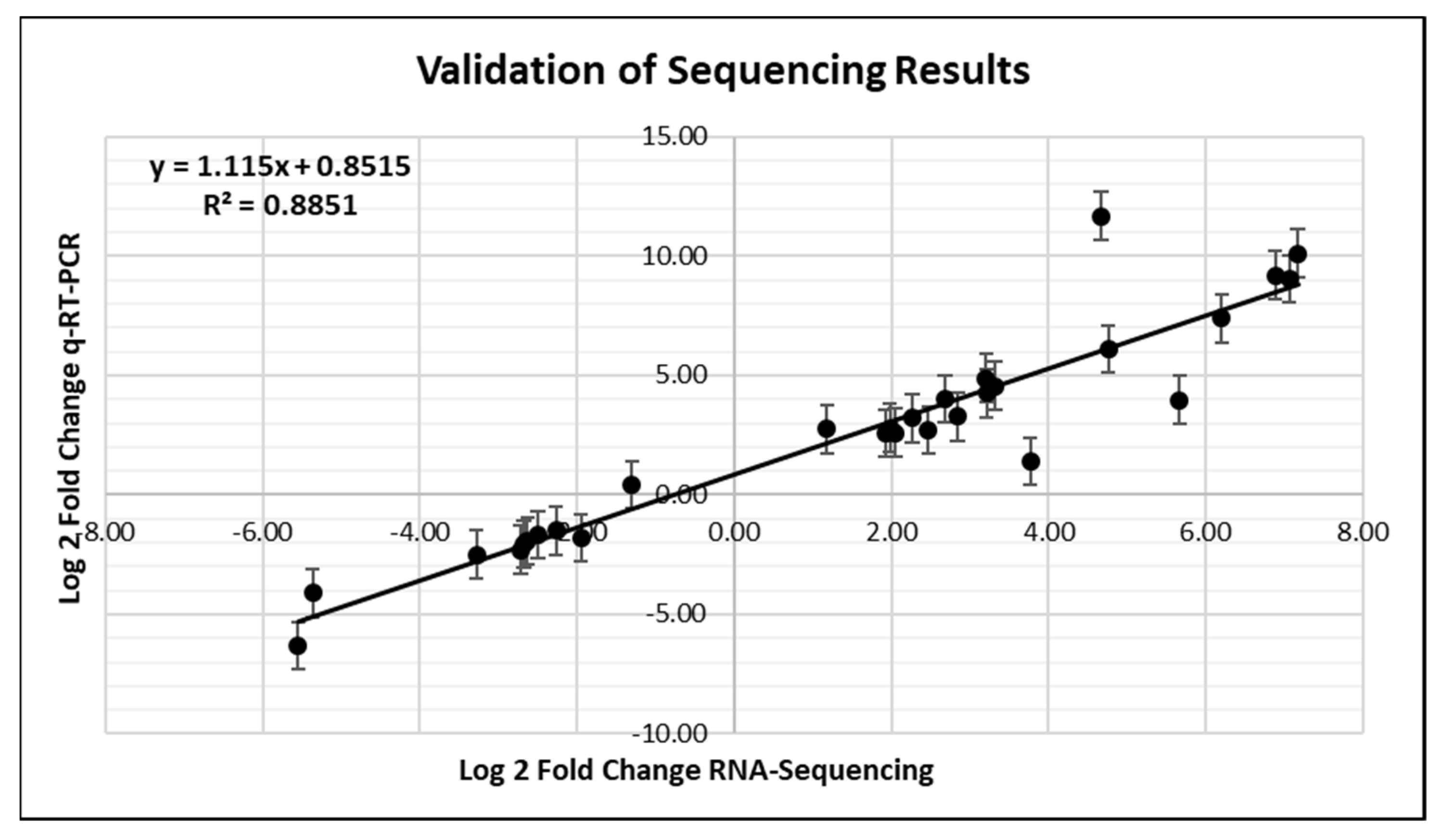
2.7. Validation of RNA-Seq with qRT-PCR
2.8. Comparison to Heat/Drought Studies in Other Species
3. Materials and Methods
3.1. Plant Materials
3.2. Growth of Lt for Experiments
3.3. Plant Treatments
3.4. Water Content Determination
3.5. RNA Sample Preparation and Illumina Sequencing
3.6. Transcriptome Alignment and Analysis
3.7. Validation of RNA-Seq with qRT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- IPCC. Climate Change 2014: Synthesis Report: Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Core Writing Team, Pachauri, R.K., Meyer, L.A., Eds.; IPCC: Geneva, Switzerland, 2014. [Google Scholar]
- NOAA. National Centers for Environmental Information (NCEI). U.S. Billion-Dollar Weather and Climate Disasters. 2021. Available online: https://www.ncdc.noaa.gov/billions/ (accessed on 8 August 2021).
