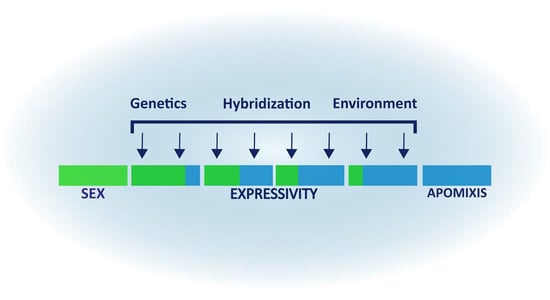
Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum
Abstract
:1. Introduction
2. Results
2.1. Reproductive Characterization of the F1 Population
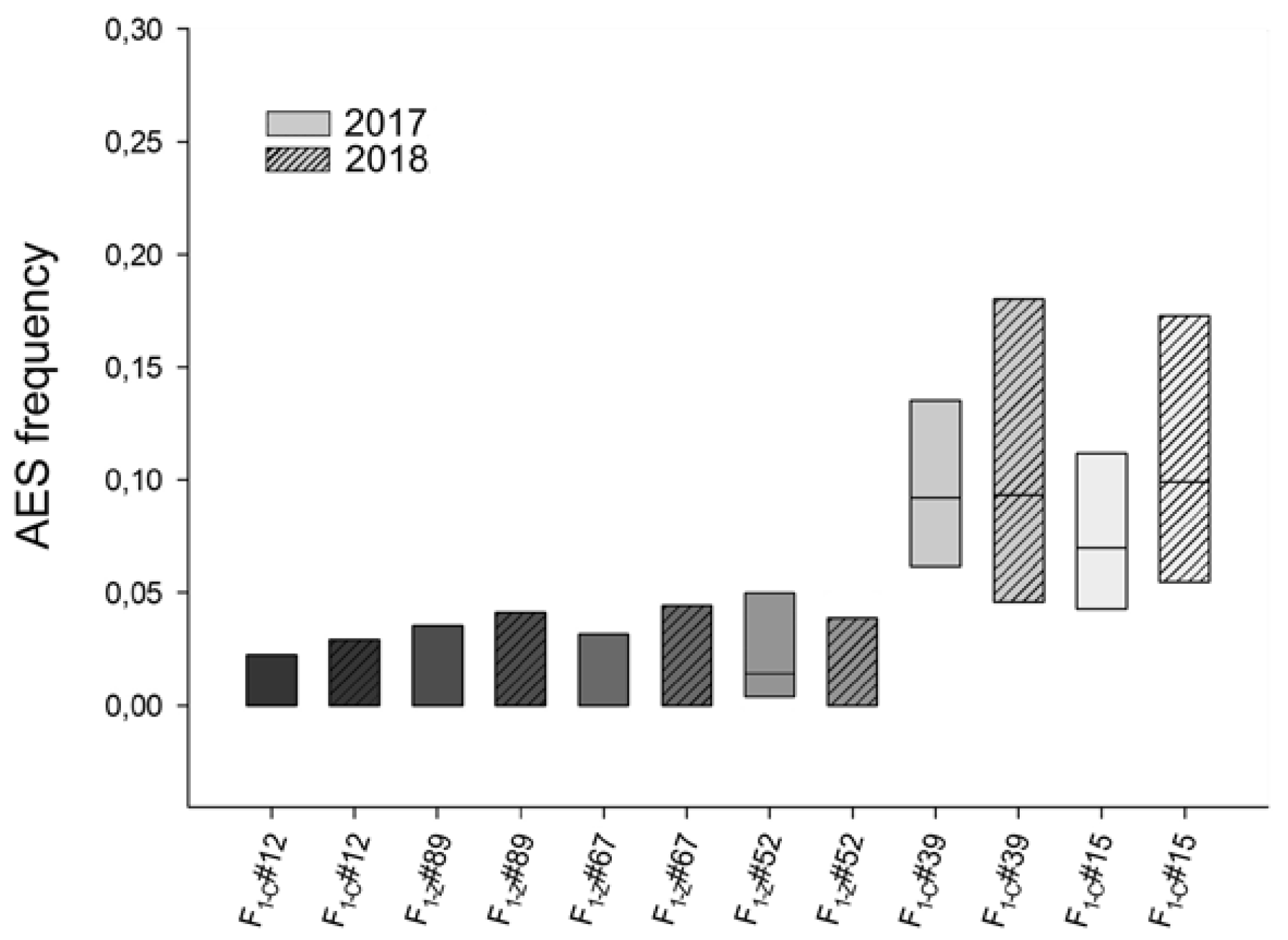
2.2. Evaluation of the Environmental Influence on Apospory Expressivity
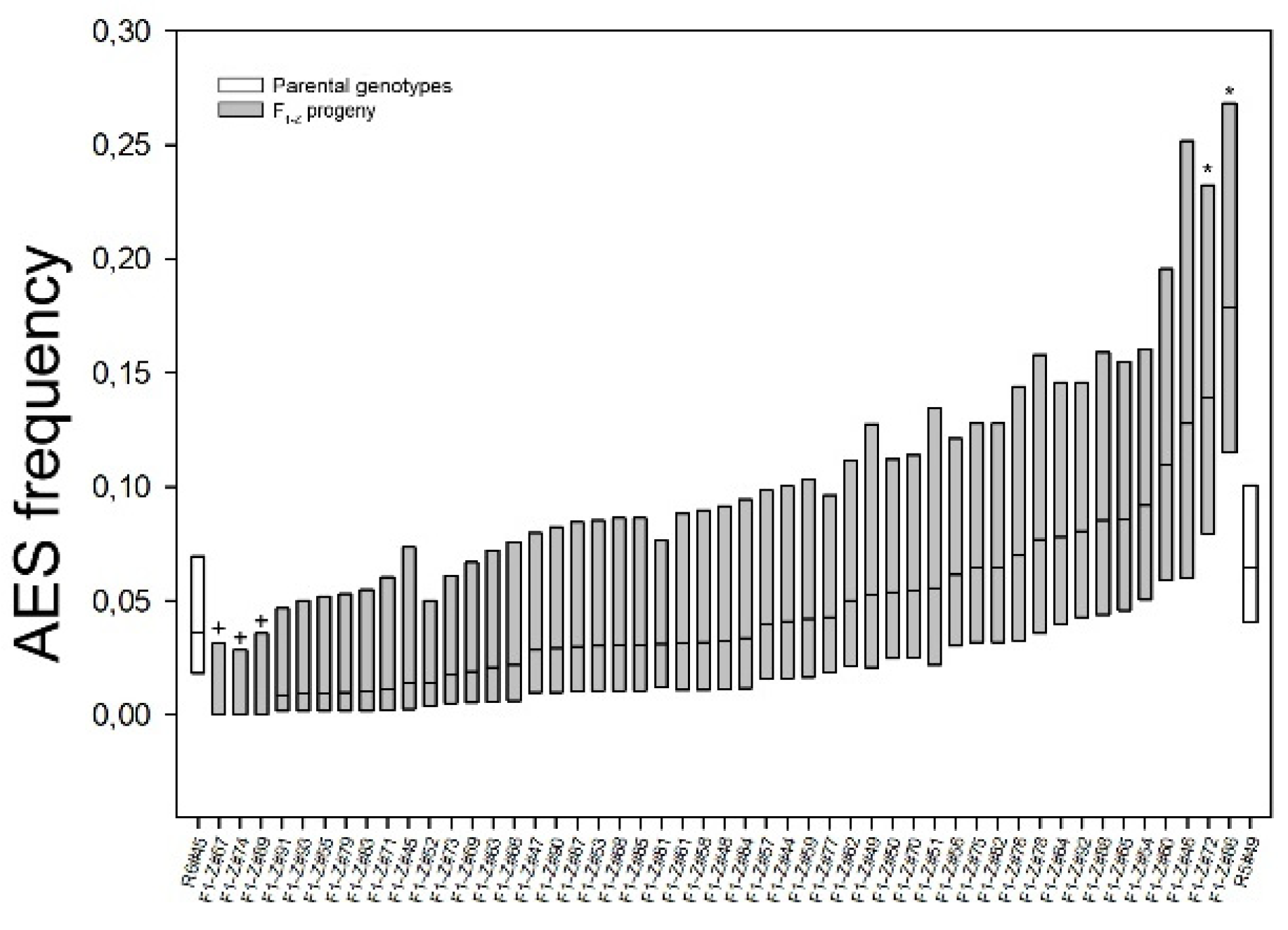
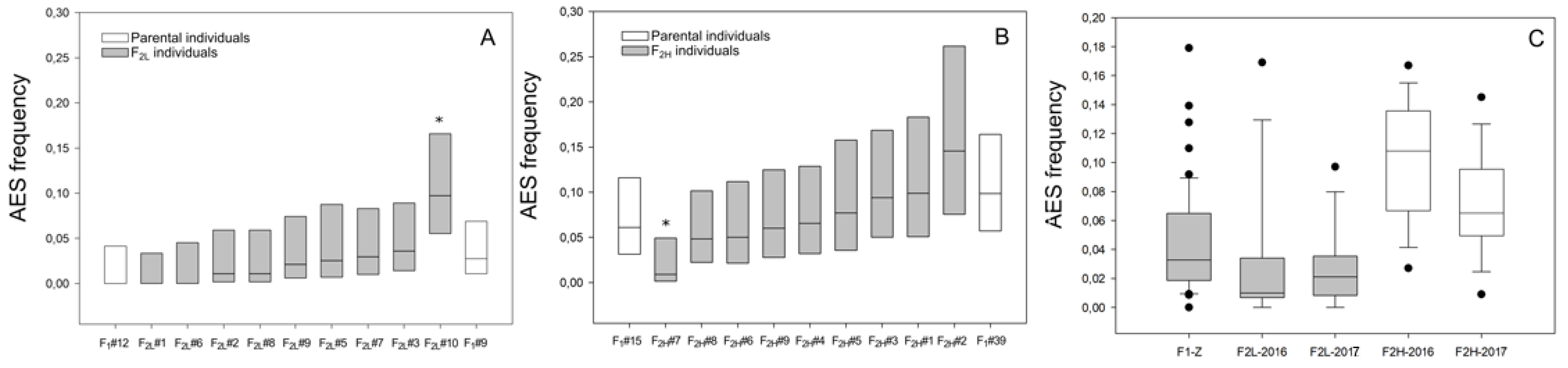
2.3. Apospory Expressivity Variation in F2 Progenies
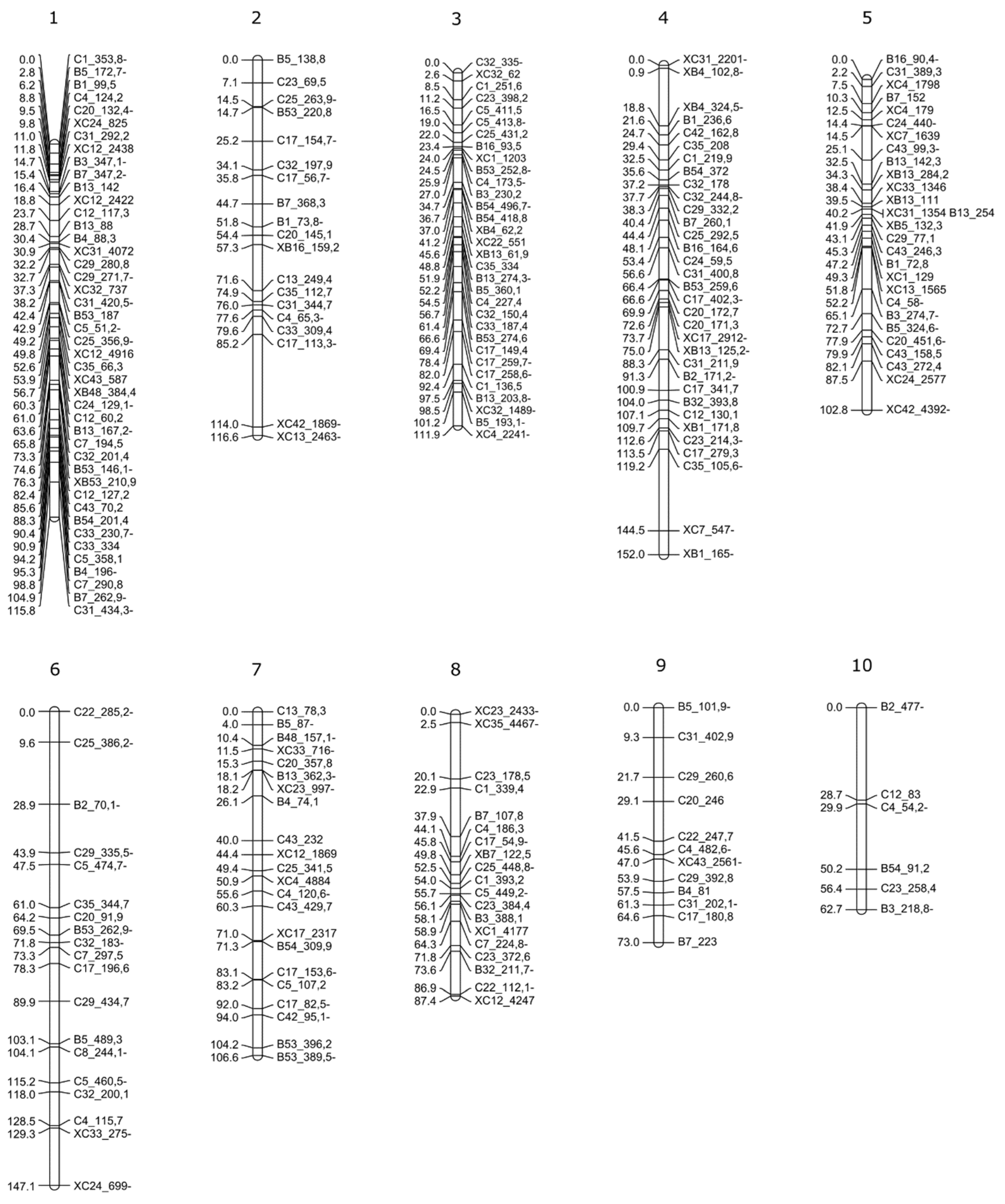
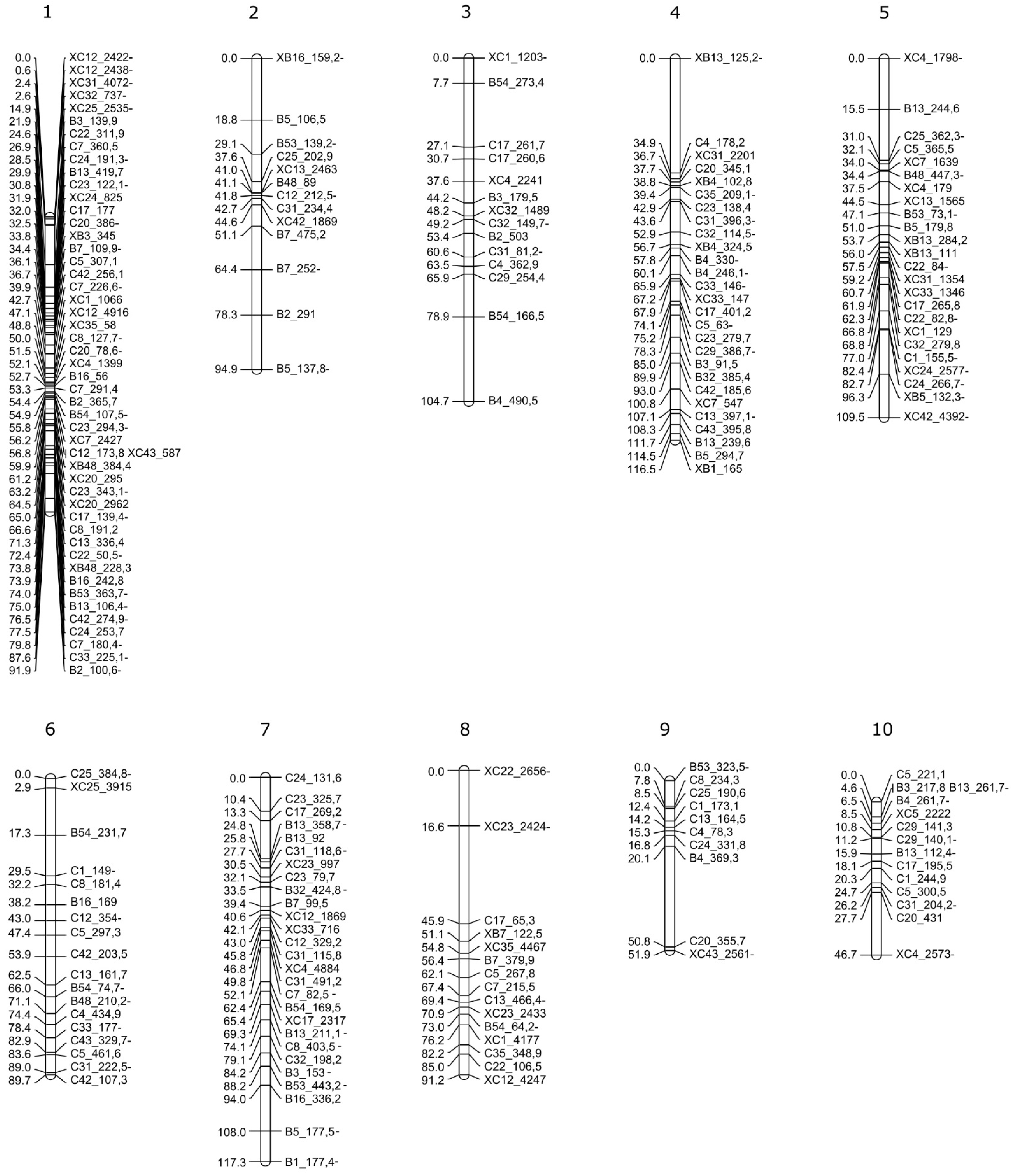
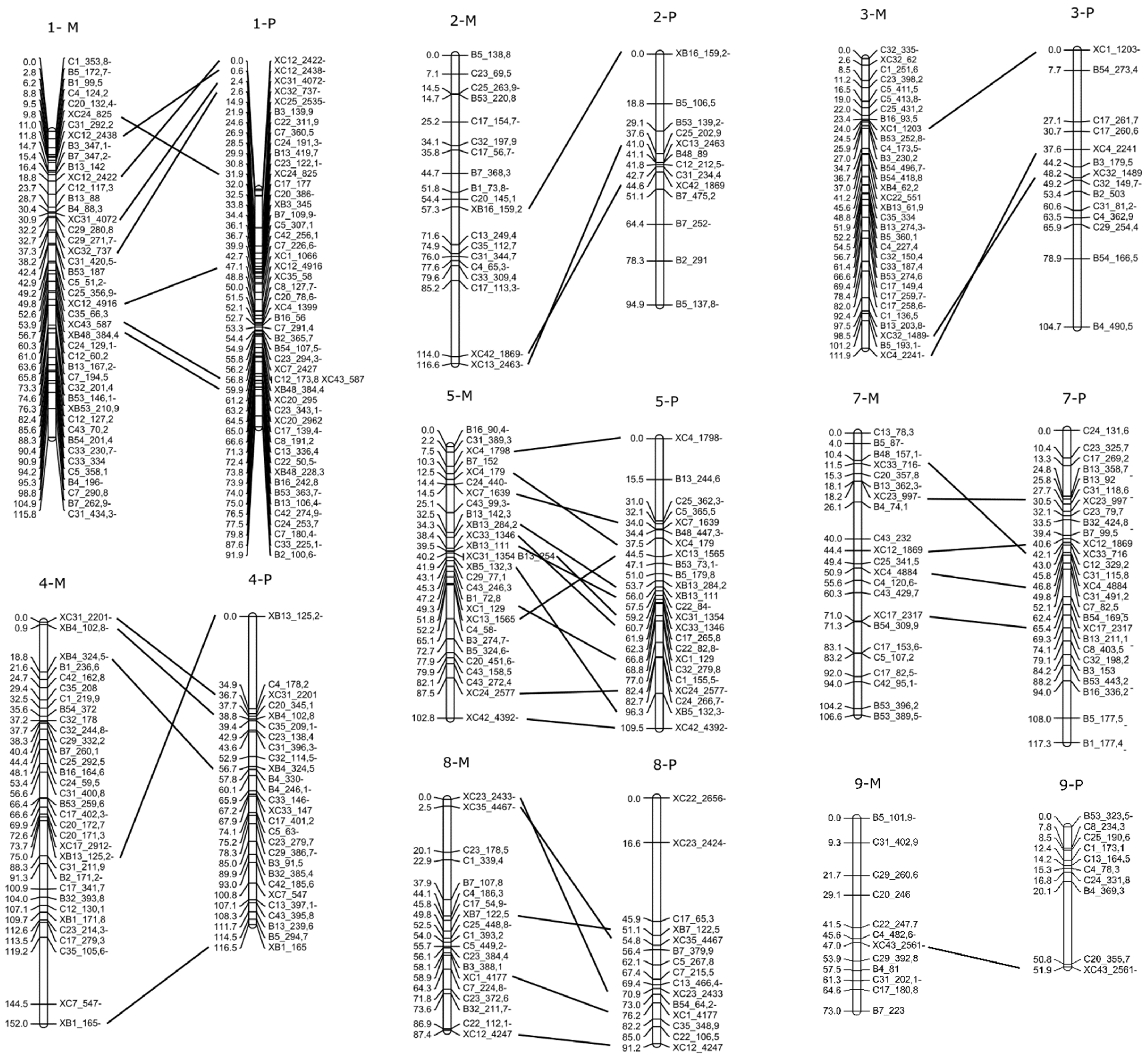
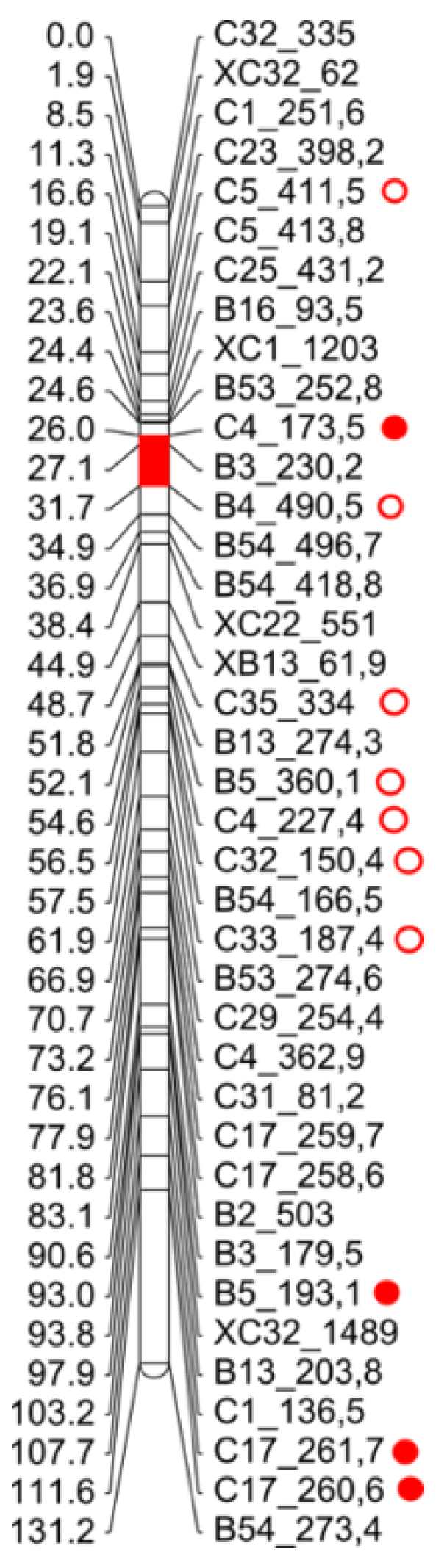
2.4. F1 Genetic Linkage Map
2.5. Identification of Genomic Regions Associated with Apospory Expressivity
3. Discussion
3.1. Apospory Inheritance
3.2. Apospory Expressivity Variation
3.3. Linkage Map
3.4. Quantitative Approach
4. Materials and Methods
4.1. Plant Material
4.2. Cytoembriological Analysis
4.3. AFLP Markers
4.4. Genetic Linkage Analysis
4.5. Linear Regression Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kumar, S. Epigenetic control of apomixis: A new perspective of an old enigma. Adv. Plants Agric. Res. 2017, 7, 10-15406. [Google Scholar] [CrossRef] [Green Version]
- Van Dijk, P.; Van Damme, J. Apomixis technology and the paradox of sex. Trends Plant Sci. 2000, 5, 81–89. [Google Scholar] [CrossRef]
- Koltunow, A.M. Apomixis: Embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 1993, 5, 1425–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toenniessen, G.H. Feeding the world in the 21st century: Plant breeding, biotechnology, and the potential role of apomixis. In The Flowering of Apomixis: From Mechanisms to Genetic Engineering; Savidan, Y., Carman, J.G., Dresselhaus, T., Eds.; CIMMYT, IRD, European Commission DG VI (FAIR): Mexico City, Mexico, 2001; pp. 1–7. [Google Scholar]
