Genome-Wide Identification, Genomic Organization, and Characterization of Potassium Transport-Related Genes in Cajanus cajan and Their Role in Abiotic Stress
Abstract
:1. Introduction
2. Results
2.1. Identification of K+ Transporters and Channels
2.1.1. Potassium Transporters
KUP/HAK/KT
Trk/HKT
KEA Family
2.1.2. Potassium Channels
Shaker Family
TPK and Kir-Like Family
2.2. Gene Structure Analysis and Chromosomal Distribution of Potassium Transport-Related Genes
2.3. Comparative Phylogenetic Analysis
2.4. Promoter Analysis
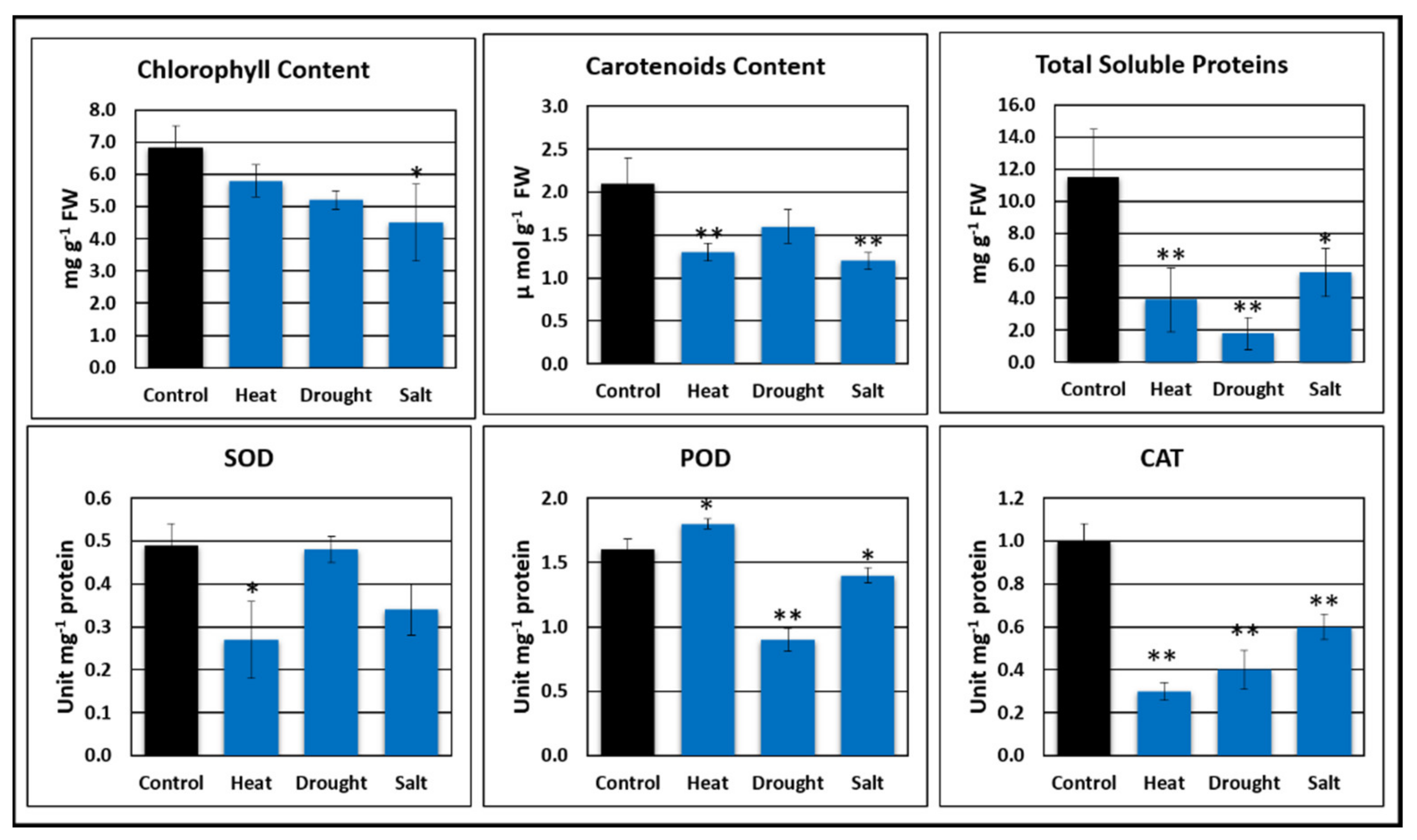
2.5. Physiological Analysis
2.5.1. Chlorophyll Content
2.5.2. Carotenoid Content
2.5.3. Total Soluble Protein Content
2.5.4. Superoxidase Dismutase (SOD), Peroxide (POD), and Catalase Activity (CAT)
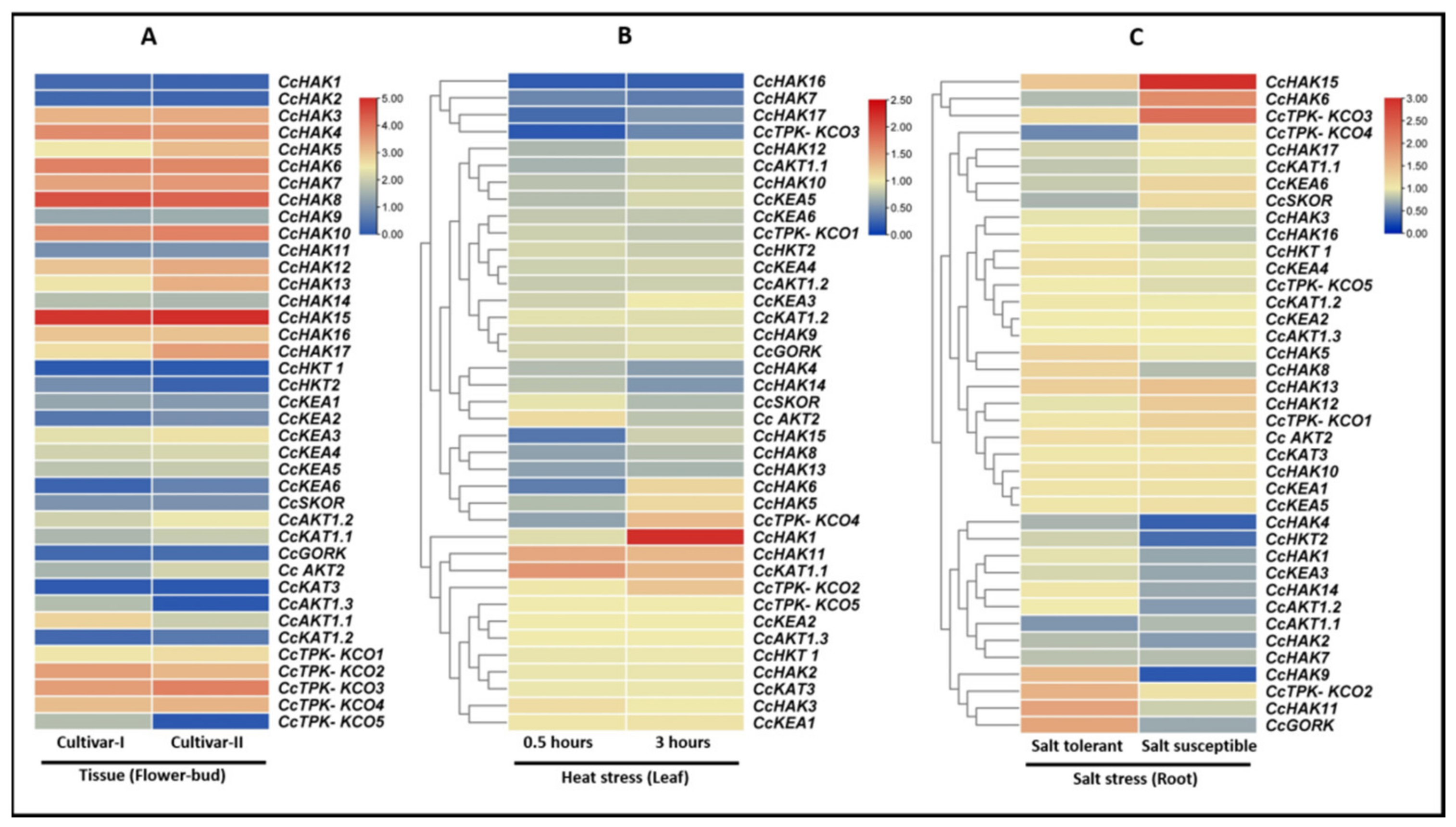
2.6. In Silico Expression Analysis
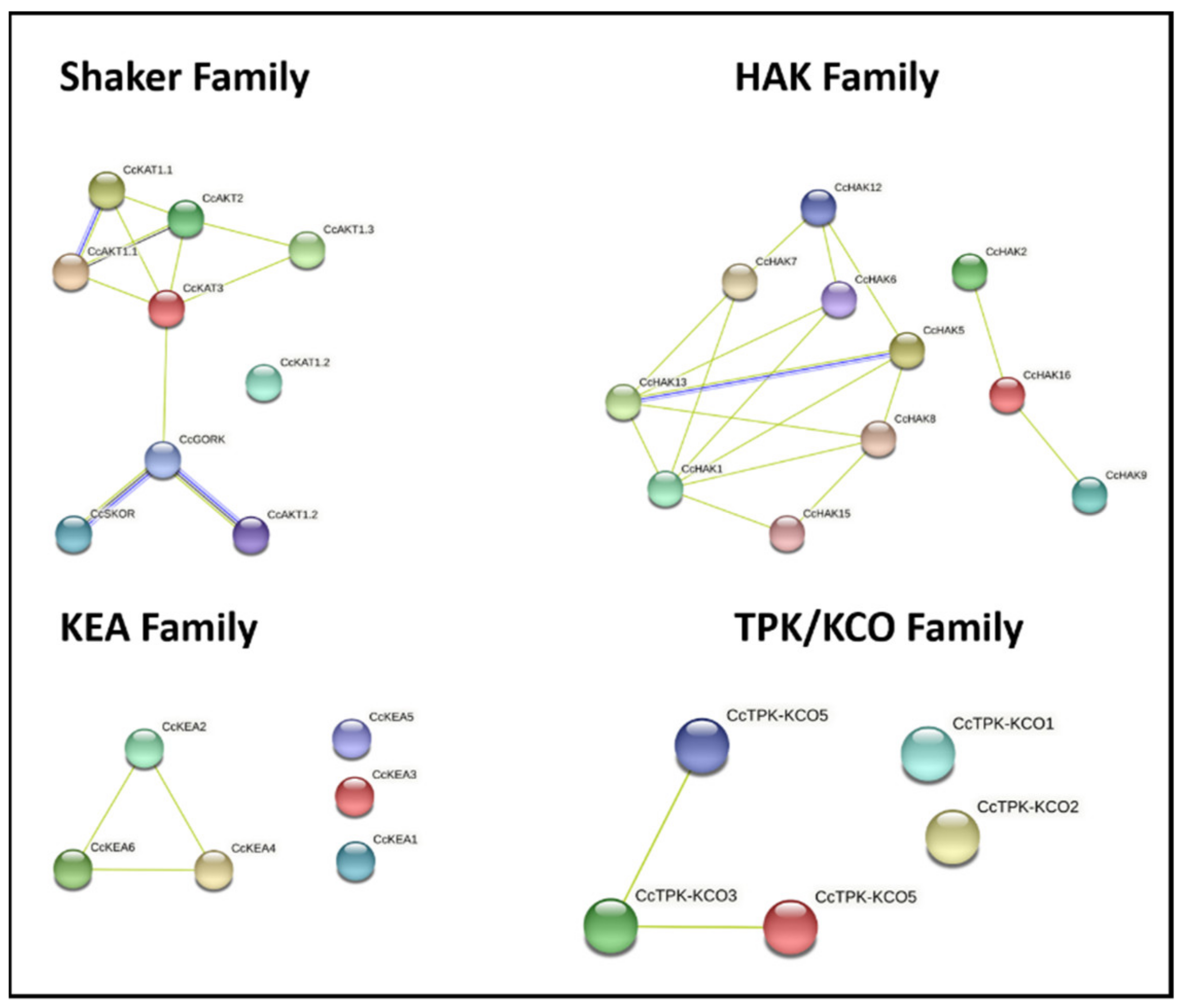
2.7. Protein–Protein Interaction
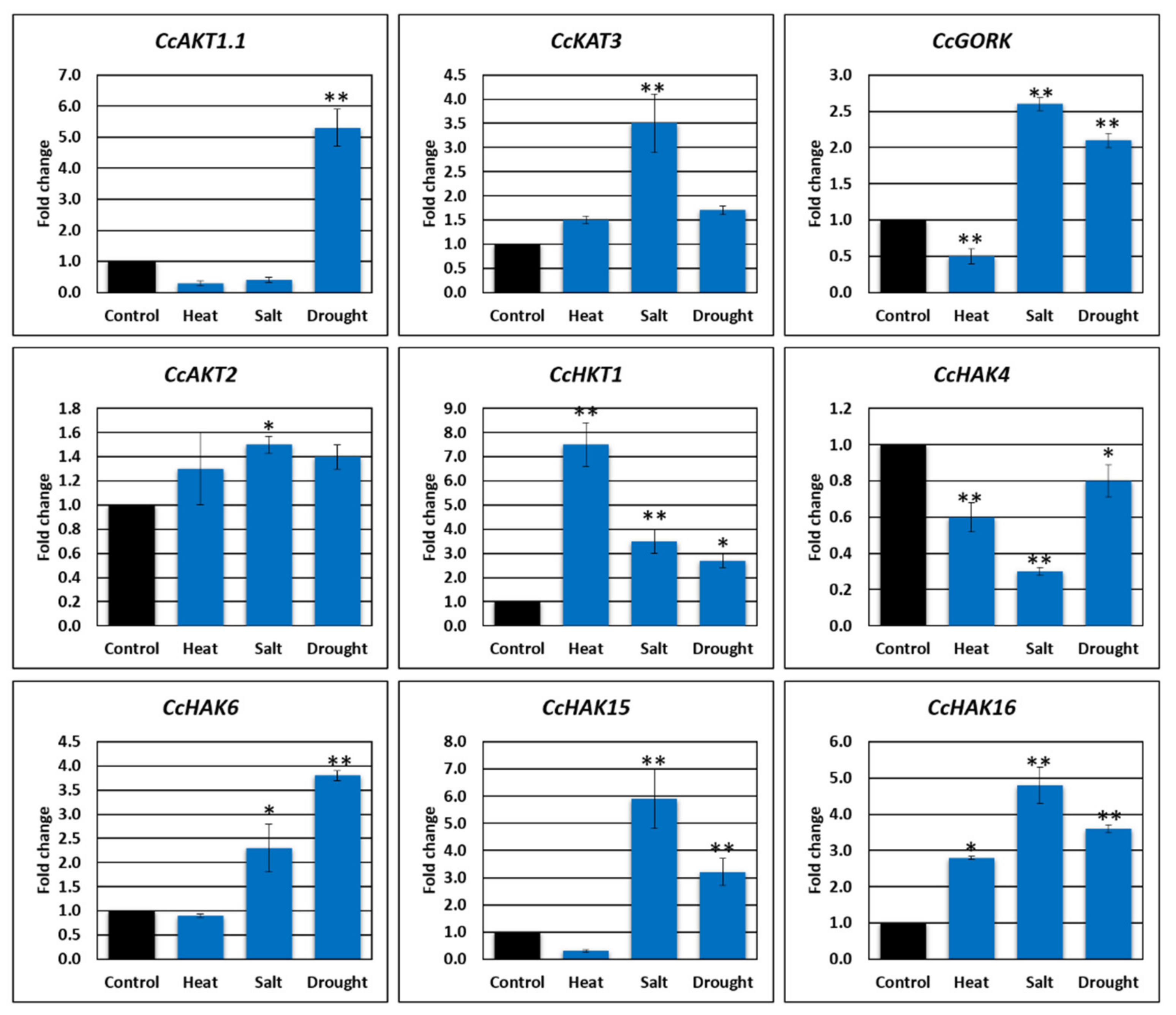
2.8. Gene Expression Analysis
3. Discussion
4. Materials and Methods
4.1. Screening of Potassium Transport System in C. cajan
4.2. Motif Recognition and Gene Structure Prediction
4.3. Multiple Sequence Alignment, Phylogenetic Analysis, and Cis-Regulatory Elements Prediction
4.4. Gene Duplication, Evolutionary Analysis, and Chromosomal Mapping in Cajanus cajan
4.5. Plant Growth and Stress Imposition
4.6. Physiological Analysis
4.6.1. Chlorophyll Content
4.6.2. Carotenoid Contents
4.6.3. Biochemical Studies
4.7. In Silico Expression Profiling and PPI Analysis of Potassium Transport Genes
4.8. RNA Isolation, Reverse Transcriptase PCR, and Quantitative Real-Time PCR (qRT-PCR)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Clarkson, D.T.; Hanson, J.B. The Mineral Nutrition of Higher Plants. Annu. Rev. Plant Physiol. 1980, 31, 239–298. [Google Scholar] [CrossRef]
- Leigh, R.A. Potassium homeostasis and membrane transport. J. Plant Nutr. Soil Sci. 2001, 164, 193–198. [Google Scholar] [CrossRef]
- Walker, D.J.; Leigh, R.A.; Miller, A.J. Potassium homeostasis in vacuolate plant cells. Proc. Natl. Acad. Sci. USA 1996, 93, 10510–10514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemoine, R.; La Camera, S.; Atanassova, R.; Dédaldéchamp, F.; Allario, T.; Pourtau, N.; Bonnemain, J.-L.; Laloi, M.; Coutos-Thévenot, P.; Maurousset, L.; et al. Source-to-sink transport of sugar and regulation by environmental factors. Front. Plant Sci. 2013, 4, 272. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.S.; Berkowitz, G.A.; Pier, P.A. Maintenance of photosynthesis at low leaf water potential in wheat: Role of potassium status and irrigation history. Plant Physiol. 1989, 89, 1358–1365. [Google Scholar] [CrossRef] [Green Version]
- Maathuis, F.J.M. Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef]