- Hassan, M.U.; Chattha, M.U.; Khan, I.; Chattha, M.B.; Barbanti, L.; Aamer, M.; Iqbal, M.M.; Nawaz, M.; Mahmood, A.; Ali, A.; et al. Heat stress in cultivated plants: Nature, impact, mechanisms, and mitigation strategies—A review. Plant Biosyst. 2021, 155, 211–234. [Google Scholar] [CrossRef]
- Wang, K.; Liu, Y.; Tian, J.; Huang, K.; Shi, T.; Dai, X.; Zhang, W. Transcriptional profiling and identification of heat-responsive genes in perennial ryegrass by RNA-sequencing. Front. Plant Sci. 2017, 8, 1032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A small peptide modulates stomatal control via abscisic acid in long-distance signaling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Xiong, L.; Schumaker, K.S.; Zhu, J.K. Cell signaling during cold, drought, and salt stress. Plant Cell 2002, 14, S165–S183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Y.; Xiong, L. General mechanisms of drought response and their application in drought resistance improvement in plants. Cell. Mol. Life Sci. 2015, 72, 673–689. [Google Scholar] [CrossRef]
- Takahashi, F.; Kuromori, T.; Urano, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Drought stress responses and resistance in plants: From cellular responses to long-distance intercellular communication. Front. Plant Sci. 2020, 11, 1407. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, Y. Identification of differentially expressed genes under drought stress in perennial ryegrass. Physiol. Plant. 2010, 139, 375–387. [Google Scholar] [CrossRef]
- Rizhsky, L.; Liang, H.; Mittler, R. The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol. 2002, 130, 1143–1151. [Google Scholar] [CrossRef] [Green Version]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol. 2004, 134, 1683–1696. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Mittler, R.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Plant adaptations to the combination of drought and high temperatures. Physiol. Plant. 2018, 162, 2–12. [Google Scholar] [CrossRef] [Green Version]
- Evans, L.T. Lolium temulentum L. A long-day plant requiring only one inductive photocycle. Nature 1958, 182, 197–198. [Google Scholar] [CrossRef]
- Baldwin, J.C.; Dombrowski, J.E. Evaluation of Lolium temulentum as a model grass species for the study of salinity stress by PCR-based subtractive suppression hybridization analysis. Plant Sci. 2006, 171, 459–469. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, J.E.; Hind, S.R.; Martin, R.C.; Stratmann, J.W. Wounding systemically activates a mitogen-activated protein kinase in forage and turf grasses. Plant Sci. 2011, 180, 686–693. [Google Scholar] [CrossRef]
- Dombrowski, J.E.; Kronmiller, B.A.; Hollenbeck, V.G.; Rhodes, A.C.; Henning, J.A.; Martin, R.C. Transcriptome analysis of the model grass Lolium temulentum exposed to green leaf volatiles. BMC Plant Biol. 2019, 19, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dombrowski, J.E.; Kronmiller, B.A.; Hollenbeck, V.; Martin, R.C. Transcriptome analysis of wounding in the model grass Lolium temulentum. Plants 2020, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Mody, T.; Bonnot, T.; Nagel, D.H. Interaction between the circadian clock and regulators of heat stress responses in plants. Genes 2020, 11, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciftci-Yilmaz, S.; Mittler, R. The zinc finger network of plants. Cell. Mol. Life Sci. 2008, 65, 1150–1160. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Jia, D.; Chen, X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Jiang, H.; Xu, Y.; Li, H.; Wu, X.; Xie, Q.; Li, C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007, 48, 1148–1158. [Google Scholar] [CrossRef]
- Guo, Y.H.; Yu, Y.P.; Wang, D.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zheng, C.C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef]
- Peng, X.; Zhao, Y.; Cao, J.; Zhang, W.; Jiang, H.; Li, X.; Ma, Q.; Zhu, S.; Cheng, B. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Z.; Lin, W.; Yu, G.; Cheng, Q.; Xu, B.; Huang, B. Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1-CBF-COR regulon and ABA-responsive genes. Biotechnol. Biofuels 2019, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.L.; Xu, Z.S.; Zhao, G.Y.; Cui, X.Y.; Chen, M.; Li, L.C.; Ma, Y.Z. Genome-wide analysis of the C3H zinc finger transcription factor family and drought responses of members in Aegilops tauschii. Plant Mol. Biol. Rep. 2014, 32, 1241–1256. [Google Scholar] [CrossRef]
- Vierstra, R.D. The ubiquitin-26S proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 2009, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, M. Research progress on plant RING-finger proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef] [Green Version]