- Whitton, J.; Sears, C.J.; Baack, E.J.; Otto, S.P. The dynamic nature of apomixis in the angiosperms. Int. J. Plant Sci. 2008, 169, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Koltunow, A.M.; Grossniklaus, U. Apomixis: A developmental perspective. Annu. Rev. Plant Biol. 2003, 54, 547–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, J.P.A.; Quarin, C.L.; Pessino, S.C.; Acuña, C.; Martínez, E.J.; Espinoza, F.; Hojsgaard, D.H.; Sartor, M.E.; Cáceres, M.E.; Pupilli, F. Harnessing apomictic reproduction in grasses: What we have learned from Paspalum. Ann. Bot. 2013, 112, 767–787. [Google Scholar] [CrossRef] [Green Version]
- Asker, S.E. Gametophytic apomixis: Elements and genetic regulation. Hereditas 1980, 93, 277–293. [Google Scholar] [CrossRef]
- Nogler, G.A. Gametophytic Apomixis. In Embryology Angiosperms; Springer: Berlin, Heidelberg, 1984; pp. 475–518. [Google Scholar] [CrossRef]
- Spillane, C.; Curtis, M.D.; Grossniklaus, U. Apomixis technology development-virgin births in farmers’ fields? Nat. Biotechnol. 2004, 22, 687–691. [Google Scholar] [CrossRef]
- Abdi, S.; Dwivedi, A.; Bhat, V. Harnessing apomixis for heterosis breeding in crop improvement. In Molecular Breeding for Sustainable Crop Improvement; Springer: Cham, Switzerland, 2016; pp. 79–99. [Google Scholar]
- Martínez, E.J.; Urbani, M.H.; Quarin, C.L.; Ortiz, J.P.A. Inheritance of apospory in bahiagrass, Paspalum notatum. Hereditas 2001, 135, 19–25. [Google Scholar] [CrossRef]
- Pupilli, F.; Labombarda, P.; Caceres, M.E.; Quarín, C.L.; Arcioni, S. The chromosome segment related to apomixis in Paspalum simplex is homoeologous to the telomeric region of the long arm of rice chromosome 12. Mol. Breed. 2001, 8, 53–61. [Google Scholar] [CrossRef]
- Sherwood, R.T.; Berg, C.C.; Young, B.A. Inheritance of apospory in Buffelgrass. Crop Sci. 1994, 34, 1490–1494. [Google Scholar] [CrossRef]
- Savidan, Y. Nature et Hérédité de L’apomixie Chez Panicum Maximum Jacq; ORSTOM: Paris, France, 1982. [Google Scholar]
- Nogler, G. Genetics of apospory in apomictic Ranunculus auricomus. V: Conclusion. Bot. Helv. 1984, 94, 411–422. [Google Scholar]
