Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro
Abstract
:Simple Summary
Abstract
1. Introduction
2. Results
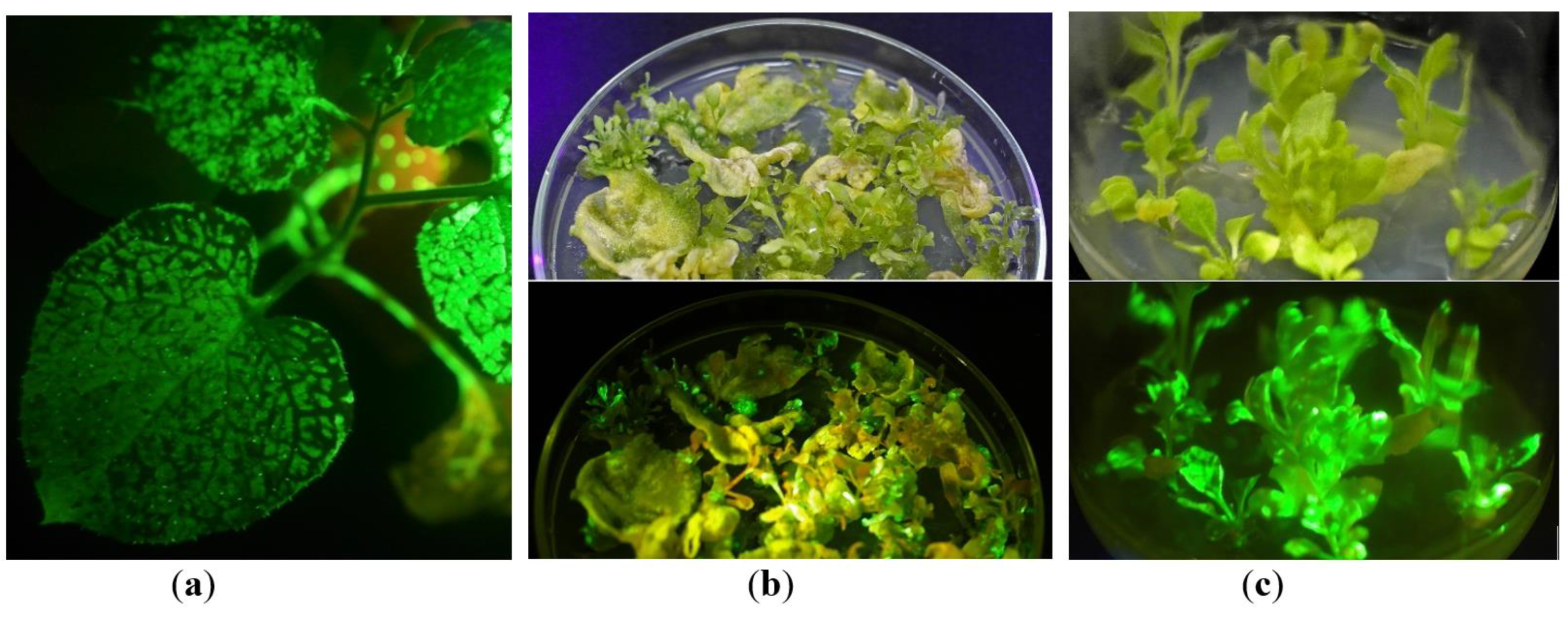
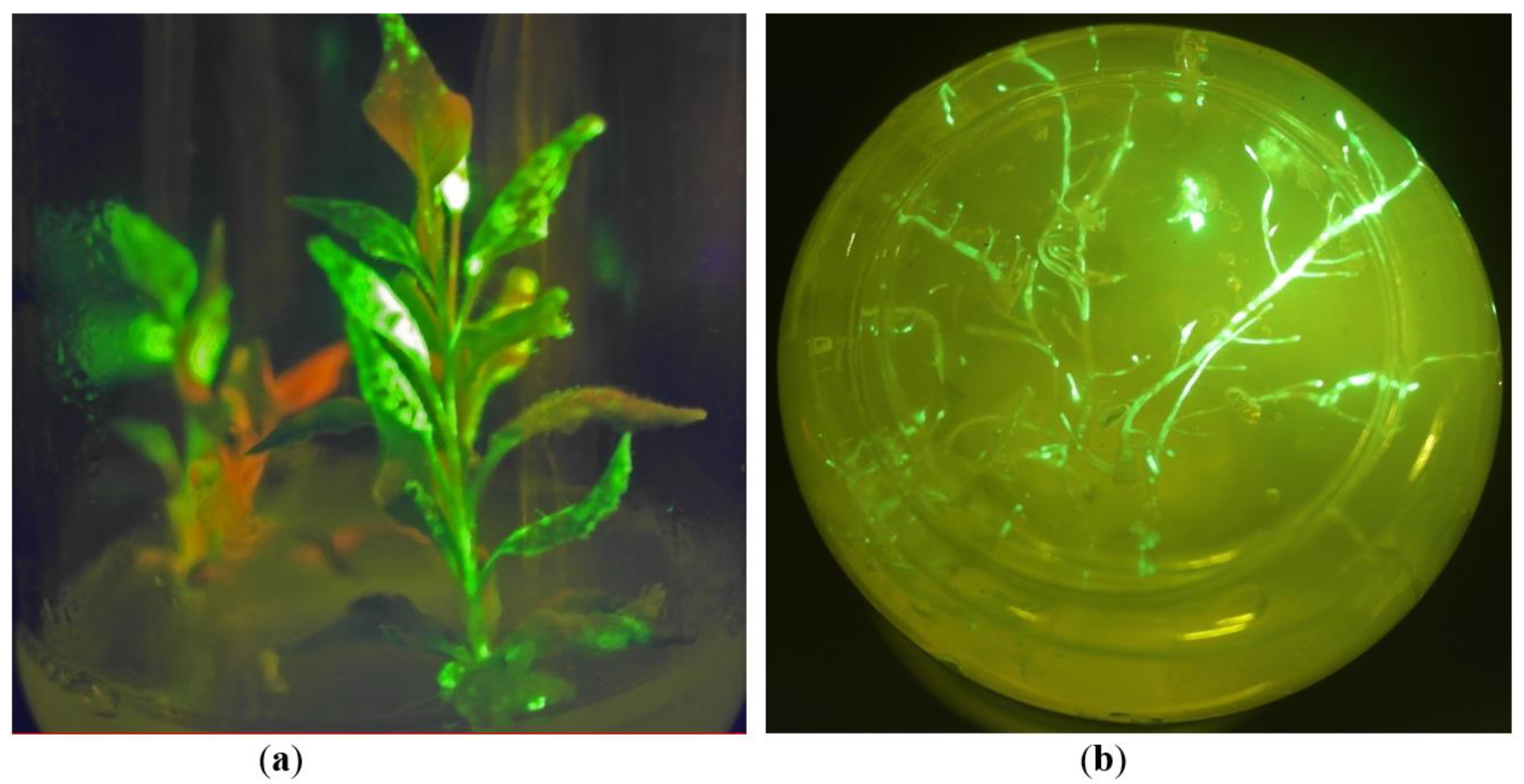
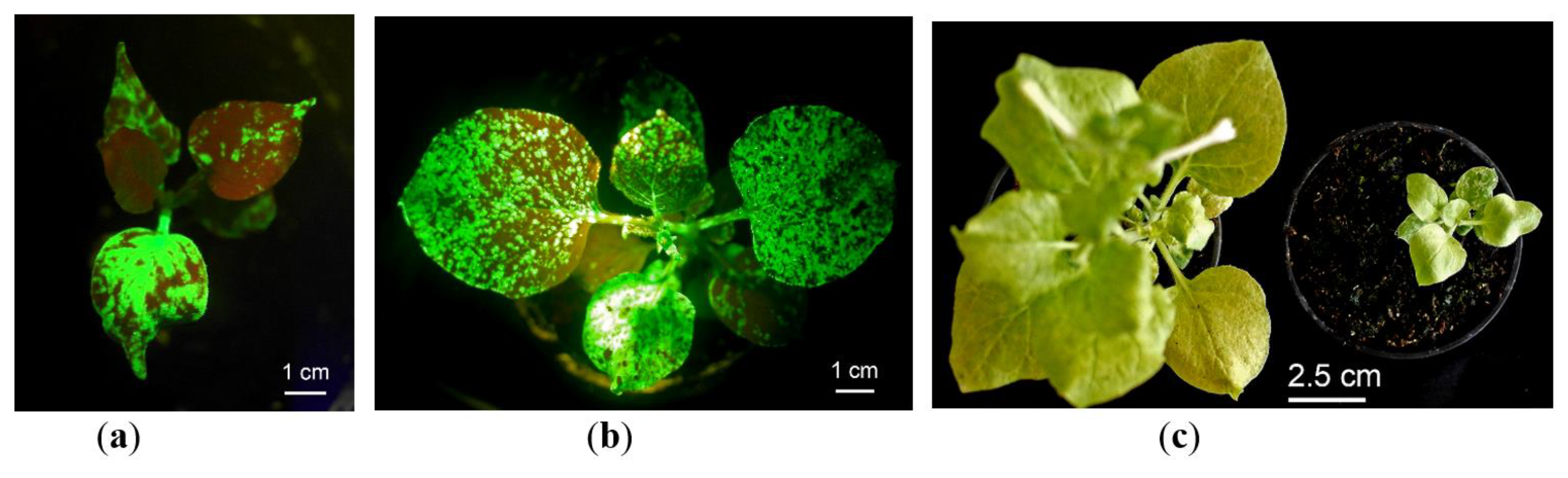
2.1. In Vitro Tissue Culture Establishment: Regeneration and Micropropagation of Plants with Long-Term Transient Expression of Recombinant Protein
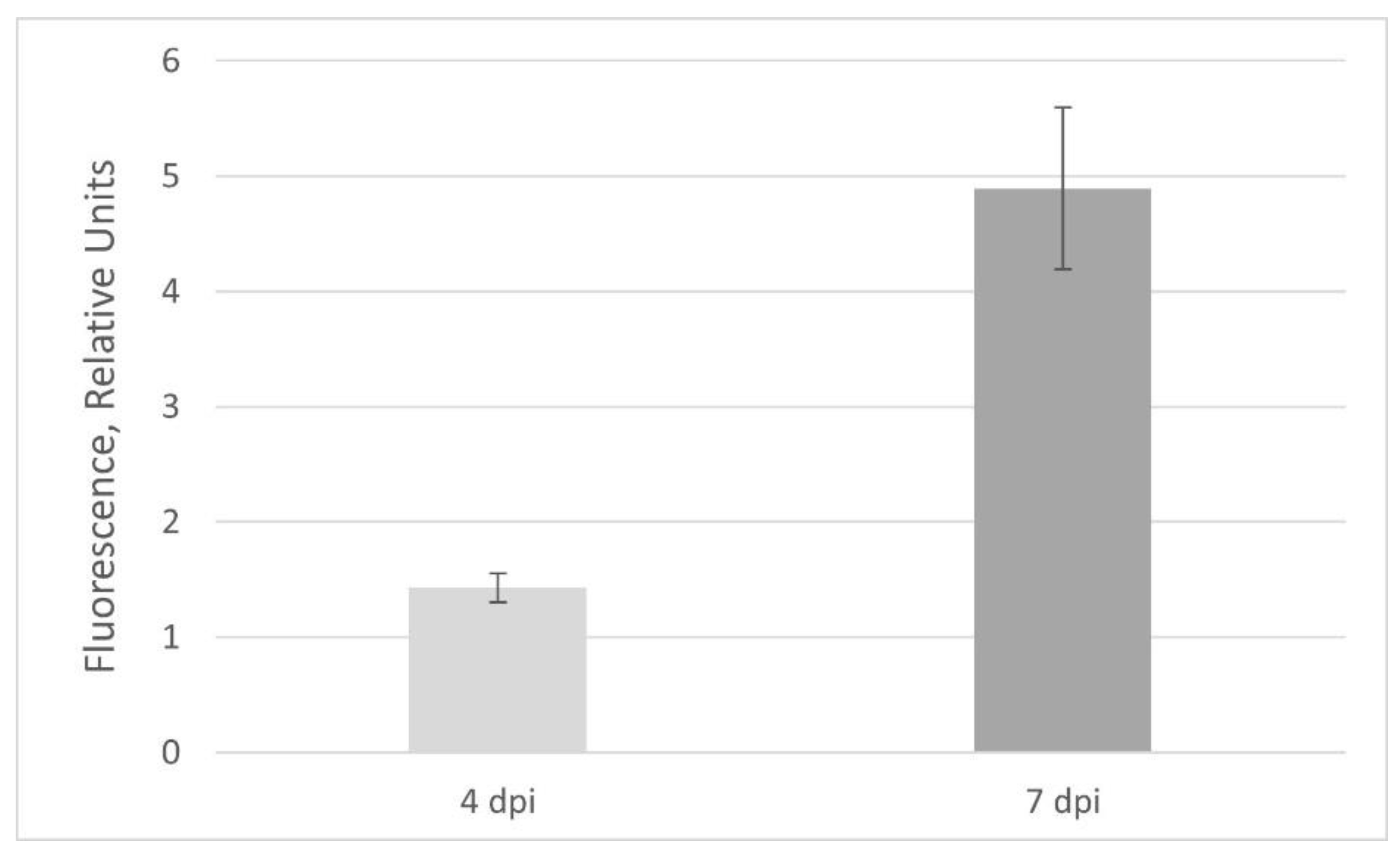
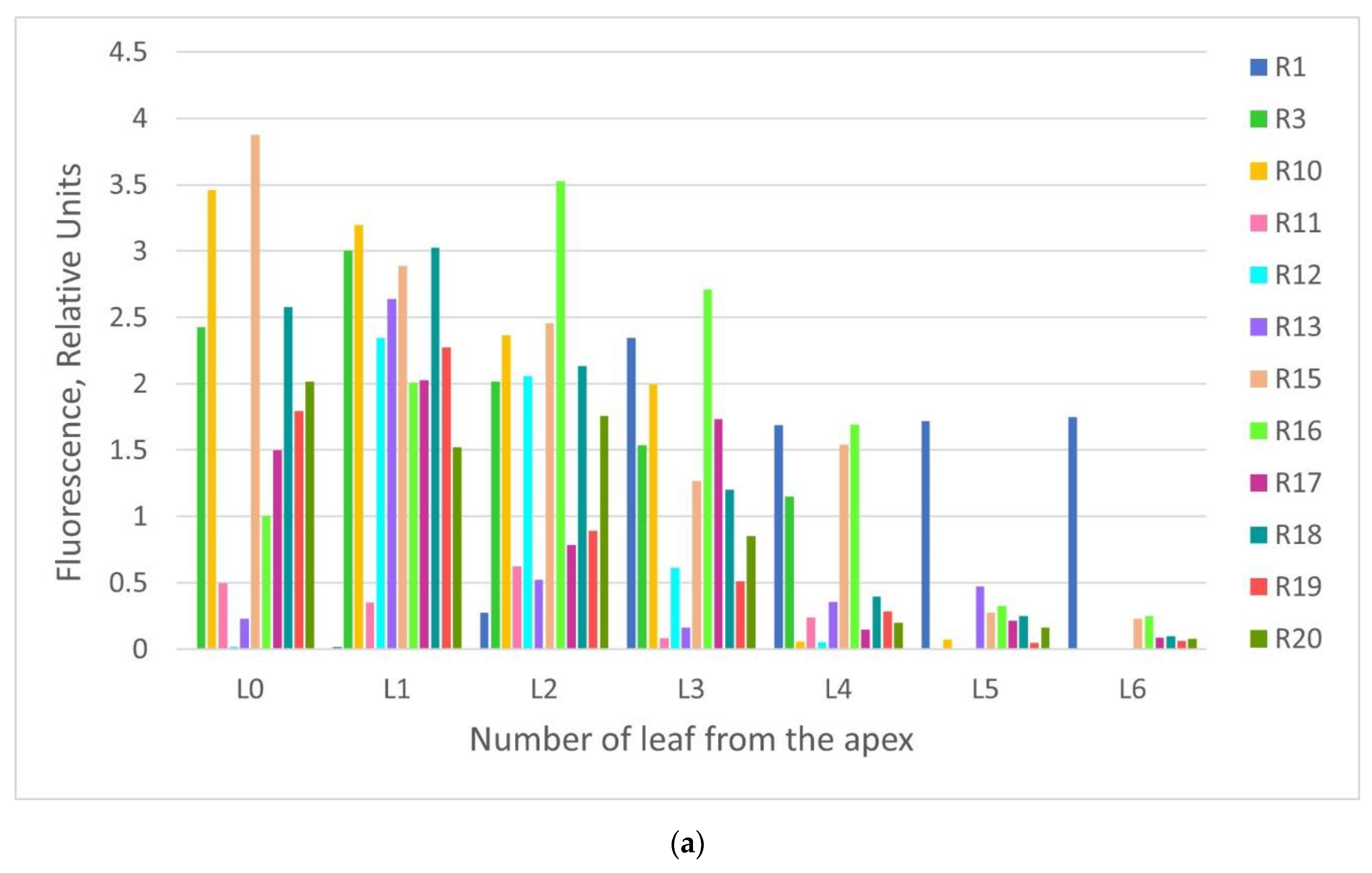
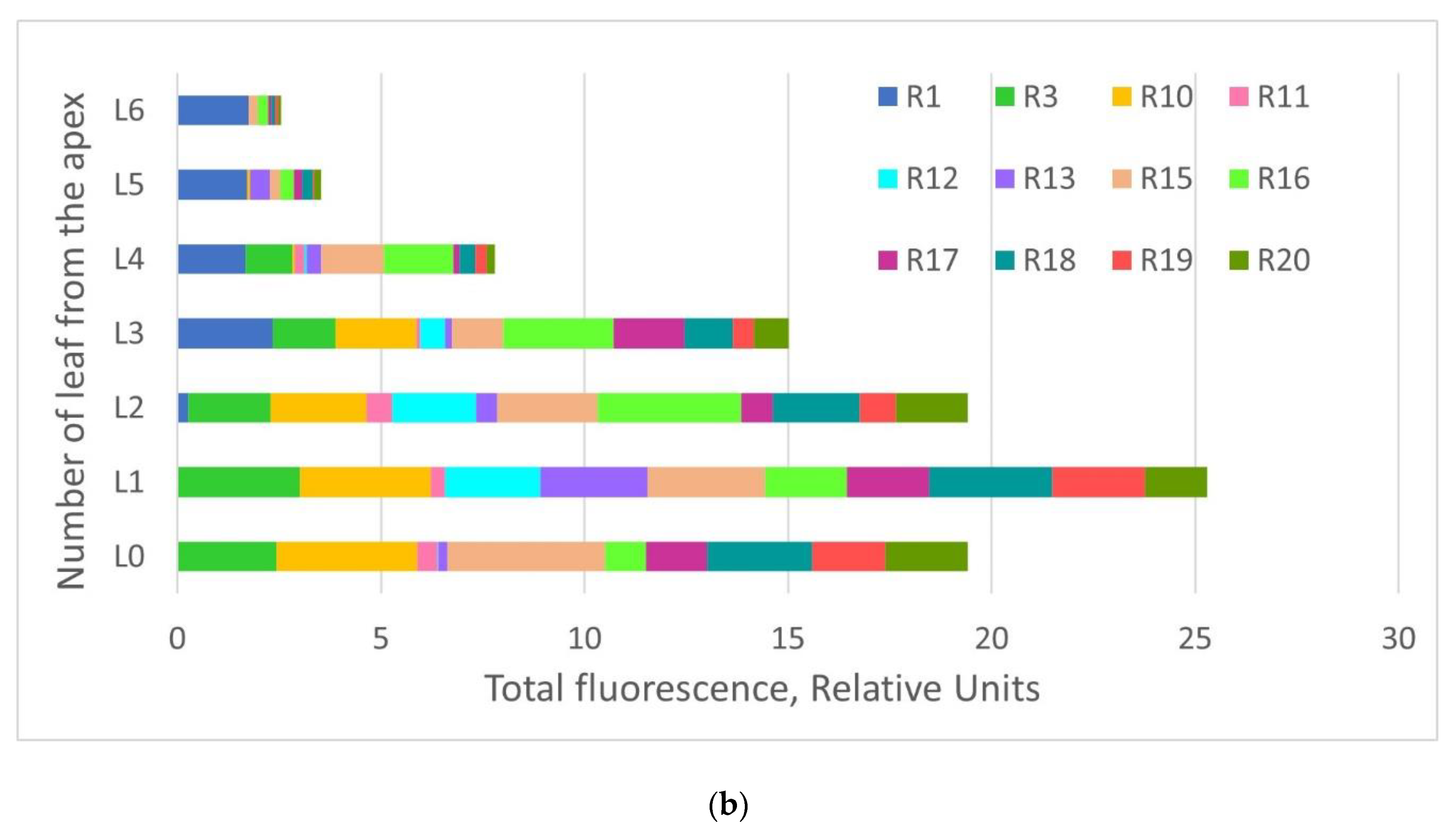
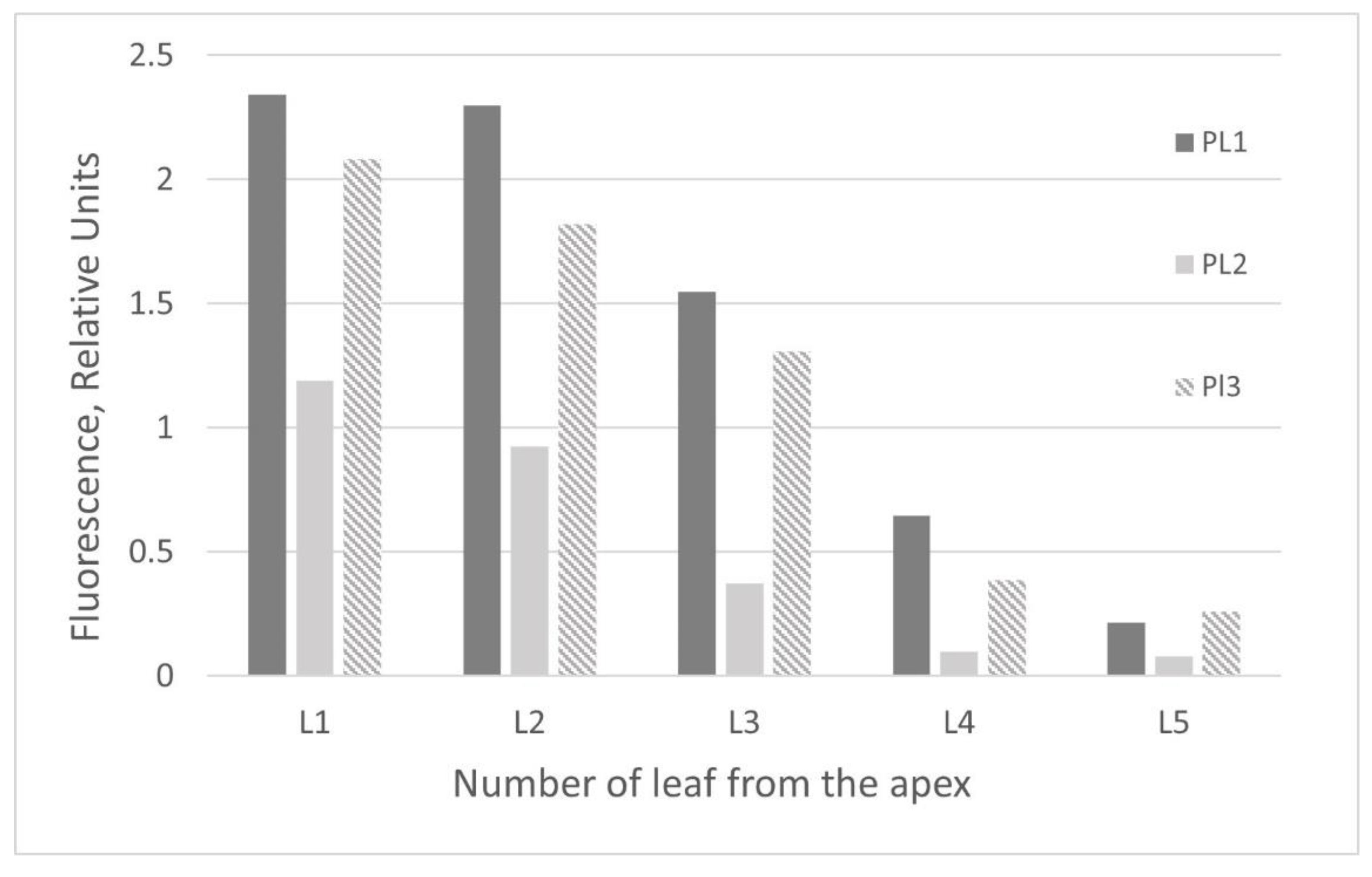
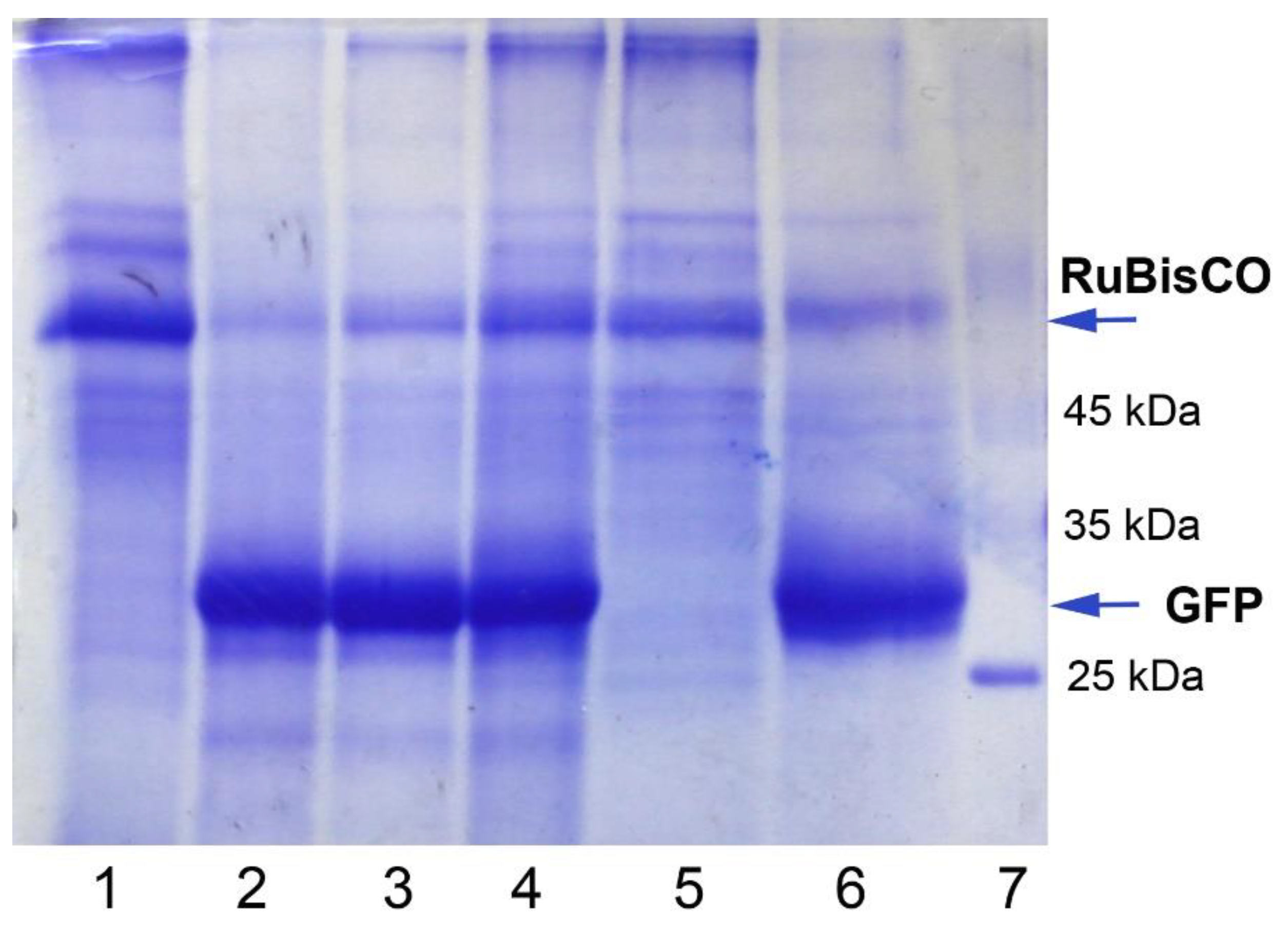
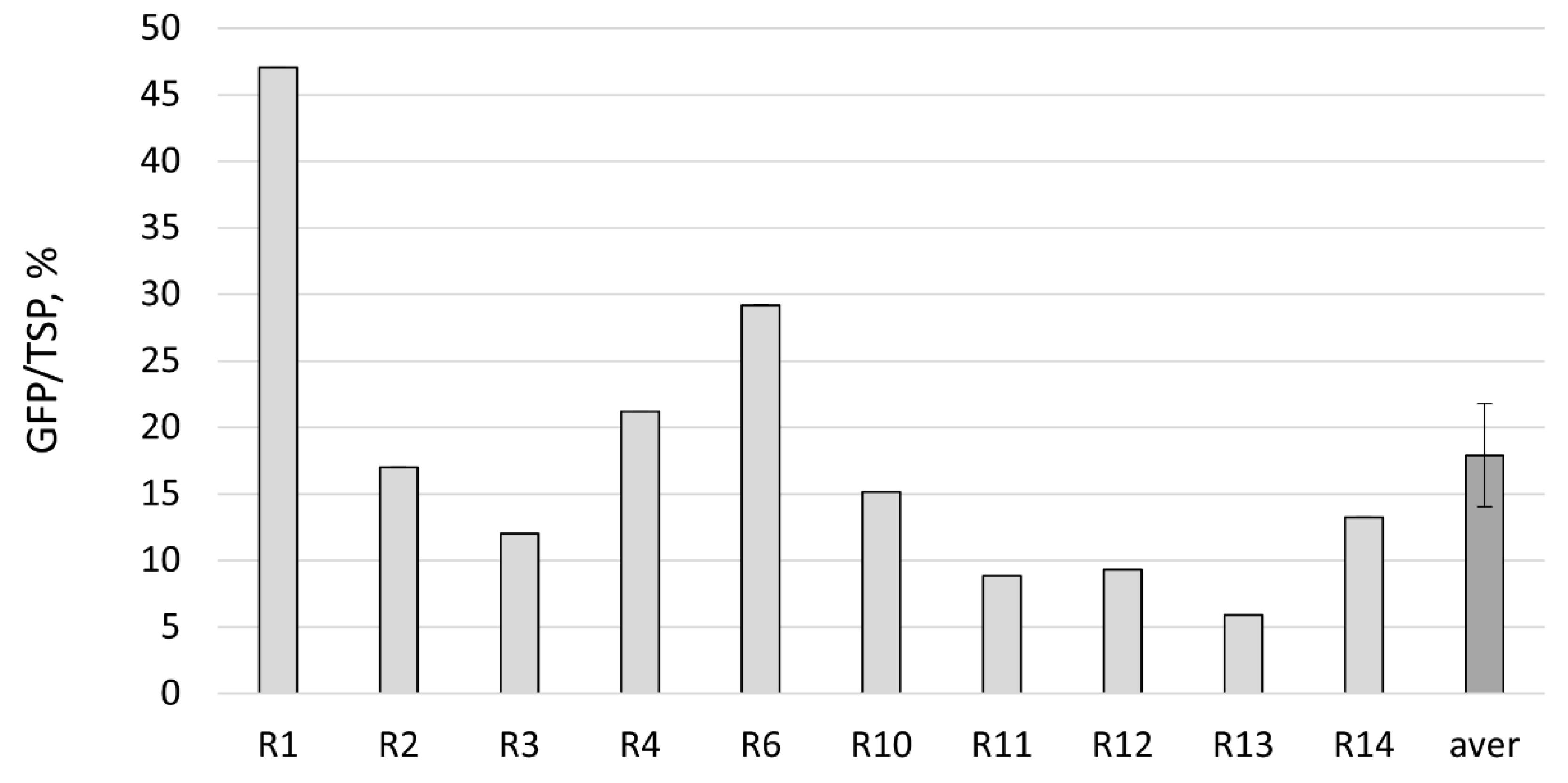
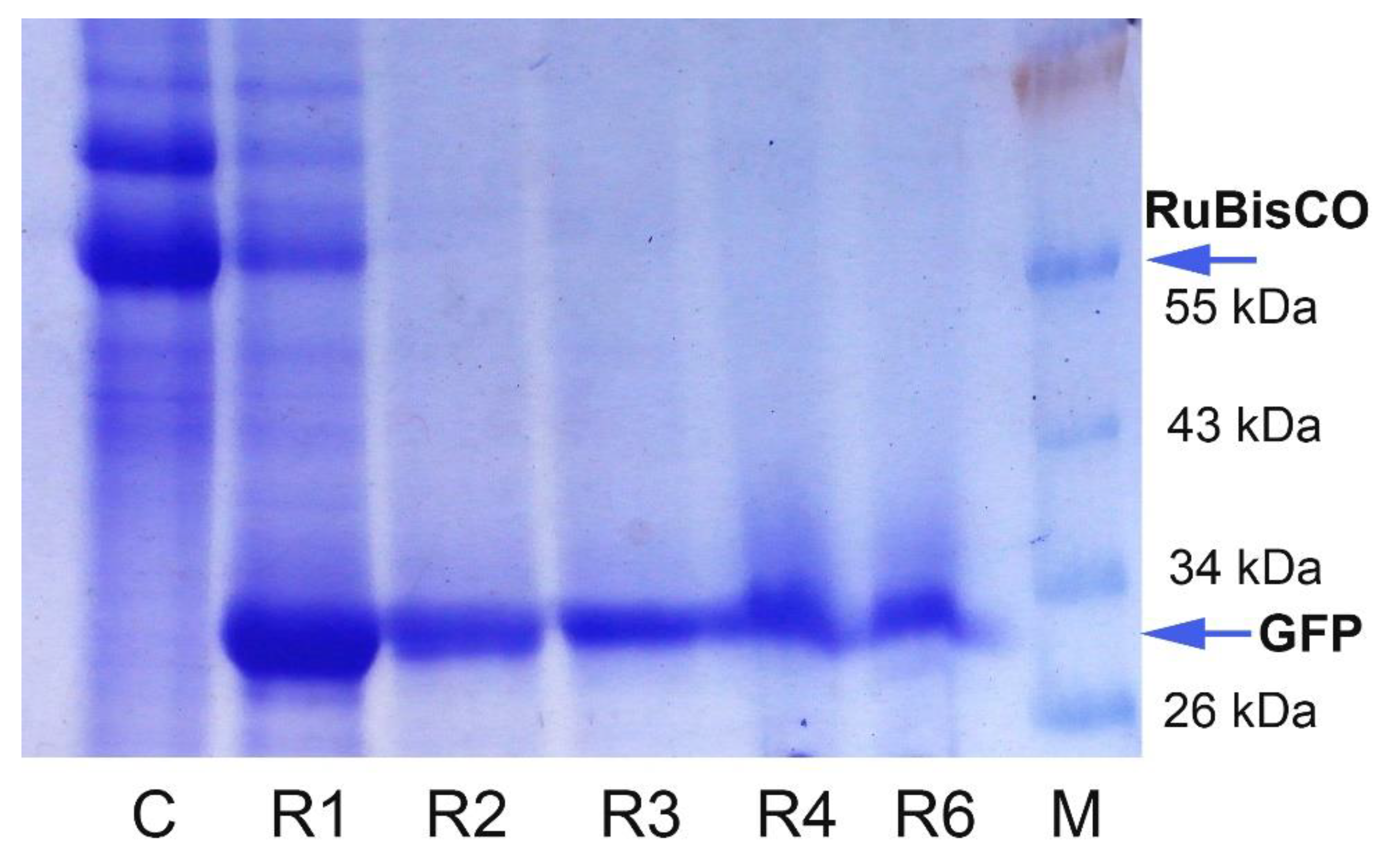
2.2. GFP Production in N. benthamiana Regenerants
3. Discussion
4. Conclusions
5. Materials and Methods
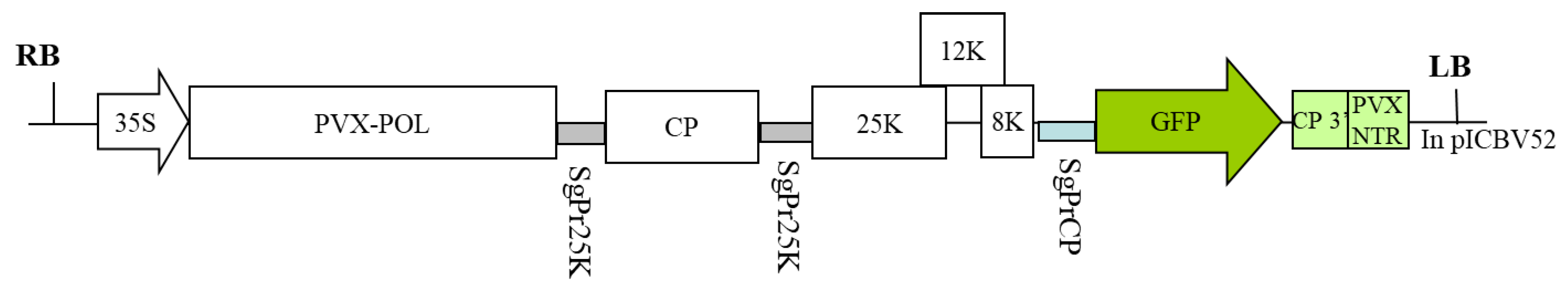
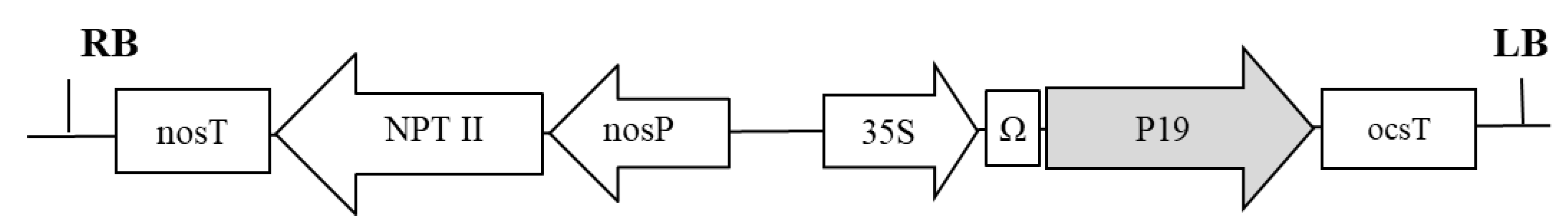
5.1. Genetic Constructions and Bacterial Strains
5.2. Plant Material and Growth Conditions in Greenhouse
5.3. Agroinfiltration Procedure
5.4. In Vitro Tissue Culture Establishment, Plant Regeneration, and Micropropagation
5.5. DNA Isolation and PCR Analysis
5.6. GFP Detection, Isolation, and Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hiatt, A.; Caffferkey, R.; Bowdish, K. Production of antibodies in transgenic plants. Nature 1989, 342, 76–78. [Google Scholar] [CrossRef]
- Marillonnet, S.; Giritch, A.; Gils, M.; Kandzia, R.; Klimyuk, V.; Gleba, Y. In Planta Engineering of Viral RNA Replicons: Efficient Assembly by Recombination of DNA Modules Delivered by Agrobacterium. Proc. Natl. Acad. Sci. USA 2004, 10, 6852–6857. [Google Scholar] [CrossRef] [Green Version]
- Sheludko, Y.V.; Sindarovska, Y.R.; Gerasymenko, I.M.; Bannikova, M.A.; Kuchuk, N.V. Comparison of Several Nicotiana Species as Hosts for High-Scale Agrobacterium-Mediated Transient Expression. Biotechnol. Bioeng. 2007, 96, 608–614. [Google Scholar] [CrossRef]
- Shi, X.; Cordero, T.; Garrigues, S.; Marcos, J.F.; Daròs, J.A.; Coca, M. Efficient Production of Antifungal Proteins in Plants Using a New Transient Expression Vector Derived from Tobacco Mosaic Virus. Plant Biotechnol. J. 2019, 17, 1069–1080. [Google Scholar] [CrossRef]
- Hahn, S.; Giritch, A.; Bartels, D.; Bortesi, L.; Gleba, Y. A Novel and Fully Scalable Agrobacterium Spray-Based Process for Manufacturing Cellulases and Other Cost-Sensitive Proteins in Plants. Plant Biotechnol. J. 2015, 13, 708–716. [Google Scholar] [CrossRef] [PubMed]