- Stone, S.L. Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 2019, 343, 65–110. [Google Scholar] [CrossRef]
- Zuluaga, A.P.; Bidzinski, P.; Chanclud, E.; Ducasse, A.; Cayrol, B.; Gomez Selvaraj, M.; Ishitani, M.; Jauneau, A.; Deslandes, L.; Kroj, T.; et al. The rice DNA-binding protein ZBED controls stress regulators and maintains disease resistance after a mild drought. Front. Plant Sci. 2020, 11, 1265. [Google Scholar] [CrossRef]
- Jin, Y.; Li, R.; Zhang, Z.; Ren, J.; Song, X.; Zhang, G. ZBED1/DREF: A transcription factor that regulates cell proliferation. Oncol. Lett. 2020, 20, 1. [Google Scholar] [CrossRef]
- Galván-Gordillo, S.V.; Martínez-Navarro, A.C.; Xoconostle-Cázares, B.; Ruiz-Medrano, R. Bioinformatic analysis of Arabidopsis reverse transcriptases with a zinc-finger domain. Biologia 2016, 71, 1223–1229. [Google Scholar] [CrossRef]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Yoon, Y.; Seo, D.H.; Shin, H.; Kim, H.J.; Kim, C.M.; Jang, G. The role of stress-responsive transcription factors in modulating abiotic stress tolerance in plants. Agronomy 2020, 10, 788. [Google Scholar] [CrossRef]
- Liao, C.; Zheng, Y.; Guo, Y. MYB30 transcription factor regulates oxidative and heat stress responses through ANNEXIN-mediated cytosolic calcium signaling in Arabidopsis. New Phytol. 2017, 216, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Zimmermann, I.M.; Heim, M.A.; Weisshaar, B.; Uhrig, J.F. Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004, 40, 22–34. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Bolitho, K.; Grafton, K.; Kortstee, A.; Karunairetnam, S.; McGhie, T.K.; Espley, R.V.; Hellens, R.P.; Allan, A.C. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC Plant Biol. 2010, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mao, B.; Ou, S.; Wang, W.; Liu, L.; Wu, Y.; Chu, C.; Wang, X. OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 2014, 84, 19–36. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, C.; Zhang, Y.; Wang, B.; Ran, Q.; Zhang, J. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 2019, 70, 5471–5486. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Lee, J.S.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Jakoby, M.; Weisshaar, B.; Dröge-Laser, W.; Vicente-Carbajosa, J.; Tiedemann, J.; Kroj, T.; Parcy, F. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002, 7, 106–111. [Google Scholar] [CrossRef]
- Xiang, Y.; Tang, N.; Du, H.; Ye, H.; Xiong, L. Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol. 2008, 148, 1938–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.; Gao, C.; Zheng, X.; Han, B. Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 2009, 229, 605–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Chen, W.; Zhou, J.; He, H.; Chen, L.; Chen, H.; Deng, X.W. Basic leucine zipper transcription factor OsbZIP16 positively regulates drought resistance in rice. Plant Sci. 2012, 193, 8–17. [Google Scholar] [CrossRef]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Tai, H.; Li, S.; Gao, W.; Zhao, M.; Xie, C.; Li, W.X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Yoon, S.; Lee, D.K.; Yu, I.J.; Kim, Y.S.; Do Choi, Y.; Kim, J.K. Overexpression of the OsbZIP66 transcription factor enhances drought tolerance of rice plants. Plant Biotechnol. Rep. 2017, 11, 53–62. [Google Scholar] [CrossRef]
- Joo, J.; Lee, Y.H.; Song, S.I. OsbZIP42 is a positive regulator of ABA signaling and confers drought tolerance to rice. Planta 2019, 249, 1521–1533. [Google Scholar] [CrossRef]
- Yang, S.; Xu, K.; Chen, S.; Li, T.; Xia, H.; Chen, L.; Liu, H.; Luo, L. A stress-responsive bZIP transcription factor OsbZIP62 improves drought and oxidative tolerance in rice. BMC Plant Biol. 2019, 19, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Howell, S.H. Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol. 2013, 64, 477–499. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, P.; Baranwal, V.K.; Khurana, P. Genome-wide analysis of bZIP transcription factors in wheat and functional characterization of a TabZIP under abiotic stress. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Dröge-Laser, W.; Weiste, C. The C/S1 bZIP network: A regulatory hub orchestrating plant energy homeostasis. Trends Plant Sci. 2018, 23, 422–433. [Google Scholar] [CrossRef]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 2013, 4, 248. [Google Scholar] [CrossRef] [Green Version]
- Bian, Z.; Gao, H.; Wang, C. NAC transcription factors as positive or negative regulators during ongoing battle between pathogens and our food crops. Int. J. Mol. Sci. 2021, 22, 81. [Google Scholar] [CrossRef]