- Bicknell, R.A.; Koltunow, A.M. Understanding apomixis: Recent advances and remaining conundrums. Plant Cell 2004, 16, S228–S245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schallau, A.; Arzenton, F.; Johnston, A.J.; Hahnel, U.; Koszegi, D.; Blattner, F.R.; Altschmied, L.; Haberer, G.; Barcaccia, G.; Baumlein, H. Identification and genetic analysis of the APOSPORY locus in Hypericum perforatum L. Plant J. 2010, 62, 773–784. [Google Scholar] [CrossRef] [PubMed]
- Martínez, E.J.; Hopp, H.E.; Stein, J.; Ortiz, J.P.A.; Quarin, C.L. Genetic characterization of apospory in tetraploid Paspalum notatum based on the identification of linked molecular markers. Mol. Breed. 2003, 12, 319–327. [Google Scholar] [CrossRef]
- Stein, J.; Quarin, C.L.; Martínez, E.J.; Pessino, S.C.; Ortiz, J.P.A. Tetraploid races of Paspalum notatum show polysomic inheritance and preferential chromosome pairing around the apospory-controlling locus. Theor. Appl. Genet. 2004, 109, 186–191. [Google Scholar] [CrossRef]
- Pupilli, F.; Martinez, E.J.; Busti, A.; Calderini, O.; Quarin, C.L.; Arcioni, S.; Martínez, E.J.; Busti, A.; Calderini, O.; Quarin, C.L. Comparative mapping reveals partial conservation of synteny at the apomixis locus in Paspalum spp. Mol. Genet. Genom. 2004, 270, 539–548. [Google Scholar] [CrossRef]
- Stein, J.; Pessino, S.C.; Martínez, E.J.; Rodriguez, M.P.; Siena, L.A.; Quarin, C.L.; Ortiz, J.P.A. A genetic map of tetraploid Paspalum notatum Flügge (bahiagrass) based on single-dose molecular markers. Mol. Breed. 2007, 20, 153–166. [Google Scholar] [CrossRef]
- Podio, M.; Siena, L.A.; Hojsgaard, D.; Stein, J.; Quarin, C.L.; Ortiz, J.P.A. Evaluation of meiotic abnormalities and pollen viability in aposporous and sexual tetraploid Paspalum notatum (Poaceae). Plant Syst. Evol. 2012, 298, 1625–1633. [Google Scholar] [CrossRef]
- Ozias-Akins, P.; Roche, D.; Hanna, W.W. Tight clustering and hemizygosity of apomixis-linked molecular markers in Pennisetum squamulatum implies genetic control of apospory by a divergent locus that may have no allelic form in sexual genotypes. Proc. Natl. Acad. Sci. USA 1998, 95, 5127–5132. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, Y.; Conner, J.A.; Goel, S.; Morishige, D.T.; Mullet, J.E.; Hanna, W.W.; Ozias-Akins, P. High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 2004, 134, 1733–1741. [Google Scholar] [CrossRef] [Green Version]
