Applying HPLC to Screening QTLs for BLB Resistance in Rice
Abstract
:1. Introduction
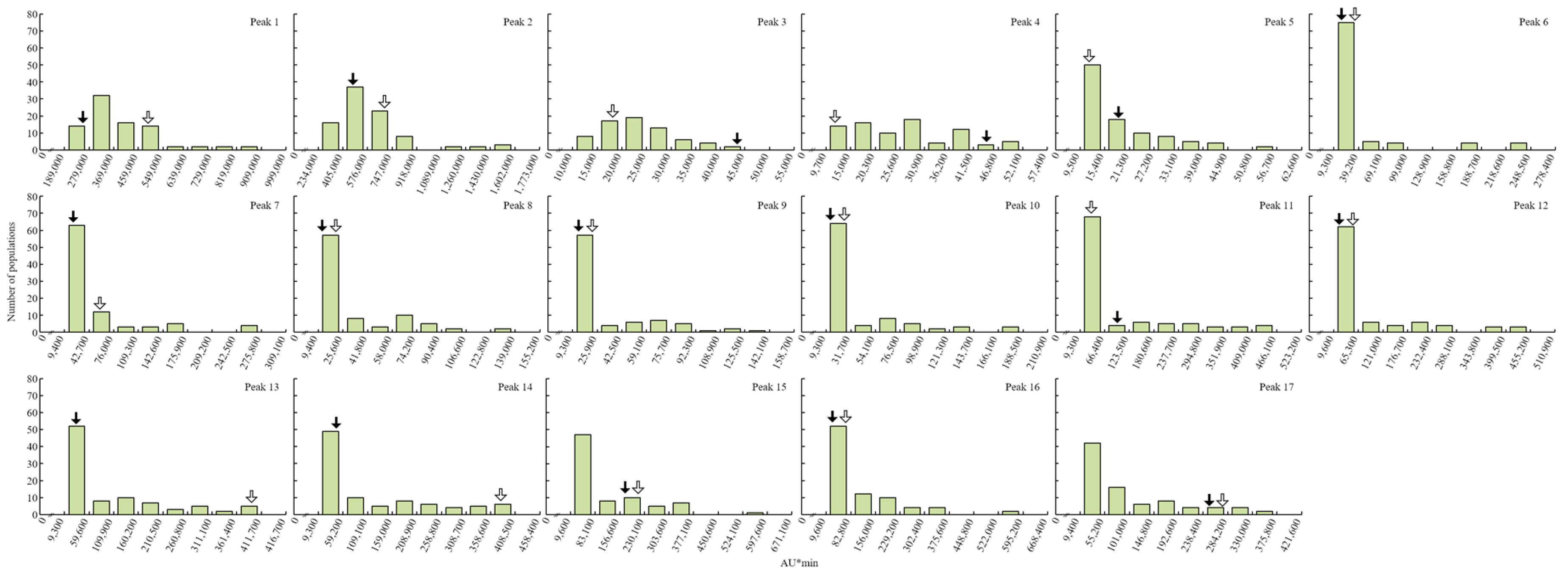
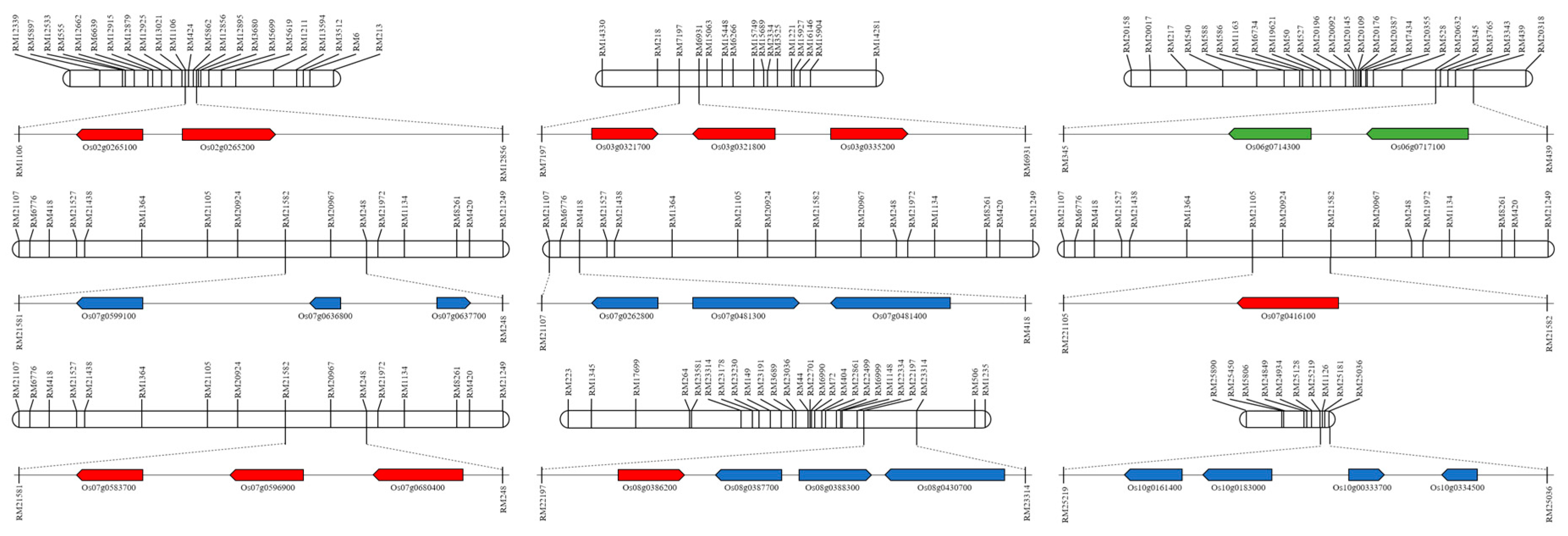
2. Results
3. Discussion
4. Materials and Methods
4.1. Rice Materials and Field Design
4.2. K3 Strain Culture Media, Inoculation, and Infection Assessment
4.3. Extraction of the Infected Rice Leaf Components and HPLC Analysis
4.4. QTL Analysis
4.5. RNA Extraction
4.6. cDNA Synthesis
4.7. Quantitative RT-PCR Analyses
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar Verma, R.; Samal, B.; Chatterjee, S. Xanthomonas oryzae pv. oryzae chemotaxis components and chemoreceptor Mcp2 are involved in the sensing of constituents of xylem sap and contribute to the regulation of virulence-associated functions and entry into rice. Mol. Plant. Pathol. 2018, 19, 2397–2415. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Sheng, L.; Zhang, G.; Gu, Q.; Zheng, K. Influence of bacterial leaf blight on the photosynthetic characteristics of resistant and susceptible rice. J. Phytopathol. 2018, 166, 547–554. [Google Scholar] [CrossRef]
- Fordjour, E.; Kim, H.H.; Aye, N.C.; Rolly, N.K.; Jummah, W.N.; Imran, Q.M.; Mun, B.G.; Hussain, A.; Yun, B.W. Screening rice (Oryza sativa) cultivars for resistance to bacterial leaf blight disease. Int. J. Agric Biol. 2020, 24, 1233–1241. [Google Scholar] [CrossRef]
- Schneider, P.; Asch, F. Rice production and food security in Asian Mega deltas—A review on characteristics, vulnerabilities and agricultural adaptation options to cope with climate change. J. Agron. Crop. Sci. 2020, 206, 491–503. [Google Scholar] [CrossRef]
- Roger, A.C.J.; Martin, J.B. Influence of climate change on plant disease infections and epidemics caused by viruses and bacteria. In Plant Sciences Reviews; CABI Publishing: Surrey, UK, 2012; pp. 73–89. [Google Scholar]
- Yasmin, S.; Zaka, A.; Imran, A.; Zahid, M.A.; Yousaf, S.; Rasul, G.; Arif, M.; Mirza, M.S. Plant growth promotion and suppression of bacterial leaf blight in rice by inoculated bacteria. PLoS ONE 2016, 11, e0160688. [Google Scholar] [CrossRef] [Green Version]
- Inoue, Y.; Matsuura, T.; Ohara, T.; Azegami, K. Bacteriophage OP1, lytic for Xanthomonas oryzae pv. oryzae, changes its host range by duplication and deletion of the small domain in the deduced tail fiber gene. J. Gen. Plant. Pathol. 2006, 72, 111–118. [Google Scholar] [CrossRef]
