Screening and Biocontrol Potential of Rhizobacteria Native to Gangetic Plains and Hilly Regions to Induce Systemic Resistance and Promote Plant Growth in Chilli against Bacterial Wilt Disease
Abstract
1. Introduction
2. Results
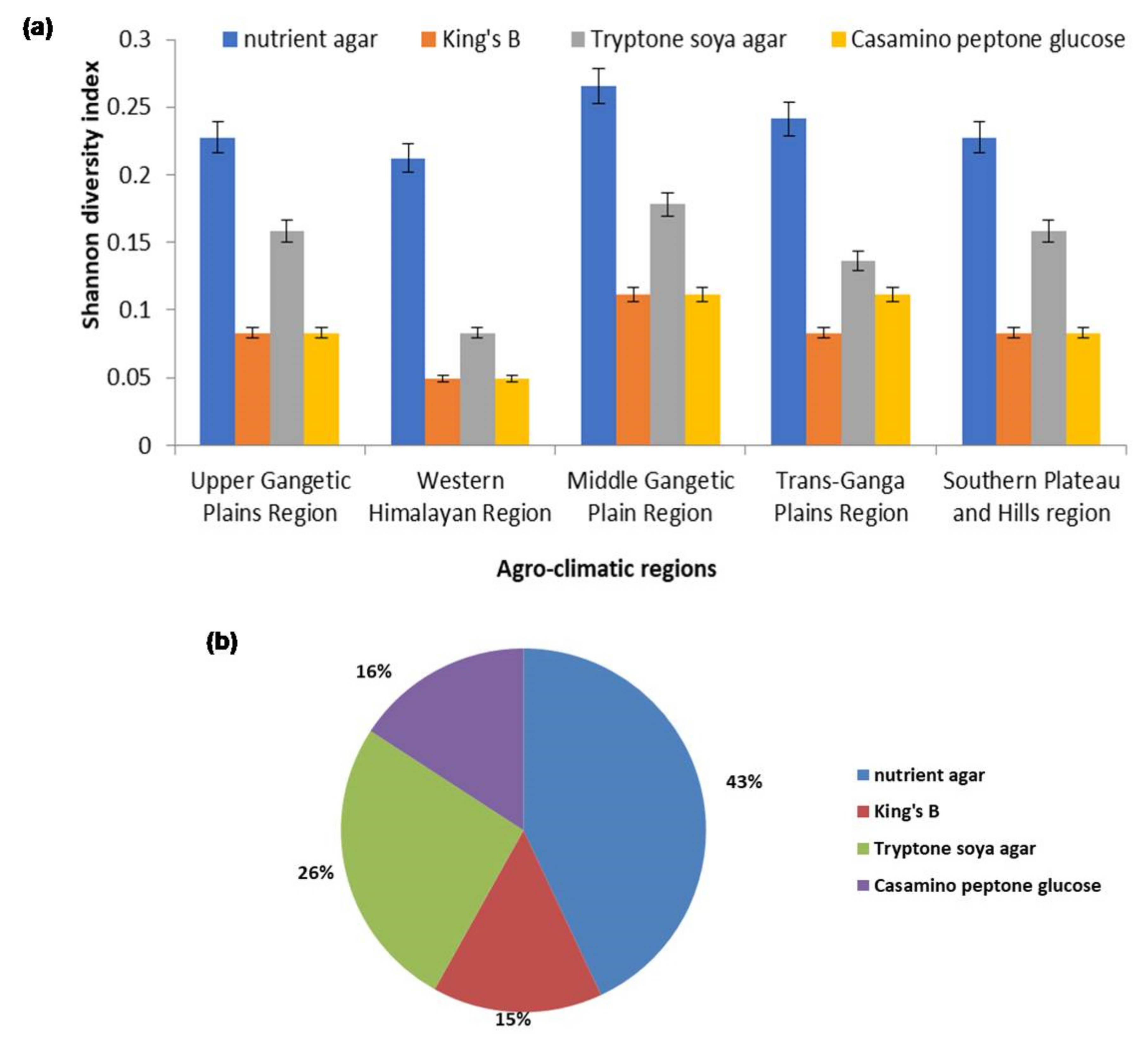
2.1. Isolation and Diversity Index of Antagonistic Rhizobacteria
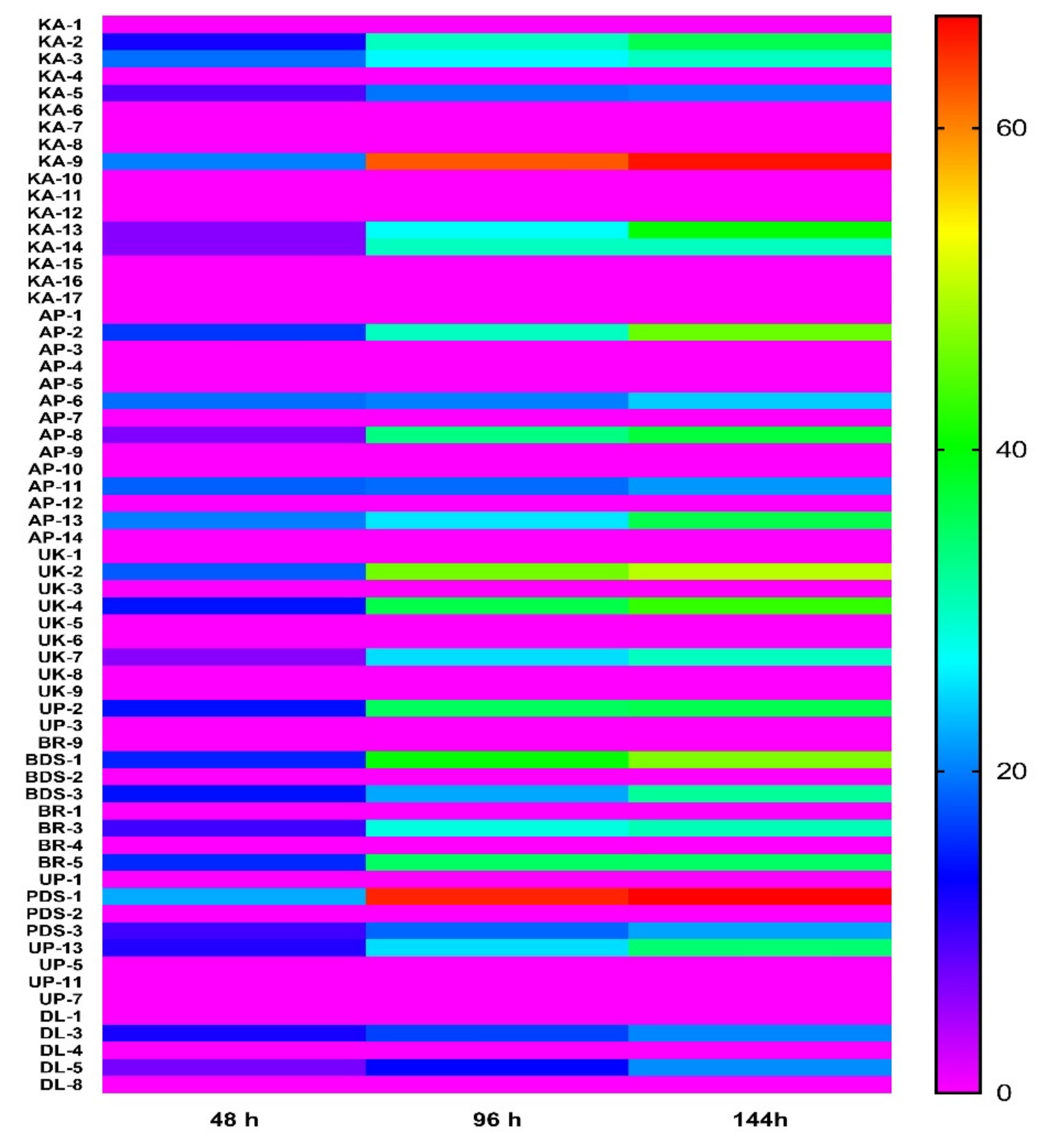
2.2. In Vitro Antagonistic Activity of Rhizobacteria against Chilli Wilt Causing Pathogen Ralstonia solanacearum
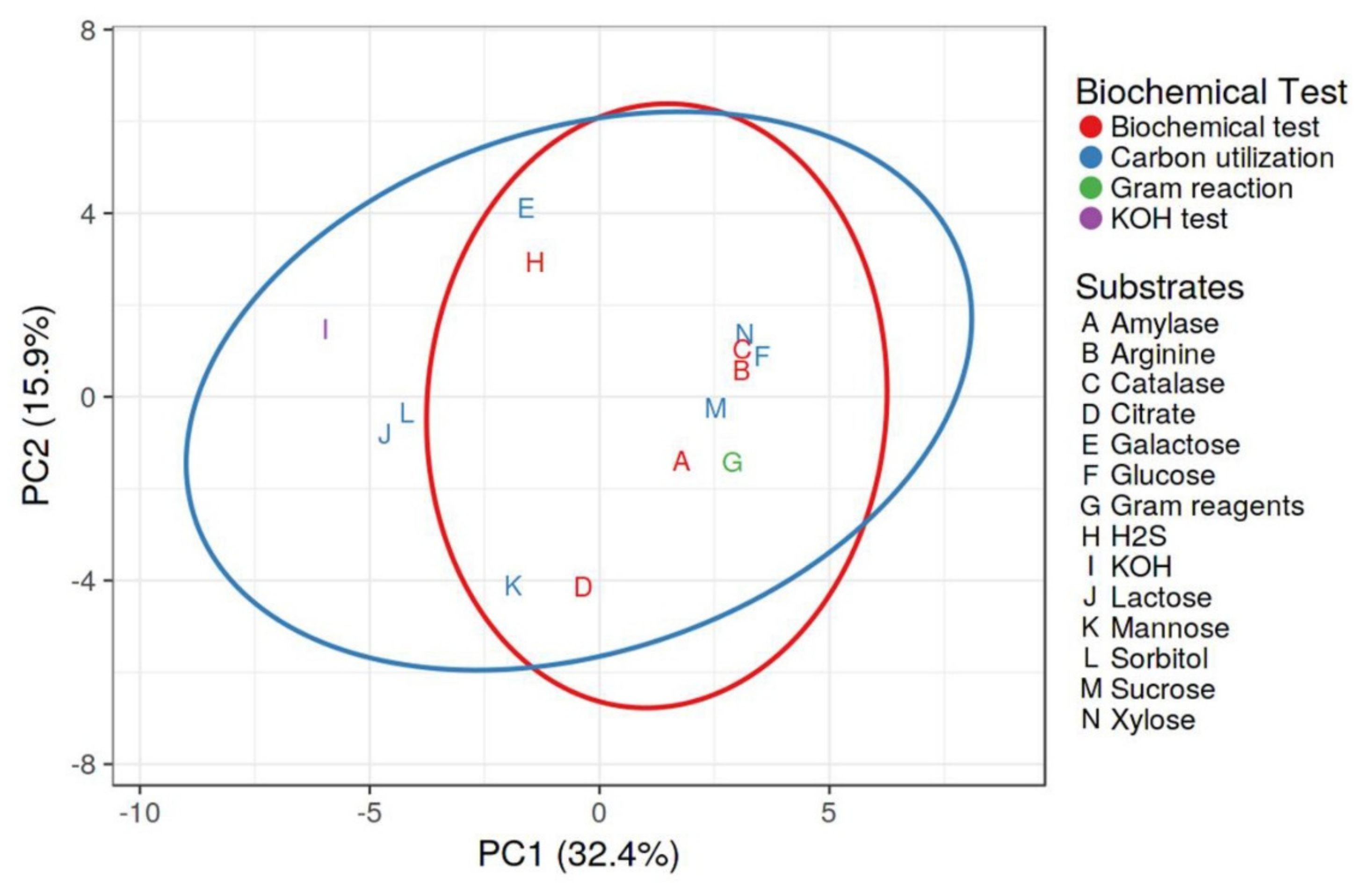
2.3. Morphological and Biochemical Characterization of Rhizobacteria
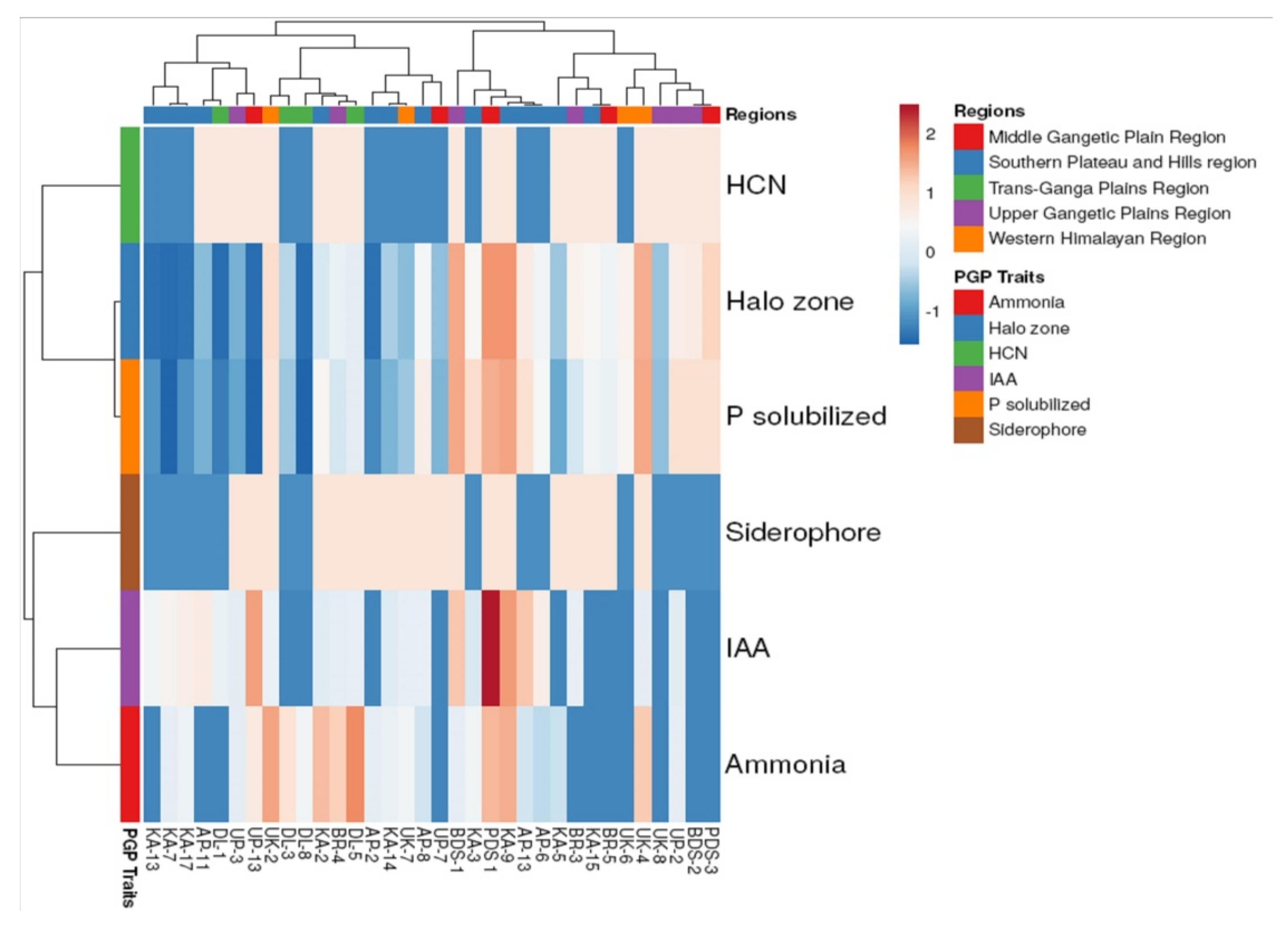
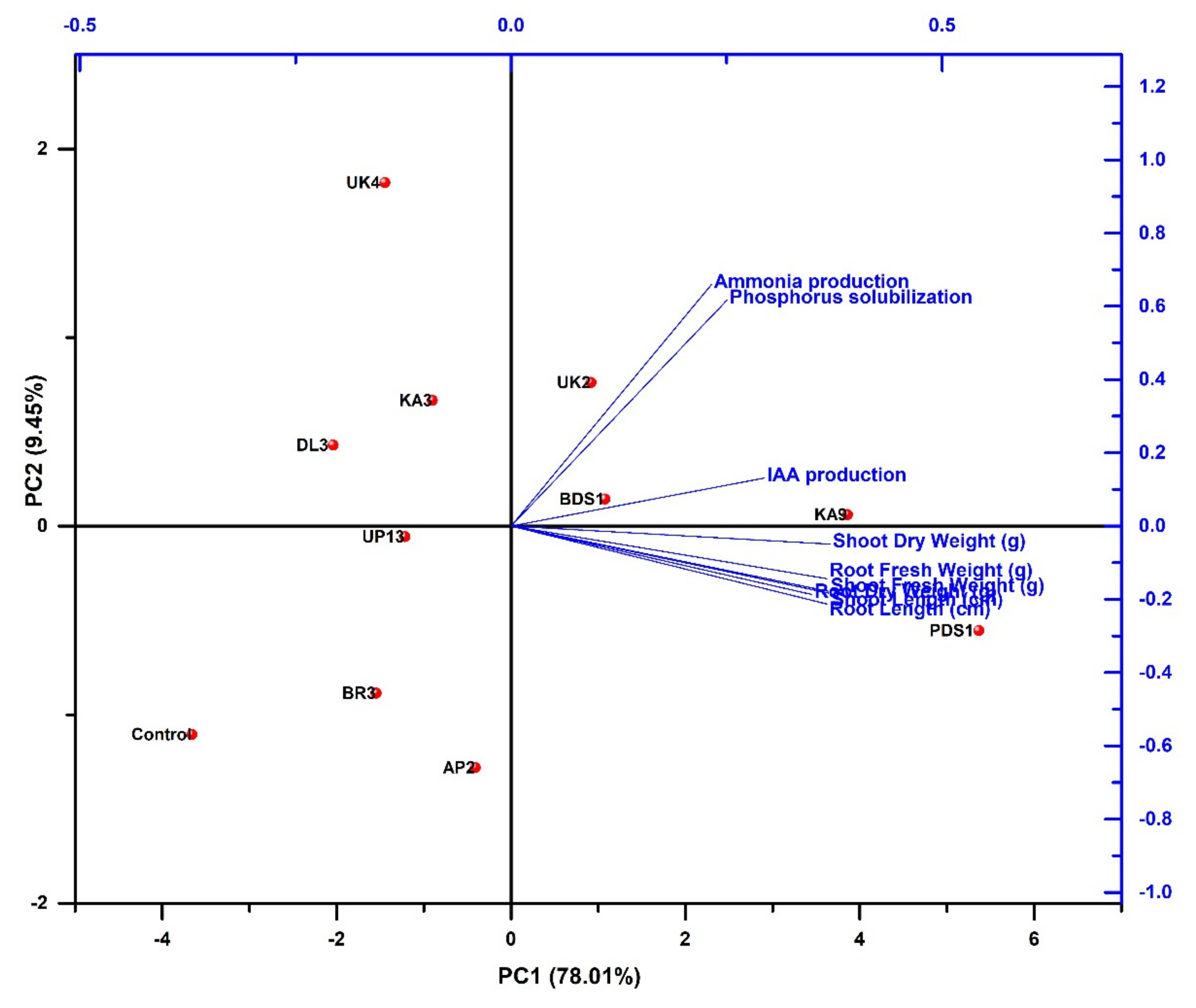
2.4. PGPR Characteristics of Rhizospheric Bacteria
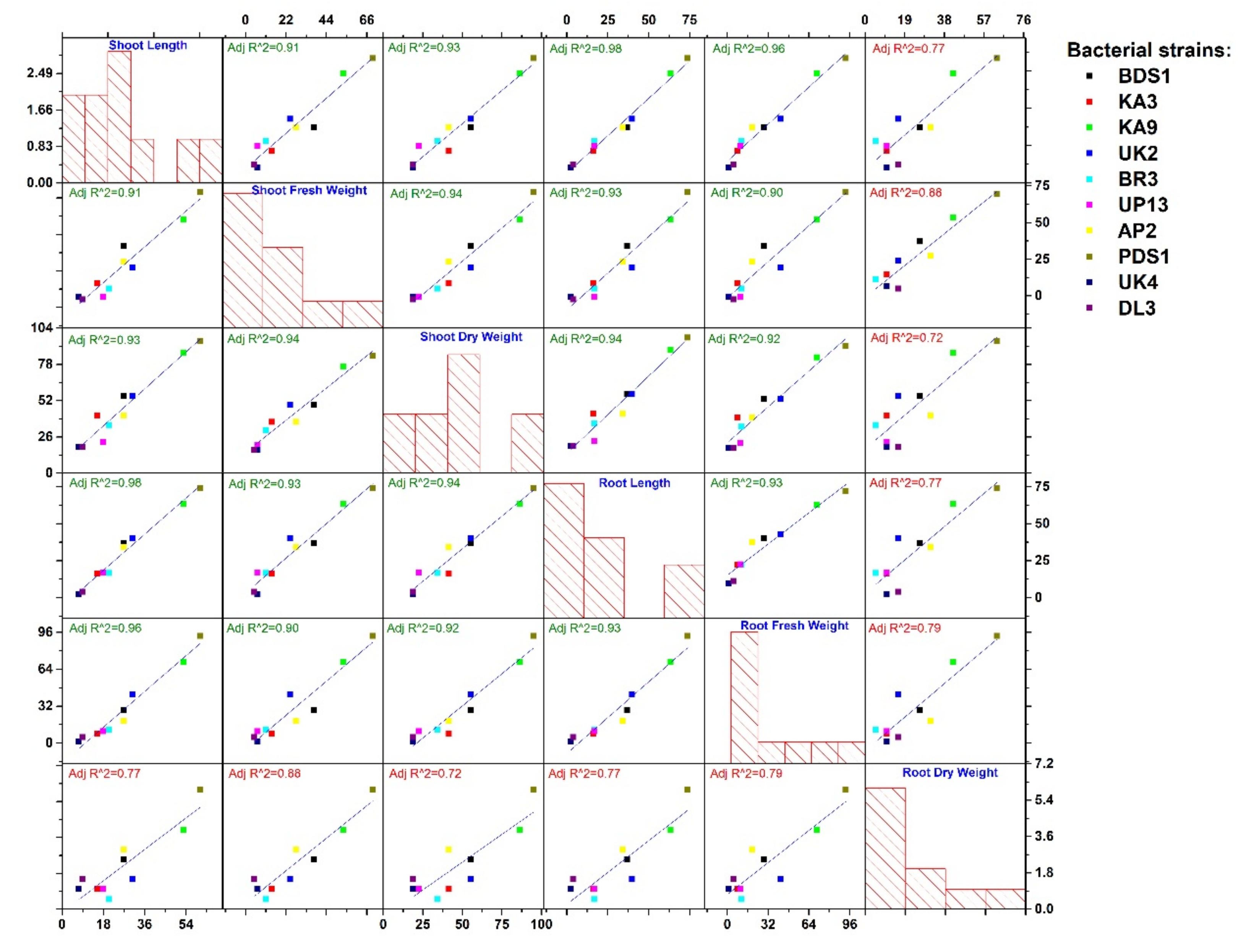
2.5. Biocontrol of Bacterial Wilt Disease and Plant Growth-Promotion Attributes
2.6. Molecular-Based Identification of Rhizobacterial Strains
2.7. Overview of Best Five Rhizobacterial Isolates Having Plant Growth Promoting (PGPs) Traits
2.8. The Effect of Rhizobacterial Strains on the Induction of Resistance in Chilli Plant against Bacterial Wilt under Glasshouse Conditions
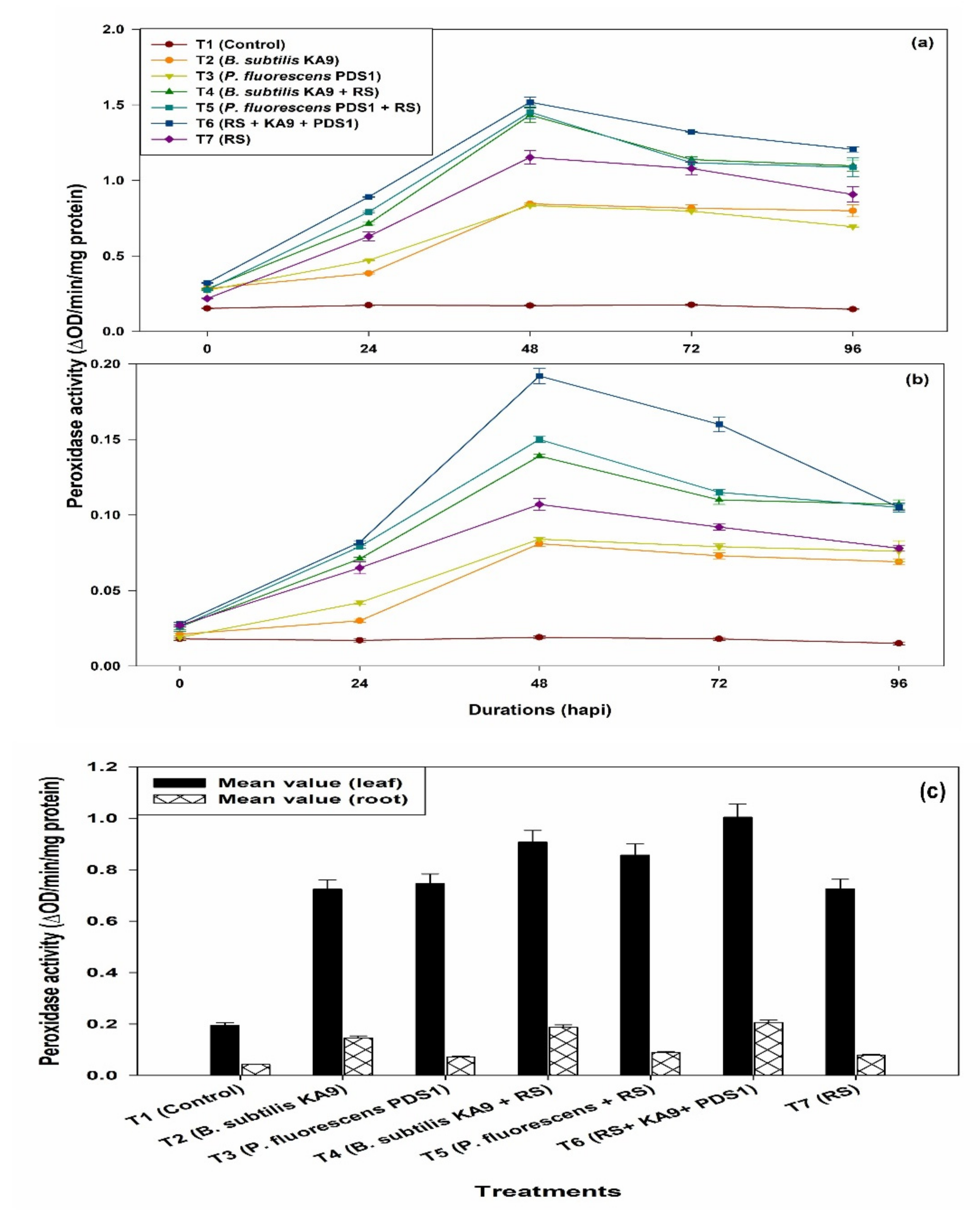
2.8.1. Peroxidase Activity (PO)
- (i)
- Leaves
- (ii)
- Roots
2.8.2. Polyphenol Oxidase Activity (PPO)
- (i)
- Leaves
- (ii)
- Roots
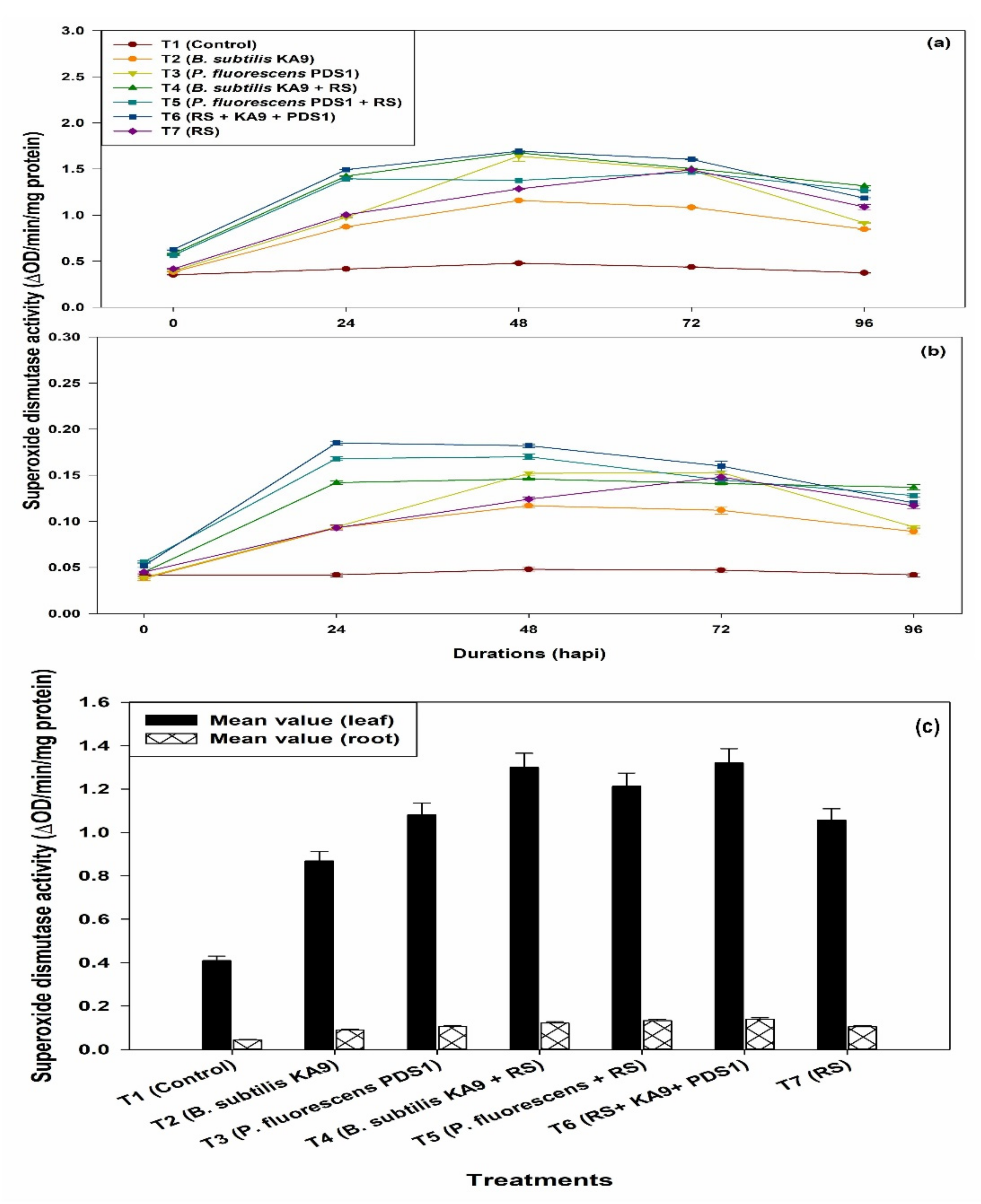
2.8.3. Superoxide Dismutase (SOD)
- (i)
- Leaves
- (ii)
- Roots
2.8.4. Phenylalanine Ammonium Lyase Activity (PAL)
- (i)
- Leaves
- (ii)
- Roots
3. Discussion
4. Materials and Methods
4.1. Isolation of Rhizobacteria from Rhizospheric Soil of Chilli Plants
4.2. Shannon Index of Microbial Diversity
4.3. Antagonistic Activity of Rhizobacteria against R. solanacearum In Vitro Conditions
4.4. Morphological and Biochemical Analysis
4.5. PGPR Characteristics of Rhizospheric Bacteria
4.5.1. Phosphorus Solubilization Assay
4.5.2. Indole Acetic Acid (IAA) Production Assay
4.5.3. Siderophore Production Assay
4.5.4. Ammonia Production Assay
4.5.5. HCN Production Assay
4.6. 16S rRNA Gene-Based Identification
Bacterial Genomic DNA Extraction and 16S rRNA PCR
4.7. Evaluation of Growth-Promoting Ability under Glasshouse Experiment
4.8. Evaluation of Biochemical Defensive Enzymes under Glasshouse Experiment
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olatunji, T.L.; Afolayan, A.J. The suitability of chili pepper (Capsicum annuum L.) for alleviating human micronutrient dietary deficiencies: A review. Food Sci. Nutr. 2018, 6, 2239–2251. [Google Scholar] [CrossRef]
- Mamphogoro, T.; Babalola, O.; Aiyegoro, O. Sustainable management strategies for bacterial wilt of sweet peppers (Capsicum annuum) and other Solanaceous crops. J. Appl. Microbiol. 2020, 129, 496–508. [Google Scholar] [CrossRef]
- Ranjan, R.; Dinesh, S.; Sharma, P.; Shri, D. Characterization and genetic diversity of Ralstonia solanacearum causing brown rot disease of potato. Ind. Phytopathol. 2015, 68, 368–374. [Google Scholar]
- Singh, S.; Singh, D.; Kumar, K.; Birah, A. Eco-friendly management modules for bacterial wilt (Ralstonia solanacearum) of tomato for protected cultivation in a tropical island ecosystem. Biol. Agric. Hort. 2014, 30, 219–227. [Google Scholar] [CrossRef]
- Kurabachew, H.; Stahl, F.; Wydra, K. Global gene expression of rhizobacteria-silicon mediated induced systemic resistance in tomato (Solanum lycopersicum) against Ralstonia solanacearum. Physiol. Mol. Plant Pathol. 2013, 84, 44–52. [Google Scholar] [CrossRef]
- Desbrosses, G.J.; Stougaard, J. Root nodulation: A paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 2011, 10, 348–358. [Google Scholar] [CrossRef]
- Santoyo, G.; Orozco-Mosqueda, M.d.C.; Govindappa, M. Mechanisms of biocontrol and plant growth-promoting activity in soil bacterial species of Bacillus and Pseudomonas: A review. Biocontrol. Sci. Technol. 2012, 22, 855–872. [Google Scholar] [CrossRef]
- Sivasakthi, S.; Usharani, G.; Saranraj, P. Biocontrol potentiality of plant growth promoting bacteria (PGPR)-Pseudomonas fluorescens and Bacillus subtilis: A review. Afr. J. Agric. Res. 2014, 9, 1265–1277. [Google Scholar]
- Borriss, R. Use of plant-associated Bacillus strains as biofertilizers and biocontrol agents in agriculture. In Bacteria in Agrobiology: Plant Growth Responses; Springer: Berlin/Heidelberg, Germany, 2011; pp. 41–76. [Google Scholar]
- Lamont, J.R.; Wilkins, O.; Bywater-Ekegärd, M.; Smith, D.L. From yogurt to yield: Potential applications of lactic acid bacteria in plant production. Soil. Biol. Biochem. 2017, 111, 1–9. [Google Scholar] [CrossRef]
- Shivlata, L.; Satyanarayana, T. Actinobacteria in agricultural and environmental sustainability. In Agro-Environmental Sustainability; Springer: Berlin/Heidelberg, Germany, 2017; pp. 173–218. [Google Scholar]