- Ozias-Akins, P.; Van Dijk, P.J. Mendelian genetics of apomixis in plants. Annu. Rev. Genet. 2007, 41, 509–537. [Google Scholar] [CrossRef]
- Yadav, C.B.; Anuj; Kumar, S.; Gupta, M.G.; Bhat, V. Genetic linkage maps of the chromosomal regions associated with apomictic and sexual modes of reproduction in Cenchrus ciliaris. Mol. Breed. 2012, 30, 239–250. [Google Scholar] [CrossRef]
- Koltunow, A.M.G.; Johnson, S.D.; Okada, T. Apomixis in hawkweed: Mendel’s experimental nemesis. J. Exp. Bot. 2011, 62, 1699–1707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotani, Y.; Henderson, S.T.; Suzuki, G.; Johnson, S.D.; Okada, T.; Siddons, H.; Mukai, Y.; Koltunow, A.M.G.G. The LOSS OF APOMEIOSIS (LOA) locus in Hieracium praealtum can function independently of the associated large-scale repetitive chromosomal structure. New Phytol. 2014, 201, 973–981. [Google Scholar] [CrossRef]
- Ebina, M.; Nakagawa, H.; Yamamoto, T.; Araya, H.; Tsuruta, S.; Takahara, M.; Nakajima, K. Co-segregation of AFLP and RAPD markers to apospory in Guineagrass (Panicum maximum Jacq.). Grassl. Sci. 2005, 51, 71–78. [Google Scholar] [CrossRef]
- Worthington, M.; Heffelfinger, C.; Bernal, D.; Quintero, C.; Zapata, Y.P.; Perez, J.G.; De Vega, J.; Miles, J.; Dellaporta, S.; Tohme, J. A parthenogenesis gene candidate and evidence for segmental allopolyploidy in apomictic Brachiaria decumbens. Genetics 2016, 203, 1117–1132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaushal, P.; Dwivedi, K.K.; Radhakrishna, A.; Srivastava, M.K.; Kumar, V.; Roy, A.K.; Malaviya, D.R. Partitioning apomixis components to understand and utilize gametophytic apomixis. Front. Plant Sci. 2019, 10, 256. [Google Scholar] [CrossRef] [PubMed]
- Pessino, S.C.; Espinoza, F.; Martínez, E.J.; Ortiz, J.P.A.; Valle, E.M.; Quarín, C.L.; Martinez, E.J.; Ortiz, J.P.A.; Valle, E.M.; Quarin, C.L. Isolation of cDNA clones differentially expressed in flowers of apomictic and sexual Paspalum notatum. Hereditas 2001, 134, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Laspina, N.V.; Vega, T.; Seijo, J.G.; González, A.M.; Martelotto, L.G.; Stein, J.; Podio, M.; Ortiz, J.P.A.; Echenique, V.C.; Quarin, C.L. Gene expression analysis at the onset of aposporous apomixis in Paspalum notatum. Plant Mol. Biol. 2008, 67, 615–628. [Google Scholar] [CrossRef]