- Jiang, H.; Feng, Y.; Qiu, L.; Gao, G.; Zhang, Q.; He, Y. Identification of Blast Resistance QTLs Based on Two Advanced Backcross Populations in Rice. Rice 2020, 13, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Du, Z.; Chen, J.; Zhang, Z. QTL mapping of wheat plant architectural characteristics and their genetic relationship with seven QTLs conferring resistance to sheath blight. PLoS ONE 2017, 12, e0174939. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.R.; Wit, M.; Hajos, T.; Wit, M.; Howren, M.B.; Insana, S.; Simonson, M.A. Quantitative Trait Locus (QTL). Encycl. Behav. Med. 2013, 1, 1609–1610. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef] [Green Version]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; de Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef] [Green Version]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef]
- Duku, C.; Sparks, A.H.; Zwart, S.J. Spatial modelling of rice yield losses in Tanzania due to bacterial leaf blight and leaf blast in a changing climate. Clim. Chang. 2016, 135, 569–583. [Google Scholar] [CrossRef]
- Noh, T.-H.; Lee, D.-K.; Park, J.-C.; Shim, H.-K.; Choi, M.-Y.; Kang, M.-H.; Kim, J.-D. Effects of Bacterial Leaf Blight Occurrence on Rice Yield and Grain Quality in Different Rice Growth Stage. Res. Plant Dis. 2007, 13, 20–23. [Google Scholar] [CrossRef] [Green Version]
- Qiang, H.; Mark, D. Bertness Extreme stresses, niches, and positive species interactions along stress gradients. Ecology 2014, 95, 1437–1443. [Google Scholar]
- Brunetti, C.; Guidi, L.; Sebastiani, F.; Tattini, M. Isoprenoids and phenylpropanoids are key components of the antioxidant defense system of plants facing severe excess light stress. Environ. Exp. Bot. 2015, 119, 54–62. [Google Scholar] [CrossRef]
- Chukwu, S.C.; Rafii, M.Y.; Ramlee, S.I.; Ismail, S.I.; Hasan, M.M.; Oladosu, Y.A.; Magaji, U.G.; Akos, I.; Olalekan, K.K. Bacterial leaf blight resistance in rice: A review of conventional breeding to molecular approach. Mol. Biol. Rep. 2019, 46, 1519–1532. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Reinke, R.F. A novel resistance gene for bacterial blight in rice, Xa43(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population. PLoS ONE 2019, 14, e0211775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, G.H.; Yun, B.W.; Kim, K.M. Analysis of QTLs associated with the rice quality related gene by double haploid populations. Int. J. Genom. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Park, J.R.; Yang, W.T.; Kim, D.H.; Kim, K.M. Identification of a novel gene, osbht, in response to high temperature tolerance at booting stage in rice. Int. J. Mol. Sci. 2020, 21, 5862. [Google Scholar] [CrossRef]
- Park, J.R.; Resolus, D.; Kim, K.M. Osbrkq1, related grain size mapping, and identification of grain shape based on qtl mapping in rice. Int. J. Mol. Sci. 2021, 22, 2289. [Google Scholar] [CrossRef]