- Kumar, M.; Kherawat, B.; Dey, P.; Saha, D.; Singh, A.; Bhatia, S.; Ghodake, G.; Kadam, A.; Kim, H.-U.; Manorama, C.; et al. Genome-Wide Identification and Characterization of PIN-FORMED (PIN) Gene Family Reveals Role in Developmental and Various Stress Conditions in Triticum aestivum L. Int. J. Mol. Sci. 2021, 22, 7396. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, H.; Liu, J.; Du, Q.; Lu, S.; Liu, C. Genome-wide identification and functional characterization of natural antisense transcripts in Salvia miltiorrhiza. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Tong, T.; Fang, Y.-X.; Zhang, Z.; Zheng, J.; Zhang, X.; Li, J.; Niu, C.; Xue, D.; Zhang, X. Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 2021, 93, 89–103. [Google Scholar] [CrossRef]
- Baruah, P.M.; Krishnatreya, D.B.; Bordoloi, K.S.; Gill, S.S.; Agarwala, N. Genome wide identification and characterization of abiotic stress responsive lncRNAs in Capsicum annuum. Plant Physiol. Biochem. 2021, 162, 221–236. [Google Scholar] [CrossRef]
- Kesawat, M.S.; Kherawat, B.S.; Singh, A.; Dey, P.; Kabi, M.; Debnath, D.; Saha, D.; Khandual, A.; Rout, S.; Manorama, C.; et al. Genome-Wide Identification and Characterization of the Brassinazole-resistant (BZR) Gene Family and Its Expression in the Various Developmental Stage and Stress Conditions in Wheat (Triticum aestivum L.). Int. J. Mol. Sci. 2021, 22, 8743. [Google Scholar] [CrossRef]
- Wang, M.; Chen, B.; Zhou, W.; Xie, L.; Wang, L.; Zhang, Y.; Zhang, Q. Genome-wide identification and expression analysis of the AT-hook Motif Nuclear Localized gene family in soybean. BMC Genom. 2021, 22, 1–26. [Google Scholar] [CrossRef]
- Mäser, P.; Thomine, S.; Schroeder, J.I.; Ward, J.M.; Hirschi, K.; Sze, H.; Talke, I.N.; Amtmann, A.; Maathuis, F.J.; Sanders, D.; et al. Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 2001, 126, 1646–1667. [Google Scholar] [CrossRef] [Green Version]
- Véry, A.-A.; Sentenac, H. Molecular mechanisms and regulation of K+ transport in higher plants. Annu. Rev. Plant Biol. 2003, 54, 575–603. [Google Scholar] [CrossRef]
- Lebaudy, A.; Véry, A.-A.; Sentenac, H. K+ channel activity in plants: Genes, regulations and functions. FEBS Lett. 2007, 581, 2357–2366. [Google Scholar] [CrossRef]
- Saxena, K.B.; Kumar, R.V.; Rao, P.V. Pigeonpea Nutrition and Its Improvement. J. Crops Prod. 2002, 5, 227–260. [Google Scholar] [CrossRef] [Green Version]
- Bohra, A.; Saxena, K.B.; Varshney, R.K.; Saxena, R.K. Genomics-assisted breeding for pigeonpea improvement. Theor. Appl. Genet. 2020, 133, 1721–1737. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The critical role of potassium in plant stress response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Porras, J.L.; Riaño-Pachón, D.M.; Benito, B.; Haro, R.; Sklodowski, K.; Rodríguez-Navarro, A.; Dreyer, I. Phylogenetic Analysis of K+ Transporters in Bryophytes, Lycophytes, and Flowering Plants Indicates a Specialization of Vascular Plants. Front. Plant Sci. 2012, 3, 167. [Google Scholar] [CrossRef] [Green Version]