- Ryu, T.H.; Go, Y.S.; Choi, S.H.; Kim, J.I.; Chung, B.Y.; Kim, J.H. SOG 1-dependent NAC 103 modulates the DNA damage response as a transcriptional regulator in Arabidopsis. Plant J. 2019, 98, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Kjaersgaard, T.; Petersen, K.; Skriver, K. NAC genes: Time-specific regulators of hormonal signaling in Arabidopsis. Plant Signal. Behav. 2010, 5, 907–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 2018, 132, 1–25. [Google Scholar] [CrossRef]
- Fang, Y.; Liao, K.; Du, H.; Xu, Y.; Song, H.; Li, X.; Xiong, L. A stress-responsive NAC transcription factor SNAC3 confers heat and drought tolerance through modulation of reactive oxygen species in rice. J. Exp. Bot. 2015, 66, 6803–6817. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; You, J.; Fang, Y.; Zhu, X.; Qi, Z.; Xiong, L. Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol. Biol. 2008, 67, 169–181. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Lee, S.B.; Lee, S.J.; Kim, S.Y. AtERF15 is a positive regulator of ABA response. Plant Cell Rep. 2015, 34, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guo, X.; Zhang, M.; Wang, X.; Zhao, Y.; Yin, Z.; Zhang, Z.; Wang, Y.; Xiong, H.; Zhang, H.; et al. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 2018, 270, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Rong, W.; Qi, L.; Wang, A.; Ye, X.; Du, L.; Liang, H.; Xin, Z.; Zhang, Z. The ERF transcription factor TaERF 3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 2014, 12, 468–479. [Google Scholar] [CrossRef]
- Liu, Q.; Kasuga, M.; Sakuma, Y.; Abe, H.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought-and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 1998, 10, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [Green Version]
- Cui, M.; Zhang, W.; Zhang, Q.; Xu, Z.; Zhu, Z.; Duan, F.; Wu, R. Induced over-expression of the transcription factor OsDREB2A improves drought tolerance in rice. Plant Physiol. Biochem. 2011, 49, 1384–1391. [Google Scholar] [CrossRef]
- Kasuga, M.; Liu, Q.; Miura, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 1999, 17, 287–291. [Google Scholar] [CrossRef]
- Zhao, J.; Ren, W.; Zhi, D.; Wang, L.; Xia, G. Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant Cell Rep. 2007, 26, 1521–1528. [Google Scholar] [CrossRef]
- Bolle, C. The role of GRAS proteins in plant signal transduction and development. Planta 2004, 218, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhou, X.; Chen, L.; Huang, W.; Yu, D. Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 2010, 29, 475–483. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Fu, Q.; Chen, L.; Huang, W.; Yu, D. Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 2011, 233, 1237–1252. [Google Scholar] [CrossRef]
- Wu, X.; Shiroto, Y.; Kishitani, S.; Ito, Y.; Toriyama, K. Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 2009, 28, 21–30. [Google Scholar] [CrossRef] [PubMed]
- He, G.H.; Xu, J.Y.; Wang, Y.X.; Liu, J.M.; Li, P.S.; Chen, M.; Ma, Y.Z.; Xu, Z.S. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Weigel, D.; Meyerowitz, E.M. The ABCs of floral homeotic genes. Cell 1994, 78, 203–209. [Google Scholar] [CrossRef]
- Castelán-Muñoz, N.; Herrera, J.; Cajero-Sánchez, W.; Arrizubieta, M.; Trejo, C.; García-Ponce, B.; Sánchez, M.D.L.P.; Álvarez-Buylla, E.R.; Garay-Arroyo, A. MADS-box genes are key components of genetic regulatory networks involved in abiotic stress and plastic developmental responses in plants. Front. Plant Sci. 2019, 10, 853. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, R.; Bate, N.J.; Sivasankar, S.; Rothstein, S.J. Footprinting of the spinach nitrite reductase gene promoter reveals the preservation of nitrate regulatory elements between fungi and higher plants. Plant Mol. Biol. 1997, 34, 465–476. [Google Scholar] [CrossRef]
- Daniel-Vedele, F.; Caboche, M. A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi. Mol. Gen. Genet. 1993, 240, 365–373. [Google Scholar] [CrossRef]
- Terzaghi, W.B.; Cashmore, A.R. Light-regulated transcription. Annu. Rev. Plant Biol. 1995, 46, 445–474. [Google Scholar] [CrossRef]
- Gupta, P.; Nutan, K.K.; Singla-Pareek, S.L.; Pareek, A. Abiotic stresses cause differential regulation of alternative splice forms of GATA transcription factor in rice. Front. Plant Sci. 2017, 8, 1944. [Google Scholar] [CrossRef] [Green Version]
- Nover, L.; Bharti, K.; Döring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones 2001, 6, 177. [Google Scholar] [CrossRef]