- Álvarez, B.; Biosca, E.G. Bacteriophage-based bacterial wilt biocontrol for an environmentally sustainable agriculture. Front. Plant Sci. 2017, 8, 1218. [Google Scholar] [CrossRef]
- Girard, L.; Höfte, M.; Mot, R.D. Lipopeptide families at the interface between pathogenic and beneficial Pseudomonas-plant interactions. Crit. Rev. Microbiol. 2020, 46, 397–419. [Google Scholar] [CrossRef] [PubMed]
- Nion, Y.A.; Toyota, K. Recent trends in control methods for bacterial wilt diseases caused by Ralstonia solanacearum. Microbes Environ. 2015, 30, 1–11. [Google Scholar]
- Shaikh, S.; Sayyed, R. Role of plant growth-promoting rhizobacteria and their formulation in biocontrol of plant diseases. In Plant Microbes Symbiosis: Applied Facets; Springer: Berlin/Heidelberg, Germany, 2015; pp. 337–351. [Google Scholar]
- Wang, X.; Li, Q.; Sui, J.; Zhang, J.; Liu, Z.; Du, J.; Xu, R.; Zhou, Y.; Liu, X. Isolation and characterization of antagonistic bacteria Paenibacillus jamilae HS-26 and their effects on plant growth. BioMed Res. Int. 2019, 2019, 3638926. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Goswami, D.; Dhandhukia, P.; Patel, P.; Thakker, J.N. Screening of PGPR from saline desert of Kutch: Growth promotion in Arachis hypogea by Bacillus licheniformis A2. Microbiol. Res. 2014, 169, 66–75. [Google Scholar] [CrossRef]
- Hayat, Q.; Hayat, S.; Irfan, M.; Ahmad, A. Effect of exogenous salicylic acid under changing environment: A review. Environ. Exp. Bot. 2010, 68, 14–25. [Google Scholar] [CrossRef]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci. 2003, 22, 107–149. [Google Scholar] [CrossRef]
- Zahir, Z.A.; Arshad, M. Perspectives in agriculture. Adv. Agron. 2004, 81, 97. [Google Scholar]
- Lagzian, A.; Saberi Riseh, R.; Khodaygan, P.; Sedaghati, E.; Dashti, H. Introduced Pseudomonas fluorescens VUPf5 as an important biocontrol agent for controlling Gaeumannomyces graminis var. tritici the causal agent of take-all disease in wheat. Arch. Phytopathol. Plant Prot. 2013, 46, 2104–2116. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, T.; Tan, C.; Vadas, P.; Qi, Z.; Wellen, C. Modeling phosphorus losses from soils amended with cattle manures and chemical fertilizers. Sci. Total Environ. 2018, 639, 580–587. [Google Scholar] [CrossRef]
- Khan, A.G. Role of soil microbes in the rhizospheres of plants growing on trace metal contaminated soils in phytoremediation. J. Trace Elem. Med. Biol. 2005, 18, 355–364. [Google Scholar] [CrossRef]
- Şahin, F.; Çakmakçi, R.; Kantar, F. Sugar beet and barley yields in relation to inoculation with N 2-fixing and phosphate solubilizing bacteria. Plant Soil 2004, 265, 123–129. [Google Scholar] [CrossRef]
- Ouhaibi-Ben Abdeljalil, N.; Vallance, J.; Gerbore, J.; Rey, P.; Daami-Remadi, M. Bio-suppression of Sclerotinia stem rot of tomato and biostimulation of plant growth using tomato-associated rhizobacteria. J. Plant Pathol. Microbiol. 2016, 7, 2. [Google Scholar] [CrossRef]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helmann, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci. Rep. 2018, 8, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. The enhancement of plant growth by free-living bacteria. Can. J. Microbiol. 1995, 41, 109–117. [Google Scholar] [CrossRef]
- Kashyap, A.S.; Pandey, V.K.; Manzar, N.; Kannojia, P.; Singh, U.B.; Sharma, P. Role of plant growth-promoting rhizobacteria for improving crop productivity in sustainable agriculture. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Springer: Berlin/Heidelberg, Germany, 2017; pp. 673–693. [Google Scholar]
- Lucy, M.; Reed, E.; Glick, B.R. Applications of free-living plant growth-promoting rhizobacteria. Antonie van Leeuwenhoek 2004, 86, 1–25. [Google Scholar] [CrossRef]
- Dey, R.; Pal, K.; Bhatt, D.; Chauhan, S. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004, 159, 371–394. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.P.; Pires, C.; Moreira, H.; Rangel, A.O.; Castro, P.M. Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil. Biol. Biochem. 2010, 42, 1229–1235. [Google Scholar] [CrossRef]
- Jacques, P. Surfactin and other lipopeptides from Bacillus spp. In Biosurfactants; Springer: Berlin/Heidelberg, Germany, 2011; pp. 57–91. [Google Scholar]
- Qiao, J.; Yu, X.; Liang, X.; Liu, Y.; Borriss, R.; Liu, Y. Addition of plant-growth-promoting Bacillus subtilis PTS-394 on tomato rhizosphere has no durable impact on composition of root microbiome. BMC Microbiol. 2017, 17, 1–12. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, M.; Yu, P.; Yu, S.; Wang, J.; Guo, H.; Zhang, J.; Zhou, H.; Chen, M.; Zeng, H. Prevalence, virulence feature, antibiotic resistance and MLST typing of Bacillus cereus isolated from retail aquatic products in China. Front. Microbiol. 2020, 11, 1513. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R. Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 1999, 17, 319–339. [Google Scholar] [CrossRef]