- Joubert, P.; Beaupere, D.; Lelievre, P.; Wadouachi, A.; Sangwan, R.S.; Sangwan-Norreel, B.S. Effects of Phenolic Compounds on Agrobacterium Vir Genes and Gene Transfer Induction a Plausible Molecular Mechanism of Phenol Binding Protein Activation. Plant Sci. 2002, 162, 733–743. [Google Scholar] [CrossRef]
- Norkunas, K.; Harding, R.; Dale, J.; Dugdale, B. Improving Agroinfiltration-Based Transient Gene Expression in Nicotiana benthamiana. Plant Methods 2018, 14, 71. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.É.C.; Silva, B.B.; Dutra, R.F.; Florean, E.O.P.T.; Menassa, R.; Guedes, M.I.F. Transient Expression of Dengue Virus NS1 Antigen in Nicotiana benthamiana for Use as a Diagnostic Antigen. Front. Plant Sci. 2020, 10, 1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleba, Y.; Klimyuk, V.; Marillonnet, S. Viral vectors for the expression of proteins in plants. Curr. Opin. Biotechnol. 2007, 18, 134–141. [Google Scholar] [CrossRef]
- Holtz, B.R.; Berquist, B.R.; Bennett, L.D.; Kommineni, V.J.M.; Munigunti, R.K.; White, E.L.; Wilkerson, D.C.; Wong, K.Y.; Ly, L.H.; Marcel, S. Commercial-Scale Biotherapeutics Manufacturing Facility for Plant-Made Pharmaceuticals. Plant Biotechnol. J. 2015, 13, 1180–1190. [Google Scholar] [CrossRef] [Green Version]
- Jin, T.; Wang, J.; Zhu, X.; Xu, Y.; Zhou, X.; Yang, L. A New Transient Expression System for Large-Scale Production of Recombinant Proteins in Plants Based on Air-Brushing an Agrobacterium Suspension. Biotechnol. Rep. 2015, 6, 36–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gils, M.; Kandzia, R.; Marillonnet, S.; Klimyuk, V.; Gleba, Y. High-Yield Production of Authentic Human Growth Hormone Using a Plant Virus-Based Expression System. Plant Biotechnol. J. 2005, 3, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Hefferon, K. Plant Virus Expression Vectors: A Powerhouse for Global Health. Biomedicines 2017, 5, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, M.C.; Hu, C.C.; Lin, N.S.; Hsu, Y.H. Production of Human IFNγ Protein in Nicotiana benthamiana Plant Through an Enhanced Expression System Based on Bamboo Mosaic Virus. Viruses 2019, 11, 509. [Google Scholar] [CrossRef] [Green Version]
- Mardanova, E.S.; Blokhina, E.A.; Tsybalova, L.M.; Peyret, H.; Lomonossoff, G.P.; Ravin, N.V. Efficient Transient Expression of Recombinant Proteins in Plants by the Novel pEff Vector Based on the Genome of Potato Virus X. Front. Plant Sci. 2017, 8, 247. [Google Scholar] [CrossRef]
- Mardanova, E.S.; Takova, K.H.; Toneva, V.T.; Zahmanova, G.G.; Tsybalova, L.M.; Ravin, N.V. A Plant-Based Transient Expression System for the Rapid Production of Highly Immunogenic Hepatitis E Virus-Like Particles. Biotechnol. Lett. 2020, 42, 2441–2446. [Google Scholar] [CrossRef]