- Capron, A.; Gourgues, M.; Neiva, L.S.; Faure, J.E.; Berger, F.; Pagnussat, G.; Krishnan, A.; Alvarez-Mejia, C.; Vielle-Calzada, J.P.; Lee, Y.R.; et al. Maternal control of male-gamete delivery in arabidopsis Involves a putative GPI-anchored protein encoded by the lorelei gene. Plant Cell 2008, 20, 3038–3049. [Google Scholar] [CrossRef] [Green Version]
- Felitti, S.A.; Seijo, G.G.; González, A.M.; Podio, M.; Laspina, N.V.; Siena, L.; Ortiz, J.P.A.; Pessino, S.C. Expression of lorelei-like genes in aposporous and sexual Paspalum notatum plants. Plant Mol. Biol. 2011, 77, 337–354. [Google Scholar] [CrossRef] [PubMed]
- Siena, L.A.; Sartor, M.E.; Quarin, C.L.; Espinoza, F.; Ortiz, J.P.A. Transference of wheat expressed sequence tag-simple sequence repeats to Paspalum species and cross-species amplification of Paspalum notatum simple sequence repeats: Potential use in phylogenetic analysis and mapping. Crop Sci. 2014, 54, 240–254. [Google Scholar] [CrossRef]
- Colono, C.; Ortiz, J.P.A.; Permingeat, H.R.; Souza Canada, E.D.; Siena, L.A.; Spoto, N.; Galdeano, F.; Espinoza, F.; Leblanc, O.; Pessino, S.C. A plant-specific TGS1 homolog influences gametophyte development in sexual tetraploid Paspalum notatum ovules. Front. Plant Sci. 2019, 10, 1566. [Google Scholar] [CrossRef] [PubMed]
- Hecht, V.; Vielle-Calzada, J.P.; Hartog, M.V.; Schmidt, E.D.L.; Boutilier, K.; Grossniklaus, U.; De Vries, S.C. Erratum: The arabidopsis somatic embryogenesis receptor kinase 1 gene is expressed in developing ovules and embryos and enhances embryogenic competence in culture. Plant Physiol. 2002, 128, 314. [Google Scholar] [CrossRef]
- Corral, J.M.; Vogel, H.; Aliyu, O.M.; Hensel, G.; Thiel, T.; Kumlehn, J.; Sharbel, T.F. A conserved apomixis-specific polymorphism is correlated with exclusive exonuclease expression in premeiotic ovules of apomictic Boechera species. Plant Physiol. 2013, 163, 1660–1672. [Google Scholar] [CrossRef] [Green Version]