- Otieno, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant growth promotion induced by phosphate solubilizing endophytic Pseudomonas isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmström, S.J. Siderophores in environmental research: Roles and applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Neilands, J. Siderophores: Structure and function of microbial iron transport compounds. J. Biol. Chem. 1995, 270, 26723–26726. [Google Scholar] [CrossRef] [PubMed]
- Höfte, M.; Seong, K.; Jurkevitch, E.; Verstraete, W. Pyoverdin production by the plant growth beneficial Pseudomonas strain 7NSK 2: Ecological significance in soil. Plant Soil 1991, 130, 249–257. [Google Scholar] [CrossRef]
- Grobelak, A.; Napora, A.; Kacprzak, M. Using plant growth-promoting rhizobacteria (PGPR) to improve plant growth. Ecol. Eng. 2015, 84, 22–28. [Google Scholar] [CrossRef]
- Rizzi, A.; Roy, S.; Bellenger, J.-P.; Beauregard, P.B. Iron homeostasis in Bacillus subtilis requires siderophore production and biofilm formation. Appl. Environ. Microbiol. 2019, 85, e02439-18. [Google Scholar] [CrossRef]
- Iqbal, M.; Wagi, S.; Ahmed, A. Phyllospheric Bacterial Treatments Improve Growth in Helianthus annuus L. RADS J. Biol. Res. Appl. Sci. 2018, 9, 30–40. [Google Scholar] [CrossRef]
- Matsuda, R.; Handayani, M.L.; Sasaki, H.; Takechi, K.; Takano, H.; Takio, S. Production of indoleacetic acid by strains of the epiphytic bacteria Neptunomonas spp. isolated from the red alga Pyropia yezoensis and the seagrass Zostera marina. Arch. Microbiol. 2018, 200, 255–265. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–16. [Google Scholar] [CrossRef]
- Iqbal, A.; Hasnain, S. Aeromonas punctata PNS-1: A promising candidate to change the root morphogenesis of Arabidopsis thaliana in MS and sand system. Acta Physiol. Plant 2013, 35, 657–665. [Google Scholar] [CrossRef]
- Shi, C.-L.; Park, H.-B.; Lee, J.S.; Ryu, S.; Ryu, C.-M. Inhibition of primary roots and stimulation of lateral root development in Arabidopsis thaliana by the rhizobacterium Serratia marcescens 90–166 is through both auxin-dependent and-independent signaling pathways. Mol. Cells 2010, 29, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014, 201, 850–861.51. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Hasnain, S. Auxins as one of the factors of plant growth improvement by plant growth promoting rhizobacteria. Pol. J. Microbiol. 2014, 63, 261. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- López-Bucio, J.; Hernández-Abreu, E.; Sánchez-Calderón, L.; Nieto-Jacobo, M.F.; Simpson, J.; Herrera-Estrella, L. Phosphate availability alters architecture and causes changes in hormone sensitivity in the Arabidopsis root system. Plant Physiol. 2002, 129, 244–256. [Google Scholar] [CrossRef]
- Tsukanova, K.; Meyer, J.; Bibikova, T. Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 2017, 113, 91–102. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef]
- Llorente, B.E.; Alasia, M.A.; Larraburu, E.E. Biofertilization with Azospirillum brasilense improves in vitro culture of Handroanthus ochraceus, a forestry, ornamental and medicinal plant. New Biotechnol. 2016, 33, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Glick, B.R.; Pasternak, J. Plant-microbial interaction under gnotobiotic conditions: A scanning electron microscope study. Curr. Microbiol. 1991, 23, 111–114. [Google Scholar] [CrossRef]
- Ruzzi, M.; Aroca, R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015, 196, 124–134. [Google Scholar] [CrossRef]
- Contesto, C.; Milesi, S.; Mantelin, S.; Zancarini, A.; Desbrosses, G.; Varoquaux, F.; Bellini, C.; Kowalczyk, M.; Touraine, B. The auxin-signaling pathway is required for the lateral root response of Arabidopsis to the rhizobacterium Phyllobacterium brassicacearum. Planta 2010, 232, 1455–1470. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.-S.; Krishnamachari, V.; Payton, P.; Sun, Y.; Grimson, M.; Farag, M.A.; Ryu, C.-M.; Allen, R.; Melo, I.S. Rhizobacterial volatile emissions regulate auxin homeostasis and cell expansion in Arabidopsis. Planta 2007, 226, 839–851. [Google Scholar] [CrossRef]
- Fahad, S.; Hussain, S.; Matloob, A.; Khan, F.A.; Khaliq, A.; Saud, S.; Hassan, S.; Shan, D.; Khan, F.; Ullah, N. Phytohormones and plant responses to salinity stress: A review. Plant Growth Regul. 2015, 75, 391–404. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Iqrar, I. Selective isolation and characterization of agriculturally beneficial endophytic bacteria from wild hemp using canola. Pak. J. Bot. 2015, 47, 1999–2008. [Google Scholar]
- Ahemad, M.; Kibret, M. Mechanisms and applications of plant growth promoting rhizobacteria: Current perspective. J. King Saud. Univ. Sci. 2014, 26, 1–20. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Shaukat, S.S.; Sheikh, I.H.; Khan, A. Role of cyanide production by Pseudomonas fluorescens CHA0 in the suppression of root-knot nematode, Meloidogyne javanica in tomato. World J. Microbiol. Biotechnol. 2006, 22, 641–650. [Google Scholar] [CrossRef]