- Shanmugaraj, B.; Bulaon, C.J.I.; Phoolcharoen, W. Plant Molecular Farming: A Viable Platform for Recombinant Biopharmaceutical Production. Plants 2020, 9, 842. [Google Scholar] [CrossRef] [PubMed]
- Sindarovska, Y.R.; Gerasymenko, I.M.; Sheludko, Y.V.; Olevinskaja, Z.M.; Spivak, N.Y.; Kuchuk, N.V. Production of Human Interferon Alfa 2b in Plants of Nicotiana excelsior by Agrobacterium-Mediated Transient Expression. Cytol Genet. 2010, 44, 313–316. [Google Scholar] [CrossRef]
- Yamada, Y.; Kidoguchi, M.; Yata, A.; Nakamura, T.; Yoshida, H.; Kato, Y.; Masuko, H.; Hizawa, N.; Fujieda, S.; Noguchi, E.; et al. High-Yield Production of the Major Birch Pollen Allergen Bet V 1 With Allergen Immunogenicity in Nicotiana benthamiana. Front. Plant Sci. 2020, 11, 344. [Google Scholar] [CrossRef]
- Peyret, H.; Lomonossoff, G.P. When Plant Virology Met Agrobacterium: The Rise of the Deconstructed Clones. Plant Biotechnol. J. 2015, 13, 1121–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lico, C.; Benvenuto, E.; Baschieri, S. The Two-Faced Potato Virus X: From Plant Pathogen to Smart Nanoparticle. Front. Plant Sci. 2015, 6, 1009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verchot-Lubicz, J. A New Cell-To-Cell Transport Model for Potexviruses. Mol. Plant Microbe Interact. 2005, 18, 283–290. [Google Scholar] [CrossRef] [Green Version]
- Batten, J.S.; Yoshinari, S.; Hemenway, C. Potato Virus X: A Model System for Virus Replication, Movement and Gene Expression. Mol. Plant Pathol. 2003, 4, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Cong, Q.Q.; Wang, Y.; Liu, J.; Lan, Y.F.; Guo, Z.K.; Yang, J.G.; Li, X.-D.; Tian, Y.P. Evaluation of Potato Virus X Mild Mutants for Cross Protection Against Severe Infection in China. Virol. J. 2019, 16, 36. [Google Scholar] [CrossRef] [PubMed]
- González-Jara, P.; Tenllado, F.; Martínez-García, B.; Atencio, F.A.; Barajas, D.; Vargas, M.; Díaz-Ruiz, J.; Díaz-Ruíz, J.R. Host-Dependent Differences During Synergistic Infection by Potyviruses with Potato Virus X. Mol. Plant. Pathol. 2004, 5, 29–35. [Google Scholar] [CrossRef]
- Sajid, I.A.; Tabassum, B.; Yousaf, I.; Khan, A.; Adeyinka, O.S.; Shahid, N.; Nasir, I.A.; Husnain, T. In Vivo Gene Silencing of Potato Virus X by Small Interference RNAs in Transgenic Potato. Potato Res. 2020, 63, 143–155. [Google Scholar] [CrossRef]