- Conner, J.A.; Mookkan, M.; Huo, H.; Chae, K.; Ozias-Akins, P. A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. USA 2015, 112, 11205–11210. [Google Scholar] [CrossRef] [Green Version]
- Conner, J.A.; Podio, M.; Ozias-Akins, P. Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 2017, 30, 41–52. [Google Scholar] [CrossRef]
- Siena, L.A.; Ortiz, J.P.A.; Calderini, O.; Paolocci, F.; Cáceres, M.E.; Kaushal, P.; Grisan, S.; Pessino, S.C.; Pupilli, F. An apomixis-linked ORC3-like pseudogene is associated with silencing of its functional homolog in apomictic Paspalum simplex. J. Exp. Bot. 2016, 67, 1965–1978. [Google Scholar] [CrossRef] [Green Version]
- Henderson, S.T.; Johnson, S.D.; Eichmann, J.; Koltunow, A.M.G. Genetic analyses of the inheritance and expressivity of autonomous endosperm formation in Hieracium with different modes of embryo sac and seed formation. Ann. Bot. 2017, 119, 1001–1010. [Google Scholar] [CrossRef]
- Acuña, C.A.; Blount, A.R.; Quesenberry, K.H.; Kenworthy, K.E.; Hanna, W.W. Bahiagrass tetraploid germplasm: Reproductive and agronomic characterization of segregating progeny. Crop Sci. 2009, 49, 581. [Google Scholar] [CrossRef]
- Zilli, A.L.; Brugnoli, E.A.; Marcón, F.; Billa, M.B.; Rios, E.F.; Martínez, E.J.; Acuña, C.A. Heterosis and expressivity of apospory in tetraploid bahiagrass hybrids. Crop Sci. 2015, 55, 1189–1201. [Google Scholar] [CrossRef]
- Aliyu, O.M.; Schranz, M.E.; Sharbel, T.F. Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 2010, 97, 1719–1731. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.R.; Okada, T.; Johnson, S.D.; Takaiwa, F.; Koltunow, A.M.G.G. Sporophytic ovule tissues modulate the initiation and progression of apomixis in Hieracium. J. Exp. Bot. 2012, 63, 3229–3241. [Google Scholar] [CrossRef] [Green Version]