- Lanteigne, C.; Gadkar, V.J.; Wallon, T.; Novinscak, A.; Filion, M. Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology 2012, 102, 967–973. [Google Scholar] [CrossRef]
- Voisard, C.; Keel, C.; Haas, D.; Dèfago, G. Cyanide production by Pseudomonas fluorescens helps suppress black root rot of tobacco under gnotobiotic conditions. EMBO J. 1989, 8, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Prakash, A.; Johri, B. Induced systemic resistance (ISR) in plants: Mechanism of action. Ind. J. Microbiol. 2007, 47, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, R.; Karaoz, U.; Volegova, M.; MacKichan, J.; Kato-Maeda, M.; Miller, S.; Nadarajan, R.; Brodie, E.L.; Lynch, S.V. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS ONE 2015, 10, e0117617. [Google Scholar] [CrossRef]
- Patel, J.B. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. J. Mol. Diagn. 2001, 6, 313–321. [Google Scholar] [CrossRef]
- Chahboune, R.; Barrijal, S.; Moreno, S.; Bedmar, E.J. Characterization of Bradyrhizobium species isolated from root nodules of Cytisus villosus grown in Morocco. Syst. Appl. Microbiol. 2011, 34, 440–445. [Google Scholar] [CrossRef]
- Singh, D.; Sinha, S.; Chaudhary, G.; Yadav, D.; Mondal, K. Genetic diversity of biovar 3 and 4 of Ralstonia solanacearum causing bacterial wilt of tomato using BOX-PCR, RAPD and hrp gene sequences. Indian J. Agric. Sci. 2014, 84, 391–395. [Google Scholar]
- Deepa, C.; Dastager, S.G.; Pandey, A. Isolation and characterization of plant growth promoting bacteria from non-rhizospheric soil and their effect on cowpea (Vigna unguiculata (L.) Walp.) seedling growth. World J. Microbiol. Biotechnol. 2010, 26, 1233–1240. [Google Scholar] [CrossRef]
- TA, P.D.; Sahoo, D.; Setti, A.; Sharma, C.; Kalita, M. Bacterial rhizosphere community profile at different growth stages of Umorok (Capsicum chinense) and its response to the root exudates. Int. Microbiol. 2019, 23, 241–251. [Google Scholar] [CrossRef]
- Hyder, S.; Gondal, A.S.; Rizvi, Z.F.; Ahmad, R.; Alam, M.M.; Hannan, A.; Ahmed, W.; Fatima, N.; Inam-ul-Haq, M. Characterization of native plant growth promoting rhizobacteria and their anti-oomycete potential against Phytophthora capsici affecting chilli pepper (Capsicum annum L.). Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Pii, Y.; Mimmo, T.; Tomasi, N.; Terzano, R.; Cesco, S.; Crecchio, C. Microbial interactions in the rhizosphere: Beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fert. Soils 2015, 51, 403–415. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. [Google Scholar] [CrossRef]
- Tan, S.; Dong, Y.; Liao, H.; Huang, J.; Song, S.; Xu, Y.; Shen, Q. Antagonistic bacterium Bacillus amyloliquefaciens induces resistance and controls the bacterial wilt of tomato. Pest. Manag. Sci. 2013, 69, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Haegeman, B.; Hamelin, J.; Moriarty, J.; Neal, P.; Dushoff, J.; Weitz, J.S. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013, 7, 1092–1101. [Google Scholar] [CrossRef] [PubMed]
- Dubey, G.; Kollah, B.; Gour, V.K.; Shukla, A.K.; Mohanty, S.R. Diversity of bacteria and archaea in the rhizosphere of bioenergy crop Jatropha curcas. 3 Biotech 2016, 6, 1–10. [Google Scholar] [CrossRef]
- Praeg, N.; Pauli, H.; Illmer, P. Microbial diversity in bulk and rhizosphere soil of Ranunculus glacialis along a high-alpine altitudinal gradient. Front. Microbiol. 2019, 10, 1429. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Li, J.; Xiao, Y.; Gu, Y.; Liu, H.; Liang, Y.; Liu, X.; Hu, J.; Meng, D.; Yin, H. An integrated insight into the relationship between soil microbial community and tobacco bacterial wilt disease. Front. Microbiol. 2017, 8, 2179. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Song, C.; Li, Z.; Kuipers, O.P. Antimicrobial activity screening of rhizosphere soil bacteria from tomato and genome-based analysis of their antimicrobial biosynthetic potential. BMC Genom. 2021, 22, 1–14. [Google Scholar] [CrossRef]
- Ramesh, R.; Phadke, G.S. Rhizosphere and endophytic bacteria for the suppression of eggplant wilt caused by Ralstonia solanacearum. Crop. Prot. 2012, 37, 35–41. [Google Scholar] [CrossRef]
- Beneduzi, A.; Ambrosini, A.; Passaglia, L.M. Plant growth-promoting rhizobacteria (PGPR): Their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 2012, 35, 1044–1051. [Google Scholar] [CrossRef]
- Singh, T.B.; Sahai, V.; Goyal, D.; Prasad, M.; Yadav, A.; Shrivastav, P.; Ali, A.; Dantu, P.K. Identification, characterization and evaluation of multifaceted traits of plant growth promoting rhizobacteria from soil for sustainable approach to agriculture. Curr. Microbiol. 2020, 77, 3633–3642. [Google Scholar] [CrossRef]