- Munro, A.W.; Ritchie, G.Y.; Lamb, A.J.; Douglas, R.M.; Booth, I.R. The cloning and DNA sequence of the gene for the glutathione-regulated potassium-efflux system KefC of Escherichia coli. Mol. Microbiol. 1991, 5, 607–616. [Google Scholar] [CrossRef]
- Pilot, G.; Gaymard, F.; Mouline, K.; Chérel, I.; Sentenac, H. Regulated expression of Arabidopsis shaker K+ channel genes involved in K+ uptake and distribution in the plant. Plant Mol. Biol. 2003, 51, 773–787. [Google Scholar] [CrossRef]
- Hedrich, R. Ion channels in plants. Physiol. Rev. 2012, 92, 1777–1811. [Google Scholar] [CrossRef]
- Moshelion, M.; Becker, D.; Czempinski, K.; Mueller-Roeber, B.; Attali, B.; Hedrich, R.; Moran, N. Diurnal and circadian regulation of putative potassium channels in a leaf moving organ. Plant Physiol. 2002, 128, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, B.; Yu, L.; Feng, D.; Wang, H.; Wang, J. Phylogenetic analysis, structural evolution and functional divergence of the 12-oxo-phytodienoate acid reductase gene family in plants. BMC Evol. Biol. 2009, 9, 90. [Google Scholar] [CrossRef] [Green Version]
- Li, W.Y.; Wang, X.; Li, R.; Li, W.Q.; Chen, K.M. Genome-wide analysis of the NADK gene family in plants. PLoS ONE 2014, 9, e101051. [Google Scholar] [CrossRef] [Green Version]
- Jones, D.M.; Vandepoele, K. Identification and evolution of gene regulatory networks: Insights from comparative studies in plants. Curr. Opin. Plant Biol. 2020, 54, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Azeem, F.; Hussain, M.; Hussain, S.; Zubair, M.; Nadeem, H.; Ali, M.A.; Afzal, M.; Siddique, M.H. Genome-wide analysis and expression profiling of potassium transport related genes in Solanum tuberosum. Pak. J. Agric. Sci. 2021, 58, 81–94. [Google Scholar]
- Azeem, F.; Ahmad, B.; Atif, R.M.; Ali, M.A.; Nadeem, H.; Hussain, S.; Manzoor, H.; Azeem, M.; Afzal, M. Genome-Wide Analysis of Potassium Transport-Related Genes in Chickpea (Cicer arietinum L.) and Their Role in Abiotic Stress Responses. Plant Mol. Biol. Rep. 2018, 36, 451–468. [Google Scholar] [CrossRef]
- Lim, C.W.; Kim, S.H.; Choi, H.W.; Luan, S.; Lee, S.C. The Shaker type potassium channel, GORK, regulates abscisic acid signaling in arabidopsis. Plant Pathol. J. 2019, 35, 684–691. [Google Scholar] [CrossRef]
- Dreyer, I.; Uozumi, N. Potassium channels in plant cells. FEBS J. 2011, 278, 4293–4303. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, Z.-H.; Liu, X.; Colmer, T.D.; Shabala, L.; Salih, A.; Zhou, M.; Shabala, S. Revealing the roles of GORK channels and NADPH oxidase in acclimation to hypoxia in Arabidopsis. J. Exp. Bot. 2016, 68, 3191–3204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ijaz, U.; Pervaiz, T.; Ahmed, T.; Seemab, R.; Shahid, M.; Noman, M.; Nadeem, M.; Azeem, F. Plant Cis-regulatory elements: Methods of identification and applications. Asian J. Agric. Biol. 2020, 8, 207–222. [Google Scholar] [CrossRef]