- Nishizawa, A.; Yabuta, Y.; Yoshida, E.; Maruta, T.; Yoshimura, K.; Shigeoka, S. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006, 48, 535–547. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.A.D.; Mittler, R.O.N. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot. 2006, 98, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, N.; Dong, L.; Deng, X.; Liu, D.; Liu, Y.; Li, M.; Hu, Y.; Yan, Y. Genome-wide identification, molecular evolution, and expression analysis of auxin response factor (ARF) gene family in Brachypodium distachyon L. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Korver, R.A.; Koevoets, I.T.; Testerink, C. Out of shape during stress: A key role for auxin. Trends Plant Sci. 2018, 23, 783–793. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Bai, Y.; Wang, S.; Zhang, S.; Wu, Y.; Chen, M.; Jiang, D.; Qi, Y. Expression profile of PIN, AUX/LAX and PGP auxin transporter gene families in Sorghum bicolor under phytohormone and abiotic stress. FEBS J. 2010, 277, 2954–2969. [Google Scholar] [CrossRef]
- Müller, A.; Guan, C.; Gälweiler, L.; Tänzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998, 17, 6903–6911. [Google Scholar] [CrossRef]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawchuk, M.G.; Edgar, A.; Scarpella, E. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, Y.; Sakakibara, H.; Martinoia, E. Cytokinin transporters: GO and STOP in signaling. Trends Plant Sci. 2017, 22, 455–461. [Google Scholar] [CrossRef] [Green Version]
- Jiang, S.Y.; Ramamoorthy, R.; Ramachandran, S. Comparative transcriptional profiling and evolutionary analysis of the GRAM domain family in eukaryotes. Dev. Biol. 2008, 314, 418–432. [Google Scholar] [CrossRef]
- Vishal, B.; Kumar, P.P. Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front. Plant Sci. 2018, 9, 838. [Google Scholar] [CrossRef]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Hauvermale, A.L.; Ariizumi, T.; Steber, C.M. The roles of the GA receptors GID1a, GID1b, and GID1c in sly1-independent GA signaling. Plant Signal. Behav. 2014, 9, 2125–2139. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Zheng, L.; Wang, X.; Hu, Z.; Zheng, Y.; Chen, Q.; Hao, X.; Xiao, X.; Wang, X.; Wang, G.; et al. Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Sci. 2019, 285, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhong, T.; Zhang, L.; Sun, S.; Zeng, H.; Han, L. Effect of localized reduction of gibberellins in different tobacco organs on drought stress tolerance and recovery. Plant Biotechnol. Rep. 2014, 8, 399–408. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, Y.; Yin, Y.; Liu, Q.; Li, N.; Li, X.; He, W.; Hao, D.; Liu, X.; Guo, C. Expression of AtGA2ox1 enhances drought tolerance in maize. Plant Growth Regul. 2019, 89, 203–215. [Google Scholar] [CrossRef]
- Achard, P.; Gong, F.; Cheminant, S.; Alioua, M.; Hedden, P.; Genschik, P. The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 2008, 20, 2117–2129. [Google Scholar] [CrossRef] [Green Version]
- Hattori, Y.; Nagai, K.; Furukawa, S.; Song, X.J.; Kawano, R.; Sakakibara, H.; Wu, J.; Matsumoto, T.; Yoshimura, A.; Kitano, H.; et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 2009, 460, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.; Imamura, A.; Watanabe, A.; Nakabayashi, K.; Okamoto, M.; Jikumaru, Y.; Hanada, A.; Aso, Y.; Ishiyama, K.; Tamura, N.; et al. High temperature-induced abscisic acid biosynthesis and its role in the inhibition of gibberellin action in Arabidopsis seeds. Plant Physiol. 2008, 146, 1368–1385. [Google Scholar] [CrossRef] [Green Version]
- Houben, M.; van de Poel, B. 1-Aminocyclopropane-1-carboxylic acid oxidase (ACO): The enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019, 10, 695. [Google Scholar] [CrossRef] [Green Version]
- Bleecker, A.B.; Kende, H. Ethylene: A gaseous signal molecule in plants. Annu. Rev. Cell Dev. Biol. 2000, 16, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Dubois, M.; van den Broeck, L.; Inzé, D. The pivotal role of ethylene in plant growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Ethylene response factors: A key regulatory hub in hormone and stress signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Pattyn, J.; Vaughan-Hirsch, J.; van de Poel, B. The regulation of ethylene biosynthesis: A complex multilevel control circuitry. New Phytol. 2021, 229, 770–782. [Google Scholar] [CrossRef]
- Srivastava, R.; Kumar, R. The expanding roles of APETALA2/ethylene responsive factors and their potential applications in crop improvement. Brief. Funct. Genom. 2019, 18, 240–254. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Wang, Y.; Chory, J. Two interacting ethylene response factors regulate heat stress response. Plant Cell 2021, 33, 338–357. [Google Scholar] [CrossRef]
- Planas-Riverola, A.; Gupta, A.; Betegón-Putze, I.; Bosch, N.; Ibañes, M.; Caño-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [Green Version]