- De Meyer, G.; Capieau, K.; Audenaert, K.; Buchala, A.; Metraux, J.-P.; Hofte, M. Nanogram amounts of salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 activate the systemic acquired resistance pathway in bean. Mol. Plant-Microbe Interact. 1999, 12, 450–458. [Google Scholar] [CrossRef]
- Arora, N.K. Plant Microbe Symbiosis: Fundamentals and Advances; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Islam, S.; Akanda, A.M.; Prova, A.; Islam, M.T.; Hossain, M.M. Isolation and identification of plant growth promoting rhizobacteria from cucumber rhizosphere and their effect on plant growth promotion and disease suppression. Front. Microbiol. 2016, 6, 1360. [Google Scholar] [CrossRef] [PubMed]
- Backer, R.; Rokem, J.S.; Ilangumaran, G.; Lamont, J.; Praslickova, D.; Ricci, E.; Subramanian, S.; Smith, D.L. Plant growth-promoting rhizobacteria: Context, mechanisms of action and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 2018, 9, 1473. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Fahad, S.; Subhan, D.; Adnan, M.; Zhen, Y.; Saud, S.; Manzer, H.; Martin, B.; Tereza, H.; Rahul, D. Sustainable management with mycorrhizae and phosphate solubilizing bacteria for enhanced phosphorus uptake in calcareous soils. Agriculture 2020, 10, 334. [Google Scholar] [CrossRef]
- Jeyanthi, V.; Ganesh, P. Production, optimization and characterization of phytohormone indole acetic acid by Pseudomonas fluorescence. Int. J. Pharm. Biol. Arch. 2013, 4, 514–520. [Google Scholar]
- Singh, D.; Yadav, D.; Sinha, S.; Upadhyay, B. Utilization of plant growth promoting Bacillus subtilis isolates for the management of bacterial wilt incidence in tomato caused by Ralstonia solanacearum race 1 biovar 3. Ind. Phytopath. 2012, 65, 18–24. [Google Scholar]
- Wagi, S.; Ahmed, A. Bacillus spp.: Potent microfactories of bacterial IAA. PeerJ 2019, 7, e7258. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef]
- Benali, S.; Mohamed, B.; Eddine, H.J. Virulence strategies of phytopathogenic bacteria and their role in plant disease pathogenesis. Afr. J. Microbiol. Res. 2014, 8, 2809–2815. [Google Scholar]
- Wang, Y.-Y.; Jing, X.-R.; Li, L.-L.; Liu, W.-J.; Tong, Z.-H.; Jiang, H. Biotoxicity evaluations of three typical biochars using a simulated system of fast pyrolytic biochar extracts on organisms of three kingdoms. ACS Sustain. Chem. Eng. 2017, 5, 481–488. [Google Scholar] [CrossRef]
- Glick, B.R. Modulating phytohormone levels. In Beneficial Plant-Bacterial Interactions; Springer: Berlin/Heidelberg, Germany, 2020; pp. 139–180. [Google Scholar]
- Ahmad, F.; Ahmad, I.; Khan, M. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008, 163, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Abbasi, M.K.; Hameed, S.; Imran, A.; Rahim, N. Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 2015, 6, 198. [Google Scholar] [CrossRef] [PubMed]
- Alam, S.I.; Bansod, S.; Goel, A.K.; Singh, L. Characterization of an environmental strain of Bacillus thuringiensis from a hot spring in western Himalayas. Curr. Microbiol. 2011, 62, 547–556. [Google Scholar] [CrossRef] [PubMed]
- Imran, A.; Hafeez, F.Y.; Frühling, A.; Schumann, P.; Malik, K.; Stackebrandt, E. Ochrobactrum ciceri sp. nov., isolated from nodules of Cicer arietinum. Int. J. Syst. Evol. Microbiol. 2010, 60, 1548–1553. [Google Scholar] [CrossRef][Green Version]
- Pliego, C.; Ramos, C.; de Vicente, A.; Cazorla, F.M. Screening for candidate bacterial biocontrol agents against soilborne fungal plant pathogens. Plant Soil 2011, 340, 505–520. [Google Scholar] [CrossRef]
- Abo-Elyousr, K.A.; Bagy, H.M.K.; Hashem, M.; Alamri, S.A.; Mostafa, Y.S. Biological control of the tomato wilt caused by Clavibacter michiganensis subsp. michiganensis using formulated plant growth-promoting bacteria. Egypt J. Biol. Pest. Control 2019, 29, 1–8. [Google Scholar] [CrossRef]
- Mekonnen, H.; Kibret, M. The roles of plant growth promoting rhizobacteria in sustainable vegetable production in Ethiopia. Chem. Biol. Technol. Agric. 2021, 8, 1–11. [Google Scholar] [CrossRef]
- Seleim, M.; Saead, F.; Abd-El-Moneem, K.; Abo-Elyousr, K. Biological control of bacterial wilt of tomato by plant growth promoting rhizobacteria. Plant Pathol. J. 2011, 10, 146–153. [Google Scholar] [CrossRef]
- Prameela, T.P.; Rajamma, S.B. Bacterial wilt of ginger (Zingiber officinale Rosc.) incited by Ralstonia pseudosolanacearum—A review based on pathogen diversity, diagnostics and management. J. Plant Pathol. 2020, 102, 709–719. [Google Scholar] [CrossRef]
- Jayapala, N.; Mallikarjunaiah, N.; Puttaswamy, H. Rhizobacteria Bacillus spp. induce resistance against anthracnose disease in chili (Capsicum annuum L.) through activating host defense response. Egypt. J. Biol. Pest Cont. 2019, 29, 45. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef]