- Ward, J.M.; Mäser, P.; Schroeder, J.I. Plant ion channels: Gene families, physiology, and functional genomics analyses. Annu. Rev. Physiol. 2009, 71, 59–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuéllar, T.; Pascaud, F.; Verdeil, J.-L.; Torregrosa, L.; Adam-Blondon, A.-F.; Thibaud, J.-B.; Sentenac, H.; Gaillard, I. A grapevine Shaker inward K(+) channel activated by the calcineurin B-like calcium sensor 1-protein kinase CIPK23 network is expressed in grape berries under drought stress conditions. Plant J. 2010, 61, 58–69. [Google Scholar] [CrossRef]
- Amrutha, R.N.; Sekhar, P.N.; Varshney, R.K.; Kishor, P.B.K. Genome-wide analysis and identification of genes related to potassium transporter families in rice (Oryza sativa L.). Plant Sci. 2007, 172, 708–721. [Google Scholar] [CrossRef] [Green Version]
- Hosy, E.; Vavasseur, A.; Mouline, K.; Dreyer, I.; Gaymard, F.; Poree, F.; Boucherez, J.; Lebaudy, A.; Bouchez, D.; Very, A.A.; et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA 2003, 100, 5549–5554. [Google Scholar] [CrossRef] [Green Version]
- Hamamoto, S.; Marui, J.; Matsuoka, K.; Higashi, K.; Igarashi, K.; Nakagawa, T.; Kuroda, T.; Mori, Y.; Murata, Y.; Nakanishi, Y.; et al. Characterization of a tobacco TPK-type K+ channel as a novel tonoplast K+ channel using yeast tonoplasts. J. Biol. Chem. 2008, 283, 1911–1920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isayenkov, S.; Isner, J.-C.; Maathuis, F.J.M. Rice two-pore K+ channels are expressed in different types of vacuoles. Plant Cell 2011, 23, 756–768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davies, C.; Shin, R.; Liu, W.; Thomas, M.R.; Schachtman, D.P. Transporters expressed during grape berry (Vitis vinifera L.) development are associated with an increase in berry size and berry potassium accumulation. J. Exp. Bot. 2006, 57, 3209–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desbrosses, G.; Kopka, C.; Ott, T.; Udvardi, M.K. Lotus japonicus LjKUP is induced late during nodule development and encodes a potassium transporter of the plasma membrane. Mol. Plant-Microbe Interact. 2004, 17, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-H.; Garvin, D.F.; Kochian, L.V. Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots. Evidence for cross talk and root/rhizosphere-mediated signals. Plant Physiol. 2002, 130, 1361–1370. [Google Scholar] [CrossRef] [Green Version]