- Zurek, D.M.; Clouse, S.D. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol. 1994, 104, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Caño-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-García, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lozano-Elena, F.; Caño-Delgado, A.I. Emerging roles of vascular brassinosteroid receptors of the BRI1-like family. Curr. Opin. Plant Biol. 2019, 51, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Tunc-Ozdemir, M.; Jones, A.M. BRL3 and AtRGS1 cooperate to fine tune growth inhibition and ROS activation. PLoS ONE 2017, 12, e0177400. [Google Scholar] [CrossRef] [Green Version]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Howe, G.A.; Major, I.T.; Koo, A.J. Modularity in jasmonate signaling for multistress resilience. Annu. Rev. Plant Biol. 2018, 69, 387–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wasternack, C.; Song, S. Jasmonates: Biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 2017, 68, 1303–1321. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, M.I.R.; Anjum, N.A.; Masood, A.; Hussain, S.J.; Khan, N.A. Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ. Exp. Bot. 2018, 145, 104–120. [Google Scholar] [CrossRef]
- Noir, S.; Bömer, M.; Takahashi, N.; Ishida, T.; Tsui, T.L.; Balbi, V.; Shanahan, H.; Sugimoto, K.; Devoto, A. Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol. 2013, 161, 1930–1951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, A.; Charagh, S.; Zahid, Z.; Mubarik, M.S.; Javed, R.; Siddiqui, M.H.; Hasanuzzaman, M. Jasmonic acid: A key frontier in conferring abiotic stress tolerance in plants. Plant Cell Rep. 2021, 40, 1513–1541. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Örvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. 2002, 31, 629–638. [Google Scholar] [CrossRef] [Green Version]
- Rea, P.A. Plant ATP-binding cassette transporters. Annu. Rev. Plant Biol. 2007, 58, 347–375. [Google Scholar] [CrossRef]
- Niño-González, M.; Novo-Uzal, E.; Richardson, D.N.; Barros, P.M.; Duque, P. More transporters, more substrates: The Arabidopsis major facilitator superfamily revisited. Mol. Plant 2019, 12, 1182–1202. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, E.S.; Schlegel, A.M.; Haswell, E.S. United in diversity: Mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 2015, 66, 113–137. [Google Scholar] [CrossRef] [Green Version]
- Stone, J.M.; Walker, J.C. Plant protein kinase families and signal transduction. Plant Physiol. 1995, 108, 451–457. [Google Scholar] [CrossRef] [Green Version]
- Dissmeyer, N.; Schnittger, A. The age of protein kinases. In Plant Kinases. Methods in Molecular Biology (Methods and Protocols); Dissmeyer, N., Schnittger, A., Eds.; Humana Press: Totowa, NJ, USA, 2011; Volume 779, pp. 7–52. [Google Scholar] [CrossRef]
- Rodriguez, P.L. Protein phosphatase 2C (PP2C) function in higher plants. Plant Mol. Biol. 1998, 38, 919–927. [Google Scholar] [CrossRef]
- Schweighofer, A.; Kazanaviciute, V.; Scheikl, E.; Teige, M.; Doczi, R.; Hirt, H.; Schwanninger, M.; Kant, M.; Schuurink, R.; Mauch, F.; et al. The PP2C-type phosphatase AP2C1, which negatively regulates MPK4 and MPK6, modulates innate immunity, jasmonic acid, and ethylene levels in Arabidopsis. Plant Cell 2007, 19, 2213–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memisoglu, G.; Eapen, V.V.; Yang, Y.; Klionsky, D.J.; Haber, J.E. PP2C phosphatases promote autophagy by dephosphorylation of the Atg1 complex. Proc. Natl. Acad. Sci. USA 2019, 116, 1613–1620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, Z.; Adamska, I.; Nakabayashi, K.; Ostersetzer, O.; Haussuhl, K.; Manuell, A.; Zheng, B.; Vallon, O.; Rodermel, S.R.; Shinozaki, K.; et al. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001, 125, 1912–1918. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Gupta, V.; Prasad, M. Plant molecular chaperones: Structural organization and their roles in abiotic stress tolerance. In Molecular Plant Abiotic Stress: Biology and Biotechnology; Roychoudhury, A., Tripathi, D., Eds.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 221–239. [Google Scholar] [CrossRef]
- Tiwari, P.; Chakrabarty, D. Dehydrin in the past four decades: From chaperones to transcription co-regulators in regulating abiotic stress response. Curr. Res. Biotechnol. 2021, 3, 249–259. [Google Scholar] [CrossRef]
- Verbančič, J.; Lunn, J.E.; Stitt, M.; Persson, S. Carbon supply and the regulation of cell wall synthesis. Mol. Plant 2018, 11, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Reem, N.T.; Chen, H.Y.; Hur, M.; Zhao, X.; Wurtele, E.S.; Li, X.; Li, L.; Zabotina, O. Comprehensive transcriptome analyses correlated with untargeted metabolome reveal differentially expressed pathways in response to cell wall alterations. Plant Mol. Biol. 2018, 96, 509–529. [Google Scholar] [CrossRef]