- Faivre-Rampant, O.; Gilroy, E.M.; Hrubikova, K.; Hein, I.; Millam, S.; Loake, G.J.; Birch, P.; Taylor, M.; Lacomme, C. Potato Virus X-Induced Gene Silencing in Leaves and Tubers of Potato. Plant. Physiol. 2004, 134, 1308–1316. [Google Scholar] [CrossRef] [Green Version]
- Qin, C.; Chen, W.; Shen, J.; Cheng, L.; Akande, F.; Zhang, K.; Yuan, C.; Li, C.; Zhang, P.; Shi, N.; et al. A Virus-Induced Assay for Functional Dissection and Analysis of Monocot and Dicot Flowering Time Genes. Plant Physiol. 2017, 174, 875–885. [Google Scholar] [CrossRef] [Green Version]
- Blokhina, E.A.; Mardanova, E.S.; Stepanova, L.A.; Tsybalova, L.M.; Ravin, N.V. Plant-Produced Recombinant Influenza a Virus Candidate Vaccine Based on Flagellin Linked to Conservative Fragments of M2 Protein and Hemagglutinin. Plants 2020, 9, 162. [Google Scholar] [CrossRef] [Green Version]
- Demurtas, O.C.; Massa, S.; Illiano, E.; De Martinis, D.; Chan, P.K.; Di Bonito, P.; Franconi, R. Antigen Production in Plant to Tackle Infectious Diseases Flare Up: The Case of SARS. Front. Plant Sci. 2016, 7, 54. [Google Scholar] [CrossRef] [Green Version]
- Giritch, A.; Marillonnet, S.; Engler, C.; van Eldik, G.; Botterman, J.; Klimyuk, V.; Gleba, Y. Rapid High-Yield Expression of Full-Size Igg Antibodies in Plants Coinfected with Noncompeting Viral Vectors. Proc. Natl. Acad. Sci. USA 2006, 103, 14701–14706. [Google Scholar] [CrossRef] [Green Version]
- Kopertekh, L.; Meyer, T.; Freyer, C.; Hust, M. Transient Plant Production of Salmonella Typhimurium Diagnostic Antibodies. Biotechnol. Rep. 2019, 21, e00314. [Google Scholar] [CrossRef]
- Mallory, A.; Parks, G.; Endres, M.; Baulcombe, D.; Bowman, L.H.; Pruss, G.J.; Vance, V.B. The Amplicon-Plus System for High-Level Expression of Transgenes in Plants. Nat. Biotechnol. 2002, 20, 622–625. [Google Scholar] [CrossRef] [PubMed]
- McMorran, J.P.; Allen, T.C. Maintenance, Symptoms and Distribution of Potato Viruses X, S, Y, A, M and Leafroll in Potato Tissue Culture Plantlets. Am. Potato J. 1983, 60, 839–847. [Google Scholar] [CrossRef]
- Schenk, G.; Neitzel, K. A Contribution to the Maintenance of Phytopathogenic Virus Strains on Tissue Culture Plantlets. Potato Res. 1985, 28, 535–538. [Google Scholar] [CrossRef]
- Kopertekh, L.; Schiemann, J. Agroinfiltration as a Tool for Transient Expression of Cre Recombinase in Vivo. Transgenic Res. 2005, 14, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Clemente, T. Nicotiana (Nicotiana tobaccum, Nicotiana benthamiana). Methods Mol. Biol. 2006, 343, 143–154. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bioassays with Tobacco Tissue Culture. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Bidabadi, S.S.; Jain, S.M. Cellular, Molecular, and Physiological Aspects of in Vitro Plant Regeneration. Plants 2020, 9, 702. [Google Scholar] [CrossRef]