- Su, H.; Golldack, D.; Zhao, C.; Bohnert, H.J. The expression of HAK-type K(+) transporters is regulated in response to salinity stress in common ice plant. Plant Physiol. 2002, 129, 1482–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Senn, M.E.; Rubio, F.; Bañuelos, M.A.; Rodríguez-Navarro, A. Comparative Functional Features of Plant Potassium HvHAK1 and HvHAK2 Transporters. J. Biol. Chem. 2001, 276, 44563–44569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garciadeblas, B.; Benito, B.; Rodríguez-Navarro, A. Molecular cloning and functional expression in bacteria of the potassium transporters CnHAK1 and CnHAK2 of the seagrass Cymodocea nodosa. Plant Mol. Biol. 2002, 50, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Bañuelos, M.A.; Garciadeblas, B.; Cubero, B.; Rodríguez-Navarro, A. Inventory and functional characterization of the HAK potassium transporters of rice. Plant Physiol. 2002, 130, 784–795. [Google Scholar] [CrossRef] [Green Version]
- Rubio, F.; Guillermo, S.M.; Rodríguez-Navarro, A. Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 2000, 109, 34–43. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, S.; Hu, Y.; Wu, F.; Hu, Q.; Chen, G.; Cai, J.; Wu, T.; Moran, N.; Yu, L.; et al. The role of a potassium transporter oshak5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014, 166, 945–959. [Google Scholar] [CrossRef] [Green Version]
- Rubio, F.; Alemán, F.; Nieves-Cordones, M.; Martínez, V. Studies on Arabidopsis athak5, atakt1 double mutants disclose the range of concentrations at which AtHAK5, AtAKT1 and unknown systems mediate K+ uptake. Physiol. Plant. 2010, 139, 220–228. [Google Scholar] [CrossRef]
- Nieves-Cordones, M.; Martínez, V.; Benito, B.; Rubio, F. Comparison between Arabidopsis and Rice for Main Pathways of K+ and Na+ Uptake by Roots. Front. Plant Sci. 2016, 7, 992. [Google Scholar] [CrossRef] [Green Version]
- Platten, J.D.; Cotsaftis, O.; Berthomieu, P.; Bohnert, H.; Davenport, R.J.; Fairbairn, D.J.; Horie, T.; Leigh, R.A.; Lin, H.-X.; Luan, S.; et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006, 11, 372–374. [Google Scholar] [CrossRef]
- Awana, M.; Jain, N.; Samota, M.K.; Rani, K.; Kumar, A.; Ray, M.; Gaikwad, K.; Praveen, S.; Singh, N.K.; Singh, A. Protein and gene integration analysis through proteome and transcriptome brings new insight into salt stress tolerance in pigeonpea (Cajanus cajan L.). Int. J. Biol. Macromol. 2020, 164, 3589–3602. [Google Scholar] [CrossRef] [PubMed]
- Tayyab; Azeem, M.; Qasim, M.; Ahmed, N.; Ahmad, R. Salt stress responses of pigeon pea (Cajanus cajan) on growth, yield and some biochemical attributes. Pak. J. Bot. 2016, 48, 1353–1360. [Google Scholar]
- Kosová, K.; Vítámvás, P.; Urban, M.O.; Prášil, I.T.; Renaut, J. Plant abiotic stress proteomics: The major factors determining alterations in cellular proteome. Front. Plant Sci. 2018, 9, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taie, H.A.A.; El-Yazal, M.A.S.; Ahmed, S.M.A.; Rady, M.M. Polyamines modulate growth, antioxidant activity, and genomic DNA in heavy metal–stressed wheat plant. Environ. Sci. Pollut. Res. 2019, 26, 22338–22350. [Google Scholar] [CrossRef]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol. Biol. 2010, 639, 292–298. [Google Scholar] [CrossRef]