- Emerson, R.; Hoover, A.; Ray, A.; Lacey, J.; Cortez, M.; Payne, C.; Karlen, D.; Birrell, S.; Laird, D.; Kallenbach, R.; et al. Drought effects on composition and yield for corn stover, mixed grasses, and Miscanthus as bioenergy feedstocks. Biofuels 2014, 5, 275–291. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Tian, N.; Huang, X.; Chen, L.Y.; Schläppi, M.; Xu, Z.Q. The tall fescue turf grass class I chitinase gene FaChit1 is activated by fungal elicitors, dehydration, ethylene, and mechanical wounding. Plant Mol. Biol. Rep. 2009, 27, 305–314. [Google Scholar] [CrossRef] [Green Version]
- Chen, R.D.; Yu, L.X.; Greer, A.F.; Cheriti, H.; Tabaeizadeh, Z. Isolation of an osmotic stress-and abscisic acid-induced gene encoding an acidic endochitinase from Lycopersicon chilense. Mol. Gen. Genet. MGG 1994, 245, 195–202. [Google Scholar] [CrossRef]
- Margis-Pinheiro, M.; Marivet, J.; Burkard, G. Bean class IV chitinase gene: Structure, developmental expression and induction by heat stress. Plant Sci. 1994, 98, 163–173. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, C.; Bauer, S.; Hématy, K.; Saxe, F.; Ibáñez, A.B.; Vodermaier, V.; Konlechner, C.; Sampathkumar, A.; Rüggeberg, M.; Aichinger, E.; et al. Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 2012, 24, 589–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jensen, P.E.; Leister, D. Chloroplast evolution, structure and functions. F1000Prime Rep. 2014, 6, 40. [Google Scholar] [CrossRef]
- Prasad, P.V.V.; Pisipati, S.R.; Momčilović, I.; Ristic, Z. Independent and combined effects of high temperature and drought stress during grain filling on plant yield and chloroplast EF-Tu expression in spring wheat. J. Agron. Crop Sci. 2011, 197, 430–441. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S.P. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ifuku, K. The PsbP and PsbQ family proteins in the photosynthetic machinery of chloroplasts. Plant Physiol. Biochem. 2014, 81, 108–114. [Google Scholar] [CrossRef] [Green Version]
- Daszkowska-Golec, A.; Collin, A.; Sitko, K.; Janiak, A.; Kalaji, H.M.; Szarejko, I. Genetic and physiological dissection of photosynthesis in barley exposed to drought stress. Int. J. Mol. Sci. 2019, 20, 6341. [Google Scholar] [CrossRef] [Green Version]
- Scafaro, A.P.; Bautsoens, N.; den Boer, B.B.; van Rie, J.; Gallé, A. A conserved sequence from heat-adapted species improves Rubisco activase thermostability in wheat. Plant Physiol. 2019, 181, 43–54. [Google Scholar] [CrossRef] [Green Version]
- Zandalinas, S.I.; Balfagón, D.; Arbona, V.; Gómez-Cadenas, A. Modulation of antioxidant defense system is associated with combined drought and heat stress tolerance in citrus. Front. Plant Sci. 2017, 8, 953. [Google Scholar] [CrossRef] [Green Version]
- Guo, K.; Li, Z.; Tian, H.; Du, X.; Liu, Z.; Huang, H.; Wang, P.; Ye, Z.; Zhang, X.; Tu, L. Cytosolic ascorbate peroxidases plays a critical role in photosynthesis by modulating reactive oxygen species level in stomatal guard cell. Front. Plant Sci. 2020, 11, 446. [Google Scholar] [CrossRef]
- Pandian, B.A.; Sathishraj, R.; Djanaguiraman, M.; Prasad, P.V.; Jugulam, M. Role of cytochrome P450 enzymes in plant stress response. Antioxidants 2020, 9, 454. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Wang, M.X.; Dai, Y.; Wang, Y.; Fan, Y.F.; Mao, P.; Ma, X.R. Identification and expression profile of CYPome in perennial ryegrass and tall fescue in response to temperature stress. Front. Plant Sci. 2017, 8, 1519. [Google Scholar] [CrossRef] [Green Version]
- Old 149 Johnson, S.M.; Lim, F.L.; Finkler, A.; Fromm, H.; Slabas, A.R.; Knight, M.R. Transcriptomic analysis of Sorghum bicolor responding to combined heat and drought stress. BMC Genom. 2014, 15, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, L.; Ma, Y.; Li, S.; Dong, S.; Zu, W. Transcriptome profilling analysis characterized the gene expression patterns responded to combined drought and heat stresses in soybean. Comput. Biol. Chem. 2018, 77, 413–429. [Google Scholar] [CrossRef]
- Sewelam, N.; Brilhaus, D.; Bräutigam, A.; Alseekh, S.; Fernie, A.R.; Maurino, V.G. Molecular plant responses to combined abiotic stresses put a spotlight on unknown and abundant genes. J. Exp. Bot. 2020, 71, 5098–5112. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, R.C.; Kronmiller, B.A.; Dombrowski, J.E. Transcriptome analysis of responses in Brachypodium distachyon overexpressing the BdbZIP26 transcription factor. BMC Plant Biol. 2020, 20, 1–18. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and ballgown. Nat. Protoc. 2016, 11, 1650. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package, v. 1.0.8. 2015. Available online: http://CRANR-Project.org/web/packages/pheatmap/index.html (accessed on 4 December 2019).