- Thorpe, T.A. History of plant tissue culture. Mol. Biotechnol. 2007, 37, 169–180. [Google Scholar] [CrossRef]
- Pathi, K.M.; Tula, S.; Tuteja, N. High Frequency Regeneration via Direct Somatic Embryogenesis and Efficient Agrobacterium-Mediated Genetic Transformation of Tobacco. Plant Signal. Behav. 2013, 8, 6. [Google Scholar] [CrossRef] [Green Version]
- Ramage, C.M.; Williams, R.R. Mineral Uptake in Tobacco Leaf Discs During Different Developmental Stages of Shoot Organogenesis. Plant. Cell Rep. 2003, 21, 1047–1053. [Google Scholar] [CrossRef]
- Rout, G.R.; Sahoo, S. In Vitro Selection and Plant Regeneration of Copper-Tolerant Plants from Leaf Explants of Nicotiana tabacum L. Cv. ‘Xanthi’. Plant Breed. 2007, 126, 403–409. [Google Scholar] [CrossRef]
- Sindarovska, Y.R.; Sheludko, Y.V.; Gerasymenko, I.M.; Bannikova, M.A.; Komarnytskyy, I.K.; Kuchuk, N.V. Transgenic Plants Regenerated from Hairy Roots of Nicotiana benthamiana: A Promising Host for Transient Expression of Foreign Proteins. Cytol. Genet. 2005, 39, 9–14. [Google Scholar]
- Stolarz, A.; Macewicz, J.; Lörz, H. Direct Somatic Embryogenesis and Plant Regeneration from Leaf Explants of Nicotiana tabacum L. J. Plant Physiol. 1991, 137, 347–357. [Google Scholar] [CrossRef]
- Cassells, A.C. Pathogen and Biological Contamination Management in Plant Tissue Culture: Phytopathogens, Vitro Pathogens, and Vitro Pests. Methods Mol. Biol. 2012, 877, 57–80. [Google Scholar] [CrossRef]
- Miguel, C.; Marum, L. An epigenetic view of plant cells cultured in vitro: Somaclonal variation and beyond. J. Exp. Bot. 2011, 62, 3713–3725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Lai, H.; Hurtado, J.; Stahnke, J.; Leuzinger, K.; Dent, M. Agroinfiltration as an Effective and Scalable Strategy of Gene Delivery for Production of Pharmaceutical Proteins. Adv. Tech. Biol. Med. 2013, 1, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galun, E. RNA Silencing in Plants. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 113–123. [Google Scholar] [CrossRef]
- Voinnet, O.; Vain, P.; Angell, S.; Baulcombe, D.C. Systemic Spread of Sequence-Specific Transgene RNA Degradation in Plants Is Initiated by Localized Introduction of Ectopic Promoterless DNA. Cell 1998, 95, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Tournier, B.; Tabler, M.; Kalantidis, K. Phloem Flow Strongly Influences the Systemic Spread of Silencing in GFP Nicotiana benthamiana Plants. Plant. J. 2006, 47, 383–394. [Google Scholar] [CrossRef]