- Chunyu, L.; Weicong, H.; Bin, P.; Yan, L.; Saifei, Y.; Yuanyuan, D.; Rong, L.; Xinyan, Z.; Biao, S.; Qirong, S. Rhizobacterium Bacillus amyloliquefaciens Strain SQRT3-Mediated Induced Systemic Resistance Controls Bacterial Wilt of Tomato. Pedosphere 2017, 27, 1135–1146. [Google Scholar]
- Ramamoorthy, V.; Raguchander, T.; Samiyappan, R. Enhancing resistance of tomato and hot pepper to Pythium disease by seed treatment with fluorescent pseudomonads. Eur. J. Plant Pathol. 2002, 108, 429–441. [Google Scholar] [CrossRef]
- Silva, H.S.A.; Romerio, R.D.S.; Macagnan, D.; Halfeld-Vieira, B.D.A.; Pereira, M.C.B.; Mounteer, A. Rhizobacterial induction of systemic resistance in tomato plants: Non-specific protection and increase in enzyme activities. Biol. Control. 2004, 29, 288–295. [Google Scholar] [CrossRef]
- Chandrasekaran, M.; Chun, S.C. Expression of PR-protein genes and induction of defense-related enzymes by Bacillus subtilis CBR05 in tomato (Solanum lycopersicum) plants challenged with Erwinia carotovora subsp. carotovora. Biosci. Biotechnol. Biochem. 2016, 80, 2277–2283. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, K.; Bowen, J.; Allan, A.; Espley, R.; Karunairetnam, S.; Ferguson, I. Differential expression within the LOX gene family in ripening kiwifruit. J. Exp. Bot. 2006, 57, 3825–3836. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Lai, Z.; Lin, Y.; Lai, G.; Lian, C. Genome-wide identification and characterization of the superoxide dismutase gene family in Musa acuminata cv. Tianbaojiao (AAA group). BMC Genom. 2015, 16, 823. [Google Scholar] [CrossRef] [PubMed]
- Vanitha, S.C.; Niranjana, S.R.; Mortensen, C.N.; Umesha, S. Bacterial wilt of tomato in Karnataka and its management by Pseudomonas fluorescens. Biocontrol 2009, 54, 685–695. [Google Scholar] [CrossRef]
- Singh, D.; Yadav, D.K.; Chaudhary, G.; Rana, V.S.; Sharma, R.K. Potential of Bacillus amyloliquefaciens for biocontrol of bacterial wilt of tomato incited by Ralstonia solanacearum. J. Plant Pathol. Microbiol. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Schaad, N.I. Initial identification of common genera. In Laboratory Guide for Identification of Plant Pathogenic Bacteria; APS Press: St. Paul, MN, USA, 2001. [Google Scholar]
- Pikovskaya, R. Mobilization of phosphorus in soil in connection with the vital activity of some microbial species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Mehta, S.; Nautiyal, C.S. An efficient method for qualitative screening of phosphate-solubilizing bacteria. Curr. Microbiol. 2001, 43, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Vikram, A.; Hamzehzarghani, H.; Alagawadi, A.; Krishnaraj, P.; Chandrashekar, B. Production of plant growth promoting substances by phosphate solubilizing bacteria isolated from vertisols. J. Plant Sci. 2007, 2, 326–333. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Bakker, A.W.; Schippers, B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp.-mediated plant growth-stimulation. Soil. Biol. Biochem. 1987, 19, 451–457. [Google Scholar] [CrossRef]
- Murray, M.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4326. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Mwaniki, P.; Birech, R.; Wagara, I.; Kinyua, Z.; Freyer, B. Distribution, Prevalence and Incidence of Potato Bacterial Wilt in Nakuru County, KENYA. Int. J. Innov. Res. Dev. 2016, 5, 435–442. [Google Scholar]
- Beauchamp, C.; Fridovich, I. Superoxide dismutase: Improved assays and an assay applicable to acryl amide gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef]
- Mayer, A.M.; Harel, E. Polyphenol oxidases in plants. Phytochemistry 1979, 18, 193–215. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Kuc, J. Enhanced peroxidase activity and lignification in the induced systemic resistance of cucumber. Phytopathology 1980, 70, 689. [Google Scholar]
- Amrhein, N.; Gödeke, K.H.; Gerhardt, J. The estimation of phenylalanine ammonia-lyase (PAL)-activity in intact cells of higher plant tissue. Planta 1976, 131, 33–40. [Google Scholar] [CrossRef] [PubMed]





| Agro-Climatic Region | Place of Collection | Geographical Coordinates | Location | Source of Isolation | Host | Annual Pluviometry (mm) | Cultivars of Chilli | Year of Collection | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Latitude | Longitude | |||||||||
| Upper Gangetic Plains Region | Saharanpur (Uttar Pradesh) | 29.57° N | 77.34° E | India | Rhizospheric soil | Chilli | 21 | 750–1500 | Jawala Pant C-1, Pahadi, Kalyanpur | 2014–15 |
| Kannauj (Uttar Pradesh) | 27.04° N | 79.91° E | India | Rhizospheric soil | Chilli | |||||
| Loni (Uttar Pradesh) | 28.72° N | 77.30° E | India | Rhizospheric soil | Chilli | |||||
| Pisawa (Uttar Pradesh) | 28.11° N | 77.76° E | India | Rhizospheric soil | Chilli | |||||
| Udham Nagar (Uttarakhand) | 28.95° N | 79.48° E | India | Rhizospheric soil | Chilli | |||||
| Western Himalayan Region | Golapar (Uttarakhand) | 29.17° N | 79.59° E | India | Rhizospheric soil | Chilli | 16 | 750–1500 | PusaSadabahar, Pusa Jwala and Pant C-1 | 2018–19 |