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The gene ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 2019, 47, D330–D338. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef] [PubMed]
- Dombrowski, J.E.; Martin, R.C. Evaluation of reference genes for quantitative RT-PCR in Lolium temulentum under abiotic stress. Plant Sci. 2009, 176, 390–396. [Google Scholar] [CrossRef]



| Time | Replicate 1 | Replicate 2 | Replicate 3 | ||||
|---|---|---|---|---|---|---|---|
| (h) | Treatment | Reads | Alignment | Reads | Alignment | Reads | Alignment |
| 12 | Control | 25998862 | 89.37% | 30688704 | 91.05% | 26382324 | 91.25% |
| 24 | Control | 21245916 | 84.90% | 20972450 | 84.05% | 30003370 | 85.26% |
| 48 | Control | 20772810 | 87.64% | 27429856 | 85.73% | 24471554 | 87.80% |
| 12 | Drought/heat | 27944440 | 85.51% | 36902414 | 86.40% | 25405614 | 84.39% |
| 24 | Drought/heat | 29130700 | 84.56% | 28372698 | 86.37% | 25201308 | 84.37% |
| 48 | Drought/heat | 27912088 | 83.78% | 25779500 | 83.51% | 21224266 | 84.53% |
| Differentially Expressed Genes * | Total UP | Total DOWN | 12-h UP | 12-h DOWN | 24-h UP | 24-h DOWN | 48-h UP | 48-h DOWN |
|---|---|---|---|---|---|---|---|---|
| Phosphatases | 161 | 81 | 77 | 30 | 112 | 38 | 111 | 70 |
| PPM type | 58 | 13 | 27 | 3 | 44 | 2 | 46 | 8 |
| Kinases | 778 | 739 | 306 | 201 | 447 | 236 | 515 | 576 |
| Calcium/Calmodulin/Calcineurin | 44 | 32 | 20 | 13 | 31 | 25 | 32 | 26 |
| Transferases | 407 | 428 | 181 | 110 | 215 | 185 | 256 | 340 |
| Transporters (Tr.) | 219 | 220 | 108 | 100 | 127 | 119 | 149 | 173 |
| ABC Tr. | 87 | 84 | 43 | 29 | 42 | 28 | 47 | 55 |
| MFS domain-containing Tr. | 59 | 34 | 25 | 7 | 20 | 13 | 27 | 24 |
| Peptide Tr. | 3 | 12 | 4 | 23 | 9 | 29 | ||
| Proteases | 106 | 45 | 58 | 14 | 68 | 24 | 82 | 37 |
| Clp | 15 | 6 | 10 | 1 | 11 | 3 | 12 | 5 |
| Ftsh | 8 | 0 | 6 | 0 | 8 | 0 | 7 | 0 |
| Protease domain-containing | 46 | 11 | 24 | 3 | 27 | 0 | 35 | 9 |
| Ubiquitin (U)-related | 147 | 70 | 66 | 15 | 80 | 27 | 97 | 54 |
| U-Transferase | 36 | 12 | 24 | 1 | 26 | 6 | 25 | 11 |
| U-Ligase | 43 | 26 | 22 | 6 | 26 | 12 | 29 | 23 |
| U-Conjugating | 5 | 7 | 3 | 4 | 3 | 6 | 3 | 5 |
| U-carboxyl-terminal hydrolase | 23 | 9 | 13 | 4 | 6 | 5 | 15 | 6 |
| Chaperones | ||||||||
| Dehydrins, LEA, DNAJ | 56 | 11 | 37 | 3 | 43 | 7 | 43 | 7 |
| HSP | 109 | 9 | 81 | 5 | 97 | 1 | 99 | 5 |
| Photosynthesis | ||||||||
| Chloroplast | 267 | 285 | 133 | 51 | 141 | 111 | 159 | 218 |
| Photosystem | 36 | 22 | 30 | 3 | 9 | 9 | 9 | 15 |
| Chlorophyll | 53 | 91 | 28 | 12 | 14 | 50 | 21 | 80 |
| Chlorophyll a-b binding protein | 23 | 82 | 18 | 10 | 7 | 45 | 9 | 76 |
| Rubisco | 12 | 41 | 10 | 7 | 7 | 20 | 9 | 34 |
| Cytochrome c biogenesis | 222 | 52 | 110 | 18 | 123 | 23 | 144 | 34 |
| Cytochrome c oxidase | 14 | 0 | 11 | 0 | 10 | 0 | 13 | 0 |
| Cytochrome P450 | 83 | 81 | 43 | 40 | 38 | 45 | 48 | 53 |
| Polyphenol oxidase | 2 | 9 | 1 | 9 | 1 | 9 | 1 | 9 |
| Peroxidase (ascorbate) | 39 (7) | 82 (0) | 20 (4) | 25 | 28 (7) | 40 | 22 (6) | 72 |
| Glutathione S-transferase | 41 | 12 | 18 | 5 | 31 | 6 | 34 | 14 |
| Thioredoxin (reductase) | 42 (7) | 11 (2) | 20 (4) | 0 | 26 (6) | 4 | 30 (7) | 10 (3) |
| Cyclin | 6 | 45 | 5 | 19 | 4 | 27 | 5 | 42 |
| Ribosome | 4 | 63 | 3 | 10 | 4 | 52 | 4 | 46 |
| Lipoxygenase | 17 | 11 | 6 | 2 | 15 | 1 | 7 | 11 |
| Lipase | 31 | 39 | 11 | 10 | 19 | 18 | 23 | 39 |
| Phospholipase | 14 | 21 | 5 | 5 | 7 | 10 | 8 | 15 |
| Phospholipid-transporting ATPase | 25 | 5 | 5 | 4 | 19 | 1 | 20 | 2 |
| Cell wall related | ||||||||
| Cellulose synthase | 9 | 35 | 3 | 7 | 5 | 12 | 4 | 32 |
| Xyloglucan endotransglucosylase | 10 | 27 | 3 | 4 | 3 | 9 | 7 | 24 |
| Expansin | 14 | 35 | 7 | 4 | 9 | 3 | 8 | 33 |
| Glucanase | 13 | 28 | 6 | 9 | 7 | 8 | 9 | 24 |
| Pectin esterase | 9 | 44 | 4 | 15 | 5 | 25 | 5 | 41 |
| Laccase | 10 | 36 | 5 | 3 | 4 | 17 | 4 | 33 |
| Chitinase | 12 | 1 | 7 | 1 | 7 | 0 | 10 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, R.C.; Kronmiller, B.A.; Dombrowski, J.E. Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress. Plants 2021, 10, 2247. https://doi.org/10.3390/plants10112247
Martin RC, Kronmiller BA, Dombrowski JE. Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress. Plants. 2021; 10(11):2247. https://doi.org/10.3390/plants10112247
Chicago/Turabian StyleMartin, Ruth C., Brent A. Kronmiller, and James E. Dombrowski. 2021. "Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress" Plants 10, no. 11: 2247. https://doi.org/10.3390/plants10112247
APA StyleMartin, R. C., Kronmiller, B. A., & Dombrowski, J. E. (2021). Transcriptome Analysis of Lolium temulentum Exposed to a Combination of Drought and Heat Stress. Plants, 10(11), 2247. https://doi.org/10.3390/plants10112247