- Li, L.; Yi, H. Effect of sulfur dioxide on ROS production, gene expression and antioxidant enzyme activity in Arabidopsis plants. Plant Physiol. Biochem. 2012, 58, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Nieves-Cordones, M.; Caballero, F.; Martínez, V.; Rubio, F. Disruption of the Arabidopsis thaliana inward-rectifier K+ channel AKT1 improves plant responses to water stress. Plant Cell Physiol. 2012, 53, 423–432. [Google Scholar] [CrossRef]
- Reintanz, B.; Szyroki, A.; Ivashikina, N.; Ache, P.; Godde, M.; Becker, D.; Palme, K.; Hedrich, R. AtKC1, a silent Arabidopsis potassium channel α-subunit modulates root hair K+ influx. Proc. Natl. Acad. Sci. USA 2002, 99, 4079–4084. [Google Scholar] [CrossRef] [Green Version]
- Rubio, F.; Gassmann, W.; Schroeder, J.I. Sodium-Driven Potassium Uptake by the Plant Potassium Transporter HKT1 and Mutations Conferring Salt Tolerance. Science 1995, 270, 1660–1663. [Google Scholar] [CrossRef] [PubMed]
- Véry, A.-A.; Nieves-Cordones, M.; Daly, M.; Khan, I.; Fizames, C.; Sentenac, H. Molecular biology of K+ transport across the plant cell membrane: What do we learn from comparison between plant species? J. Plant Physiol. 2014, 171, 748–769. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Beyer, W.F.; Fridovich, I. Assaying for superoxide dismutase activity: Some large consequences of minor changes in conditions. Anal. Biochem. 1987, 161, 559–566. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 764–775. [Google Scholar] [CrossRef]
- Kar, M.; Mishra, D. Catalase, Peroxidase, and Polyphenoloxidase Activities during Rice Leaf Senescence. Plant Physiol. 1976, 57, 315–319. [Google Scholar] [CrossRef] [Green Version]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Sinha, P.; Singh, V.K.; Suryanarayana, V.; Krishnamurthy, L.; Saxena, R.K.; Varshney, R.K. Evaluation and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under drought stress conditions. PLoS ONE 2015, 10, e0122847. [Google Scholar] [CrossRef] [PubMed]



| Sr.# | Gene Name | * Protein ID | TM Domains | Domains | Protein Length | Chromosome Number | Exons | Isoelectric Point | Molecular Weight (kDa) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | CcHAK1 | XP 020213425 | 12 | K_trans | 784 | 3 | 8 | 8.56 | 87.6626 |
| 2 | CcHAK2 | XP 020212862 | 12 | K_trans | 804 | 3 | 10 | 8.83 | 89.3791 |
| 3 | CcHAK3 | XP 020214355 | 12 | PLN00148 | 784 | 4 | 9 | 8.79 | 87.4835 |
| 4 | CcHAK4 | XP 029127280 | 9 | K_trans | 706 | 5 | 10 | 8.69 | 78.7490 |
| 5 | CcHAK5 | XP 020218079 | 13 | K_trans | 778 | 6 | 8 | 8.45 | 86.9579 |