- Rajeevkumar, S.; Anunanthini, P.; Sathishkumar, R. Epigenetic Silencing in Transgenic Plants. Front. Plant Sci. 2015, 6, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johansen, L.K.; Carrington, J.C. Silencing on the Spot. Induction and Suppression of RNA Silencing in the Agrobacterium-Mediated Transient Expression System. Plant. Physiol. 2001, 126, 930–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manske, U.; Landsmann, J.; Dietz-Pfeilstetter, A. Comparison of Different Methods for the Establishment of RNA Silencing in Plants. Plant. Biotechnol. Rep. 2017, 11, 115–125. [Google Scholar] [CrossRef]
- Voinnet, O.; Lederer, C.; Baulcombe, D.C. A Viral Movement Protein Prevents Spread of the Gene Silencing Signal in Nicotiana benthamiana. Cell 2000, 103, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Jay, F.; Vitel, M.; Brioudes, F.; Louis, M.; Knobloch, T.; Voinnet, O. Chemical Enhancers of Posttranscriptional Gene Silencing in Arabidopsis. RNA 2019, 25, 1078–1090. [Google Scholar] [CrossRef] [PubMed]
- Liang, D.; White, R.G.; Waterhouse, P.M. Mobile Gene Silencing in Arabidopsis Is Regulated by Hydrogen Peroxide. PeerJ 2014, 2, e701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stam, M.; Mol, J.N.M.; Kooter, J.M. The Silence of Genes in Transgenic Plants. Ann. Bot. 1997, 79, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Lai, T.; Zhang, P.; Zhang, X.; Yuan, C.; Jin, Z.; Li, H.; Yu, Z.; Qin, C.; Tör, M.; et al. Mini Review: Revisiting Mobile RNA Silencing in Plants. Plant Sci. 2019, 278, 113–117. [Google Scholar] [CrossRef]
- Angell, S.M.; Baulcombe, D.C. Consistent Gene Silencing in Transgenic Plants Expressing a Replicating Potato Virus X RNA. EMBO J. 1997, 16, 3675–3684. [Google Scholar] [CrossRef] [Green Version]
- Sindarovska, Y.R.; Gerasymenko, I.M.; Kuchuk, M.V.; Sheludko, Y.V. Influence of Exogenous Phytohormones, Methyl Jasmonate and Suppressors of Jasmonate Biosynthesis on Agrobacterium-Mediated Transient Expression in Nicotiana Excelsior. Biopolym. Cell 2012, 28, 357–362. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Kahmann, R. Manipulation of Phytohormone Pathways by Effectors of Filamentous Plant Pathogens. Front. Plant Sci. 2019, 10, 822. [Google Scholar] [CrossRef] [Green Version]
- Naseem, M.; Dandekar, T. The Role of Auxin-Cytokinin Antagonism in Plant-Pathogen Interactions. PLoS Pathog. 2012, 8, e1003026. [Google Scholar] [CrossRef]