- Shulaev, V.; Cortes, D.; Miller, G.; Mittler, R. Metabolomics for plant stress response. Physiol. Plant. 2008, 132, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Naika, M.; Shameer, K.; Mathew, O.K.; Gowda, R.; Sowdhamini, R. STIFDB2: An updated version of plant stress-responsive transcription factor database with additional stress signals, stress-responsive transcription factor binding sites and stress-responsive genes in arabidopsis and rice. Plant. Cell Physiol. 2013, 54, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Song, Y.; Li, S.; Zhang, L.; Zou, C.; Yu, D. The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta Gene Regul. Mech. 2012, 1819, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, R.; Jiang, S.Y.; Kumar, N.; Venkatesh, P.N.; Ramachandran, S. A comprehensive transcriptional profiling of the WRKY gene family in rice under various abiotic and phytohormone treatments. Plant. Cell. Physiol. 2008, 49, 865–879. [Google Scholar] [CrossRef]
- Ali, S.; Ganai, B.A.; Kamili, A.N.; Bhat, A.A.; Mir, Z.A.; Bhat, J.A.; Tyagi, A.; Islam, S.T.; Mushtaq, M.; Yadav, P.; et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018, 212–213, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant. Biol. 2016, 16, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Bakshi, M.; Oelmüller, R. Wrky transcription factors jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [Green Version]
- Dilla-Ermita, C.J.; Tandayu, E.; Juanillas, V.M.; Detras, J.; Lozada, D.N.; Dwiyanti, M.S.; Vera Cruz, C.; Mbanjo, E.G.N.; Ardales, E.; Diaz, M.G.; et al. Genome-wide Association Analysis Tracks Bacterial Leaf Blight Resistance Loci In Rice Diverse Germplasm. Rice 2017, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S.D. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef]
- Akhtar, M.A.; Rafi, A.; Hameed, A. Comparison of methods of inoculation of Xanthomonas Oryzae pv. oryzae in rice cultivars. Pak. J. Bot. 2008, 40, 2171–2175. [Google Scholar]
- Pallant, J. A Step by Step Guide to Data Analysis Using SPSS Version 15 SPSS Survival Manual, 3rd ed.; Open University Press: London, UK, 2007. [Google Scholar]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Du, X.-X.; Park, J.-R.; Wang, X.-H.; Jang, Y.-H.; Kim, E.-G.; Lee, G.-S.; Kim, K.-M. Applying HPLC to Screening QTLs for BLB Resistance in Rice. Plants 2021, 10, 2145. https://doi.org/10.3390/plants10102145
Du X-X, Park J-R, Wang X-H, Jang Y-H, Kim E-G, Lee G-S, Kim K-M. Applying HPLC to Screening QTLs for BLB Resistance in Rice. Plants. 2021; 10(10):2145. https://doi.org/10.3390/plants10102145
Chicago/Turabian StyleDu, Xiao-Xuan, Jae-Ryoung Park, Xiao-Han Wang, Yoon-Hee Jang, Eun-Gyeong Kim, Gang-Seob Lee, and Kyung-Min Kim. 2021. "Applying HPLC to Screening QTLs for BLB Resistance in Rice" Plants 10, no. 10: 2145. https://doi.org/10.3390/plants10102145
APA StyleDu, X.-X., Park, J.-R., Wang, X.-H., Jang, Y.-H., Kim, E.-G., Lee, G.-S., & Kim, K.-M. (2021). Applying HPLC to Screening QTLs for BLB Resistance in Rice. Plants, 10(10), 2145. https://doi.org/10.3390/plants10102145