| Chargaliya (Uttarakhand) | 29.12° N | 79.70° E | India | Rhizospheric soil | Chilli | |||||
| Chaffi (Hill region of Uttarakhand,) | 29.39° N | 79.55° E | India | Rhizospheric soil | Chilli | |||||
| Middle Gangetic Plain Region1 | Mau (Eastern Uttar Pradesh) | 25.89° N | 83.48° E | India | Rhizospheric soil | Chilli | 25 | 1000–2000 | MotiMirchi, Chittee, NP-46, Unknown | 2014–15 |
| Varanasi (Eastern Uttar Pradesh) | 25.44° N | 82.85° E | India | Rhizospheric soil | Chilli | |||||
| Bhojpur (Bihar) | 25.58° N | 84.12° E | India | Rhizospheric soil | Chilli | |||||
| Buxar (Bihar) | 25.55° N | 83.95° E | India | Rhizospheric soil | Chilli | |||||
| Trans-Ganga Plains Region | Delhi | 28.64° N | 77.16° E | India | Rhizospheric soil | Chilli | 5 | 650–1250 | Pusa Jwala | 2018–19 |
| Haryana | 29.96° N | 76.89° E | India | Rhizospheric soil | Chilli | |||||
| Southern Plateau and Hills region | Warangal (Telangana) | 17.93° N | 79.57° E | India | Rhizospheric soil | Chilli | 23 | 500–1000 | Maduru, Karakulu, Sannalu, Jawala, Bayadgi | 2014–15 |
| Guntur (Andhra Pradesh) | 16.32° N | 80.45° E | India | Rhizospheric soil | Chilli | |||||
| Raichur (Karnataka) | 16.22° N | 77.36° E | India | Rhizospheric soil | Chilli | |||||
| Chikkamagaluru, (Karnataka) | 13.63° N | 75.73° E | India | Rhizospheric soil | Chilli | |||||
| Dharwad (Karnataka) | 15.43° N | 74.98° E | India | Rhizospheric soil | Chilli | |||||
| Agroclimatic Zone Region | Strains | Phosphate Solubilization | Indole-3-Acetic Acid | Siderophore | Ammonia | HCN | Root Colonization | |
|---|---|---|---|---|---|---|---|---|
| HD (mm) | mg/mL | µg/mL | Q | (mol/mL) | Q | |||
| Southern Plateau and Hills region | KA2 | 5.73 ± 0.32 gh | 3.89 ± 0.12 cd | 2.53 ± 0.15 fg | + | 4.59 ± 0.16 ab | + | + |
| KA3 | 8.13 ± 0.24 cde | 4.89 ± 0.11 b | 2.32 ± 0.12 g | − | 2.82 ± 0.25 e | − | + | |
| KA5 | 3.89 ± 0.37 ijk | 1.35 ± 0.03 ghijk | 0.000 | + | 1.70 ± 0.15 f | + | + | |
| KA7 | 0.000 | 0.000 | 3.52 ± 0.22 de | − | 2.44 ± 0.22 e | − | + | |
| KA9 | 13.40 ± 0.61 a | 5.92 ± 0.07 a | 5.65 ± 0.19 b | + | 4.80 ± 0.09 bc | + | + | |
| KA13 | 0.08 ± 0.01 | 0.93 ± 0.02 ijkl | 3.09 ± 0.10 ef | − | 0.000 | − | + | |
| KA14 | 4.25 ± 0.30 ij | 1.54 ± 0.047 ghij | 2.49 ± 0.18 fg | + | 2.58 ± 0.20 e | − | + | |
| KA15 | 8.25 ± 0.21 cde | 3.63 ± 0.26 cd | 0.000 | + | 0.000 | + | + | |
| KA17 | 0.05 ± 0.02 | 0.87 ± 0.03 ijkl | 3.72 ± 0.16 de | − | 2.67 ± 0.17 e | − | + | |
| AP8 | 7.83 ± 0.32 def | 4.19 ± 0.23 bc | 2.81 ± 0.20 fg | + | 1.84 ± 0.08 f | − | + | |
| AP13 | 9.31 ± 0.32 c | 4.74 ± 0.27 b | 4.89 ± 0.17 c | − | 1.85 ± 0.04 f | + | ||
| AP2 | 0.040 ± 0.017 | 0.70 ± 0.10 ijkl | 0.000 | + | 2.45 ± 0.13 e | − | + | |
| AP6 | 7.760 ± 0.378 def | 3.75 ± 0.32 cd | 3.72 ± 0.14 de | − | 1.57 ± 0.16 f | − | + | |
| AP11 | 3.403 ± 0.265 jk | 1.51 ± 0.03 ghij | 3.77 ± 0.28 d | − | 0.000 | + | + | |
| Western Himalayan Region | UK2 | 10.64 ± 0.58 b | 4.64 ± 0.24 b | 2.83 ± 0.20 fg | + | 4.98 ± 0.08 ab | + | + |
| UK6 | 8.73 ± 0.31 cd | 3.93 ± 0.30 cd | 0.000 | − | 0.000 | − | + | |
| UK4 | 12.53 ± 0.66 a | 5.78 ± 0.24 a | 2.65 ± 0.35 fg | + | 4.37 ± 0.16 c | + | + | |
| UK7 | 3.34 ± 0.52 ijk | 1.92 ± 0.13 gh | 2.76 ± 0.22 fg | + | 2.78 ± 0.19 e | − | + | |
| Upper Gangetic Plains Region | UK8 | 3.77 ± 0.24 ijk | 1.78 ± 0.06 gh | 0.000 | − | 0.000 | + | + |
| UP2 | 9.10 ± 0.06 c | 4.73 ± 0.26 b | 2.52 ± 0.22 fg | − | 2.46 ± 0.18 e | + | + | |
| BDS1 | 12.74 ± 0.39 a | 5.84 ± 0.26 a | 4.92 ± 0.13 c | + | 2.42 ± 0.16 e | + | + | |
| UP3 | 2.87 ± 0.36 | 1.24 ± 0.11 hijk | 2.63 ± 0.19 fg | + | 2.4 ± 0.14 e | + | + | |
| BDS2 | 9.30 ± 0.32 c | 4.76 ± 0.23 b | 0.000 | − | 0.000 | + | + | |
| BR3 | 8.81 ± 0.46 cd | 2.69 ± 0.22 f | 2.75 ± 0.28 fg | + | 0.000 | + | + | |
| BR4 | 7.22 ± 0.34 ef | 2.72 ± 0.32 f | 2.673 ± 0.23 fg | + | 4.33 ± 0.12 c | + | + | |
| Middle Gangetic Plain Region | BR5 | 7.61 ± 0.69 def | 3.40 ± 0.42 de | 0.000 | + | 0.000 | + | + |
| PDS1 | 13.39 ± 0.50 a | 5.77 ± 0.28 a | 7.26 ± 0.32 a | ++ | 4.70 ± 0.27 ab | + | + | |
| UP13 | 0.000 | 0.000 | 5.55 ± 0.28 b | + | 3.57 ± 0.19 d | + | + | |
| PDS3 | 11.20 ± 0.67 b | 4.74 ± 0.23 b | 0.000 | − | 0.000 | + | + | |
| UP7 | 3.49 ± 0.35 jk | 1.59 ± 0.25 ghi | 0.000 | + | 0.000 | − | + | |
| Trans-Ganga Plains Region | DL3 | 4.67 ± 0.50 hi | 1.97 ± 0.13 g | 0.000 | − | 3.81 ± 0.21 d | − | + |
| DL5 | 6.67 ± 0.32 fg | 3.157 ± 0.386 ef | 2.747 ± 0.226 fg | + | 5.33 ± 0.21 a | + | + | |
| DL1 | 0.01 ± 0.01 | 0.523 ± 0.187 l | 2.840 ± 0.149 fg | − | 0.000 | + | + | |
| DL8 | 0.000 | 0.000 | 0.000 | − | 2.84 ± 0.07 e | − | + | |