- O’Brien, J.A.; Benková, E. Cytokinin Cross-Talking During Biotic and Abiotic Stress Responses. Front. Plant Sci. 2013, 4, 451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berger, S.; Papadopoulos, M.; Schreiber, U.; Kaiser, W.; Roitsch, T. Complex Regulation of Gene Expression, Photosynthesis and Sugar Levels by Pathogen Infection in Tomato. Physiol. Plant 2004, 122, 419–428. [Google Scholar] [CrossRef]
- Stevens, L.H.; Stoopen, G.M.; Elbers, I.J.; Molthoff, J.W.; Bakker, H.A.C.; Lommen, A.; Bosch, D.; Jordi, W. Effect of Climate Conditions and Plant Developmental Stage on the Stability of Antibodies Expressed in Transgenic Tobacco. Plant Physiol. 2000, 124, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Wydro, M.; Kozubek, E.; Lehmann, P. Optimization of Transient Agrobacterium-Mediated Gene Expression System in Leaves of Nicotiana benthamiana. Acta Biochim. Pol. 2006, 53, 289–298. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V. The Molecular Biology of Leaf Senescence. J. Exp. Bot. 1997, 48, 181–199. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Sun, H.; Lai, H.; Hurtado, J.; Chen, Q. Plant-Produced Zika Virus Envelope Protein Elicits Neutralizing Immune Responses that Correlate with Protective Immunity against Zika Virus in Mice. Plant Biotechnol. J. 2018, 16, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Hoshikawa, K.; Ezura, K.; Okazawa, R.; Fujita, S.; Takaoka, M.; Mason, H.S.; Ezura, H.; Miura, K. Improvement of the Transient Expression System for Production of Recombinant Proteins in Plants. Sci. Rep. 2018, 8, 4755. [Google Scholar] [CrossRef] [Green Version]
- Fujiuchi, N.; Matoba, N.; Matsuda, R. Environment Control to Improve Recombinant Protein Yields in Plants Based on Agrobacterium-Mediated Transient Gene Expression. Front. Bioeng. Biotechnol. 2016, 4, 23. [Google Scholar] [CrossRef] [Green Version]
- Patil, B.L.; Fauquet, C.M. Light Intensity and Temperature Affect Systemic Spread of Silencing Signal in Transient Agroinfiltration Studies. Mol. Plant. Pathol. 2015, 16, 484–494. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.E.; Jones, R.A.C.; Coutts, R.H.A. Effect of Temperature on Potato Virus X Infection in Potato Cultivars Carrying Different Combinations of Hypersensitivity Genes. Plant Pathol. 1986, 35, 517–526. [Google Scholar] [CrossRef]
- Draper, M.D.; Pasche, J.S.; Gudmestad, N.C. Factors Influencing PVY Development and Disease Expression in Three Potato Cultivars. Am. J. Pot. Res. 2002, 7, 155–165. [Google Scholar] [CrossRef]