- Shirasawa, K.; Hand, M.L.; Henderson, S.T.; Okada, T.; Johnson, S.D.; Taylor, J.M.; Spriggs, A.; Siddons, H.; Hirakawa, H.; Isobe, S.; et al. A reference genetic linkage map of apomictic Hieracium species based on expressed markers derived from developing ovule transcripts. Ann. Bot. 2015, 115, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Zappacosta, D.; Gallardo, J.; Carballo, J.; Meier, M.; Rodrigo, J.M.; Gallo, C.A.; Selva, J.P.; Stein, J.; Ortiz, J.P.A.; Albertini, E.; et al. A high-density linkage map of the forage grass Eragrostis curvula and localization of the diplospory locus. Front. Plant Sci. 2019, 10, 918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asker, S.E.; Jerling, L. Apomixis in Plants; CRC Press: Boca Raton, FL, USA, 1992; ISBN 9781351466318. [Google Scholar]
- Hojsgaard, D.H.; Martínez, E.J.; Quarin, C.L.; Martinez, E.J.; Quarin, C.L. Competition between meiotic and apomictic pathways during ovule and seed development results in clonality. New Phytol. 2013, 197, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Quarin, C.L.; Lombardo, E.P. Niveles de ploidía y distribución geográfica de Paspalum quadrifarium (Gramineae). Mendeliana 1986, 7, 101–107. [Google Scholar]
- Quarin, C.L.; Norrmann, G.A. Cytology and reproductive behavior of Paspalum equitans, P. ionanthum, and their hybrids with diploid and tetraploid cytotypes of P. cromyorrhizon. Bot. Gaz. 1987, 148, 386–391. [Google Scholar] [CrossRef]
- Norrmann, G.A.; Quarin, C.L.; Burson, B.L. Cytogenetics and reproductive behavior of different chromosome races in six Paspalum species. J. Hered. 1989, 80, 24–28. [Google Scholar] [CrossRef]
- Quarin, C.L.; Espinoza, F.; Martinez, E.J.; Pessino, S.C.; Bovo, O.A. A rise of ploidy level induces the expression of apomixis in Paspalum notatum. Sex. Plant Reprod. 2001, 13, 243–249. [Google Scholar] [CrossRef]
- Galdeano, F.; Urbani, M.H.; Sartor, M.E.; Honfi, A.I.; Espinoza, F.; Quarin, C.L. Relative DNA content in diploid, polyploid, and multiploid species of Paspalum (Poaceae) with relation to reproductive mode and taxonomy. J. Plant Res. 2016, 129, 697–710. [Google Scholar] [CrossRef]