| 6 | CcHAK6 | XP 020220430 | 14 | K_trans | 792 | 8 | 9 | 7.63 | 88.4431 |
| 7 | CcHAK7 | XP 020222401 | 13 | PLN00151 | 841 | 9 | 10 | 6.93 | 93.4439 |
| 8 | CcHAK8 | XP 020222257 | 13 | PLN00151 | 844 | 9 | 10 | 5.97 | 93.9803 |
| 9 | CcHAK9 | XP 020222600 | 12 | K_trans | 713 | 9 | 8 | 8.93 | 79.5610 |
| 10 | CcHAK10 | XP 020223374 | 10 | K_trans | 790 | 10 | 9 | 9.30 | 88.1669 |
| 11 | CcHAK11 | XP 020225584 | 12 | K_trans | 773 | 11 | 8 | 8.79 | 86.3152 |
| 12 | CcHAK12 | XP 020226102 | 12 | PLN00151 | 841 | 11 | 10 | 8.50 | 93.5142 |
| 13 | CcHAK13 | XP 029130105 | 14 | K_trans/ | 790 | Unknown | 8 | 8.13 | 88.5552 |
| 14 | CcHAK14 | XP 020207687 | 13 | PLN00149 | 773 | Unknown | 8 | 7.29 | 86.9488 |
| 15 | CcHAK15 | XP 020232137 | 12 | PLN00149 | 781 | Unknown | 8 | 7.09 | 87.4695 |
| 16 | CcHAK16 | XP 020234883 | 11 | K_trans | 760 | Unknown | 9 | 6.24 | 84.7841 |
| 17 | CcHAK17 | XP 020236481 | 12 | K_trans | 776 | Unknown | 9 | 6.89 | 86.3213 |
| 18 | CcHKT 1 | XP 020220422 | 10 | TrkG | 527 | 8 | 4 | 9.35 | 59.3970 |
| 19 | CcHKT2 | XP 020222038 | 9 | N/A | 507 | 8 | 3 | 9.43 | 57.4761 |
| 20 | CcKEA1 | XP 020227261 | 12 | Na_H_Exchanger | 638 | 2 | 21 | 7.23 | 69.2591 |
| 21 | CcKEA2 | XP 020215520 | 11 | Na_H_Exchanger | 586 | 5 | 21 | 5.90 | 63.0490 |
| 22 | CcKEA3 | XP 020218588 | 10 | SMC_N- PRK03562 | 1193 | 7 | 21 | 4.97 | 128.0696 |
| 23 | CcKEA4 | XP 020220606 | 10 | Na_H_Exchanger | 574 | 8 | 20 | 6.33 | 62.3534 |
| 24 | CcKEA5 | XP 020225548 | 0 | PRK03562 | 815 | 11 | 20 | 5.75 | 88.8492 |
| 25 | CcKEA6 | XP 020233144 | 10 | PRK03562 - | 1200 | Unknown | 21 | 4.98 | 129.4016 |
| 26 | CcSKOR | XP 020202567 | 5 | ANK-Ank_2 | 837 | Unknown | 13 | 6.68 | 95.9104 |
| 27 | CcAKT1.2 | XP 020205453 | 5 | ANK-Ank_2 | 879 | Unknown | 12 | 6.02 | 99.3995 |
| 28 | CcKAT1.1 | XP 020208418 | 5 | Ank_2 | 717 | Unknown | 11 | 7.02 | 83.3877 |
| 29 | CcGORK | XP 020215636 | 5 | ANK/Ank_2 | 808 | 5 | 11 | 8.71 | 92.3245 |
| 30 | Cc AKT2 | XP 020220877 | 5 | ANK/Ank_2 | 825 | 8 | 11 | 6.96 | 94.5856 |
| 31 | CcKAT3 | XP 020225577 | 5 | KHA/Ank_2 | 622 | 11 | 13 | 7.02 | 71.5766 |
| 32 | CcAKT1.3 | XP 020227900 | 5 | N/A | 864 | 2 | 13 | 6.50 | 97.5863 |
| 33 | CcAKT1.1 | XP 020231629 | 5 | ANK/Ank_2 | 878 | Unknown | 12 | 6.77 | 87.5007 |
| 34 | CcKAT1.2 | XP 029125234 | 4 | Ank_2 | 759 | Unknown | 11 | 8.56 | 99.6863 |
| 35 | CcTPK- KCO1 | XP 020205106 | 5 | Dockerin_like/ Ion_trans_2 | 359 | 2 | 2 | 8.79 | 40.0668 |
| 36 | CcTPK- KCO2 | XP 020222684 | 5 | Ion_trans_2-EFh_CREC | 348 | 2 | 4 | 5.91 | 38.8132 |
| 37 | CcTPK- KCO3 | XP 029126956 | 5 | Ion_trans | 324 | 3 | 2 | 9.39 | 35.9705 |
| 38 | CcTPK- KCO4 | XP 020232959 | 5 | Ion_trans | 423 | Unknown | 3 | 8.72 | 46.8026 |
| 39 | CcTPK- KCO5 | XP 020230725 | 5 | Ion_trans_2 | 340 | Unknown | 3 | 5.40 | 38.1138 |