- Daniell, H. Production of biopharmaceuticals and vaccines in plants via the chloroplast genome. Biotechnol. J. 2006, 1, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Denkovskienė, E.; Paškevičius, Š.; Werner, S.; Gleba, Y.; Ražanskienė, A. Inducible Expression of Agrobacterium Virulence Gene VirE2 for Stringent Regulation of T-DNA Transfer in Plant Transient Expression Systems. Mol. Plant-Microbe Interact. 2015, 28, 1247–1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marillonnet, S.; Thoeringer, C.; Kandzia, R.; Klim, V.; Gleba, Y. Systemic Agrobacterium tumefaciens–mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005, 23, 718–723. [Google Scholar] [CrossRef]
- Weigel, D.; Glazebrook, J. Quick Miniprep for Plant DNA Isolation. Cold Spring Harb. Protoc. 2009, 3, pdb.prot5179. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantification of Microgram Quantities of Protein Utilizing the Principle of Protein Due Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
| Stage | Time Required (Days) |
|---|---|
| Plant growing prior to infection | 35–55 |
| An agroinfiltration procedure | 1–2 |
| System infection and transient gene expression in the upper leaves | 5–10 |
| Transferring leaves to in vitro conditions | 1 |
| Shoots regeneration | 14–45 |
| Roots appearing | 14–30 |
| Cycle step to “rejuvenate” plant tissue culture: leaves of regenerants growing in vitro can be put on a regeneration medium to obtain the young regenerants. | |
| It is possible to postpone the next step and maintain the regenerants in vitro for up to 6 months. | |
| Transferring the regenerants to soil and yielding the recombinant protein from the whole plant | 35–50 |
| Total time required to obtain the recombinant protein | Minimum—2 months (when tissue culture is established)Maximum—6–7 months (starting from the plant growing stage) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sindarovska, Y.; Kuchuk, M. Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro. Plants 2021, 10, 2187. https://doi.org/10.3390/plants10102187
Sindarovska Y, Kuchuk M. Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro. Plants. 2021; 10(10):2187. https://doi.org/10.3390/plants10102187
Chicago/Turabian StyleSindarovska, Yana, and Mykola Kuchuk. 2021. "Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro" Plants 10, no. 10: 2187. https://doi.org/10.3390/plants10102187
APA StyleSindarovska, Y., & Kuchuk, M. (2021). Long-Term Potato Virus X (PVX)-Based Transient Expression of Recombinant GFP Protein in Nicotiana benthamiana Culture In Vitro. Plants, 10(10), 2187. https://doi.org/10.3390/plants10102187