| Strains Treatment/Growth Trait | Wilt Disease Incidence (%) | Biocontrol Efficacy (%) | Shoot Length (cm) | GPE (%) | Shoot Fresh Weight (g) | GPE (%) | Shoot Dry Weight (g) | GPE (%) | Root Length (cm) | GPE (%) | Root Fresh Weight (g) | GPE (%) | Root Dry Weight (g) | GPE (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BDS1 | 29.70 i | 58.75 d | 15.60 d | 26.82 | 8.50 c | 37.09 | 0.90 c | 55.17 | 9.61 c | 36.89 | 1.26 d | 28.57 | 0.24 d | 26.31 |
| KA3 | 38.20 f | 46.94 f | 14.20 g | 15.44 | 7.10 f | 14.51 | 0.82 d | 41.37 | 8.16 f | 16.23 | 1.06 g | 8.16 | 0.21 f | 10.52 |
| KA9 | 24.50 j | 65.97 b | 18.80 b | 52.84 | 9.50 b | 53.22 | 1.08 b | 86.20 | 11.46 b | 63.24 | 1.67 b | 70.40 | 0.27 b | 42.10 |
| UK2 | 28.80 h | 60.00 c | 16.10 c | 30.89 | 7.70 e | 24.19 | 0.90 c | 55.17 | 9.82 c | 39.88 | 1.39 c | 41.83 | 0.22 e | 15.78 |
| BR3 | 41.80 e | 41.94 g | 14.80 e | 20.32 | 6.90 g | 11.29 | 0.78 e | 34.48 | 8.19 g | 16.66 | 1.09 f | 11.22 | 0.20 g | 5.26 |
| UP13 | 47.20 c | 34.44 i | 14.50 f | 17.88 | 6.60 h | 6.45 | 0.71 f | 22.41 | 8.21 e | 16.95 | 1.08 f | 10.20 | 0.21 f | 10.52 |
| AP2 | 42.40 d | 41.10 h | 15.60 d | 26.82 | 7.90 d | 27.41 | 0.82 d | 41.37 | 9.42 d | 34.18 | 1.17 e | 19.38 | 0.25 c | 31.57 |
| PDS1 | 20.80 k | 71.11 a | 19.70 a | 60.16 | 10.50 a | 69.35 | 1.13 a | 94.82 | 12.2 a | 73.78 | 1.89 a | 92.85 | 0.31 a | 63.15 |
| UK4 | 34.60 g | 51.93 e | 13.20 h | 7.31 | 6.60 h | 6.45 | 0.69 g | 18.96 | 7.18 i | 2.27 | 0.99 i | 1.02 | 0.21 f | 10.52 |
| DL3 | 50.00 b | 30.56 j | 13.40 i | 8.94 | 6.50 i | 4.83 | 0.69 g | 18.96 | 7.30 h | 3.98 | 1.03 h | 5.10 | 0.22 e | 15.78 |
| Control | 72.00 a | - | 12.30 j | - | 6.20 j | - | 0.58 h | - | 7.02 j | - | 0.98 j | 0.19 h | - |
| Characters | Rhizobacteria | ||||
|---|---|---|---|---|---|
| Pseudomonas fluorescens Strain PDS1 | Bacillus subtilis Strain BDS1 | Bacillus cereus Strain UK4 | Bacillus amyloliquefaciens Strain UK2 | Bacillus subtilis Strain KA9 | |
| Accession no. | MN368159 | MN395039 | MT491099 | MT491100 | MT491101 |
| Morphological characters | |||||
| Gram’s reactions | Negative | Positive | Positive | Positive | Positive |
| Pigmentation | Cream white | Gray-white | White | Light brown | Gray-white |
| Appearance | Rod-shaped | Bacilli | Bacilli | Bacilli | Bacilli |
| Colony characters | |||||
| Size | Small | Medium | Medium | Medium | Medium |
| Shape | Uniform | Round | Round | Round | Round |
| Margin | Entire round | Thick ridges | Undulate | Entire round | Thick ridges |
| Opacity | Opaque | Opaque | Opaque | Semi-transparent | Opaque |
| Elevation | Convex | Convex | Convex | Convex | Convex |
| Texture | Smooth, shiny | Smooth, moist | Irregular | Smooth | Smooth, moist |
| Biochemical characters | |||||
| H2S | + | − | + | − | + |
| Arginine | + | + | + | + | + |
| Catalase | + | + | + | + | + |
| Amylase | + | + | + | + | + |
| Glucose | + | + | + | + | + |
| Galactose | − | − | − | − | − |
| Lactose | − | − | − | − | − |
| Sorbitol | − | − | − | − | − |
| Mannose | + | + | + | + | + |
| Xylose | + | + | + | + | + |
| Sucrose | + | + | + | + | + |
| Citrate | + | + | + | + | + |
| PGP traits | |||||
| IAA Production | + | ++ | + | + | +++ |
| Phosphorus Solubilization | + | + | + | + | + |
| Ammonia Production | ++ | + | ++ | ++ | ++ |
| HCN Production | − | − | − | − | − |
| Siderophore Production | ++ | + | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashyap, A.S.; Manzar, N.; Rajawat, M.V.S.; Kesharwani, A.K.; Singh, R.P.; Dubey, S.C.; Pattanayak, D.; Dhar, S.; Lal, S.K.; Singh, D. Screening and Biocontrol Potential of Rhizobacteria Native to Gangetic Plains and Hilly Regions to Induce Systemic Resistance and Promote Plant Growth in Chilli against Bacterial Wilt Disease. Plants 2021, 10, 2125. https://doi.org/10.3390/plants10102125
Kashyap AS, Manzar N, Rajawat MVS, Kesharwani AK, Singh RP, Dubey SC, Pattanayak D, Dhar S, Lal SK, Singh D. Screening and Biocontrol Potential of Rhizobacteria Native to Gangetic Plains and Hilly Regions to Induce Systemic Resistance and Promote Plant Growth in Chilli against Bacterial Wilt Disease. Plants. 2021; 10(10):2125. https://doi.org/10.3390/plants10102125
Chicago/Turabian StyleKashyap, Abhijeet Shankar, Nazia Manzar, Mahendra Vikram Singh Rajawat, Amit Kumar Kesharwani, Ravinder Pal Singh, S. C. Dubey, Debasis Pattanayak, Shri Dhar, S. K. Lal, and Dinesh Singh. 2021. "Screening and Biocontrol Potential of Rhizobacteria Native to Gangetic Plains and Hilly Regions to Induce Systemic Resistance and Promote Plant Growth in Chilli against Bacterial Wilt Disease" Plants 10, no. 10: 2125. https://doi.org/10.3390/plants10102125
APA StyleKashyap, A. S., Manzar, N., Rajawat, M. V. S., Kesharwani, A. K., Singh, R. P., Dubey, S. C., Pattanayak, D., Dhar, S., Lal, S. K., & Singh, D. (2021). Screening and Biocontrol Potential of Rhizobacteria Native to Gangetic Plains and Hilly Regions to Induce Systemic Resistance and Promote Plant Growth in Chilli against Bacterial Wilt Disease. Plants, 10(10), 2125. https://doi.org/10.3390/plants10102125