| Regulatory Element | Core Sequence | CcAKT1.1 | CcAKT1.2 | CcAKT1.3 | CcAKT2 | CcKAT1.1 | CcKAT1.2 | CcKAT3 | CcGORK | CcHKT1 | CcHKT2 | Function |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABRE | CACGTG | 1 | 1 | 1 | 1 | Response to Abscisic acid signals | ||||||
| ACGTG | 1 | 2 | 1 | 3 | 3 | 1 | ||||||
| MYB | TAACCA | 2 | 4 | 2 | 4 | 2 | 2 | 2 | Response to drought stress and ABA signals | |||
| CAACCA | 1 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | ||||
| MYC | CATTTG | 2 | 2 | 2 | 3 | 2 | 3 | 3 | 2 | 3 | Response to drought, ABA, and cold signals | |
| W-box | TTGACC | 1 | 1 | 1 | Response to SA, GA, and pathogenesis signals | |||||||
| GT-1 motif | GGTTAA | 1 | 1 | 1 | 3 | 4 | 1 | 1 | 1 | Light responsive element | ||
| G-box | CACGTG | 1 | 3 | 3 | 3 | 1 | Involved in light response | |||||
| GARE | TCTGTTG | 1 | Gibberellin-responsive element | |||||||||
| MBS | CAACTG | 1 | 2 | Involved in drought-inducibility | ||||||||
| ARE | AAACCA | 1 | 1 | 1 | 1 | 4 | 1 | Essential for the anaerobic induction | ||||
| TCA-element | CCATCTTTTT | 2 | 1 | 1 | 1 | Response to salicylic acid | ||||||
| TC-rich repeats | ATTCTCTAAC | 2 | 2 | 2 | Involved in defense and stress response |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Siddique, M.H.; Babar, N.I.; Zameer, R.; Muzammil, S.; Nahid, N.; Ijaz, U.; Masroor, A.; Nadeem, M.; Rashid, M.A.R.; Hashem, A.; et al. Genome-Wide Identification, Genomic Organization, and Characterization of Potassium Transport-Related Genes in Cajanus cajan and Their Role in Abiotic Stress. Plants 2021, 10, 2238. https://doi.org/10.3390/plants10112238
Siddique MH, Babar NI, Zameer R, Muzammil S, Nahid N, Ijaz U, Masroor A, Nadeem M, Rashid MAR, Hashem A, et al. Genome-Wide Identification, Genomic Organization, and Characterization of Potassium Transport-Related Genes in Cajanus cajan and Their Role in Abiotic Stress. Plants. 2021; 10(11):2238. https://doi.org/10.3390/plants10112238
Chicago/Turabian StyleSiddique, Muhammad Hussnain, Naeem Iqbal Babar, Roshan Zameer, Saima Muzammil, Nazia Nahid, Usman Ijaz, Ashir Masroor, Majid Nadeem, Muhammad Abdul Rehman Rashid, Abeer Hashem, and et al. 2021. "Genome-Wide Identification, Genomic Organization, and Characterization of Potassium Transport-Related Genes in Cajanus cajan and Their Role in Abiotic Stress" Plants 10, no. 11: 2238. https://doi.org/10.3390/plants10112238
APA StyleSiddique, M. H., Babar, N. I., Zameer, R., Muzammil, S., Nahid, N., Ijaz, U., Masroor, A., Nadeem, M., Rashid, M. A. R., Hashem, A., Azeem, F., & Fathi Abd_Allah, E. (2021). Genome-Wide Identification, Genomic Organization, and Characterization of Potassium Transport-Related Genes in Cajanus cajan and Their Role in Abiotic Stress. Plants, 10(11), 2238. https://doi.org/10.3390/plants10112238