- Sartor, M.E.; Quarin, C.L.; Urbani, M.H.; Espinoza, F. Ploidy levels and reproductive behaviour in natural populations of five Paspalum species. Plant Syst. Evol. 2011, 293, 31–41. [Google Scholar] [CrossRef]
- Grimanelli, D.; Leblanc, O.; Perotti, E.; Grossniklaus, U. Developmental genetics of gametophytic apomixis. Trends Genet. 2001, 17, 597–604. [Google Scholar] [CrossRef]
- Carman, J.G. Asynchonous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 1997, 61, 51–94. [Google Scholar] [CrossRef]
- Quarin, C.L. The nature of apomixis and its origin in Panicoid grasses. Apomixis Newsl. 1992, 5, 7–15. [Google Scholar]
- Quarín, C.L.; Norrmann, G.A.; Espinoza, F. Evidence for autoploidy in apomictic Paspalum rufum. Hereditas 1998, 129, 119–124. [Google Scholar] [CrossRef]
- Hojsgaard, D.; Hörandl, E. The rise ofapomixis in natural plant populations. Front. Plant Sci. 2019, 10, 358. [Google Scholar] [CrossRef]
- Quarin, C.L. Seasonal changes in the incidence of apomixis of diploid, triploid, and tetraploid plants of Paspalum cromyorrhizon. Euphytica 1986, 35, 515–522. [Google Scholar] [CrossRef]
- Rebozzio, R.N.; Sartor, M.E.; Quarin, C.L.; Espinoza, F. Residual sexuality and its seasonal variation in natural apomictic Paspalum notatum accessions. Biol. Plant. 2011, 55, 391–395. [Google Scholar] [CrossRef]
- Acuña, C.A.; Martínez, E.J.; Zilli, A.L.; Brugnoli, E.A.; Espinoza, F.; Marcón, F.; Urbani, M.H.; Quarin, C.L. Reproductive systems in Paspalum: Relevance for germplasm collection and conservation, breeding techniques, and adoption of released cultivars. Front. Plant Sci. 2019, 10, 1377. [Google Scholar] [CrossRef] [Green Version]
- Siena, L.A.; Sartor, M.E.; Espinoza, F.; Quarin, C.L.; Ortiz, J.P.A. Genetic and embryological evidences of apomixis at the diploid level in Paspalum rufum support recurrent auto-polyploidization in the species. Sex. Plant Reprod. 2008, 21, 205–215. [Google Scholar] [CrossRef]
- Delgado, L.; Galdeano, F.; Sartor, M.E.; Quarin, C.L.; Espinoza, F.; Ortiz, J.P.A. Analysis of variation for apomictic reproduction in diploid Paspalum rufum. Ann. Bot. 2014, 113, 1211–1218. [Google Scholar] [CrossRef]
- Delgado, L.; Sartor, M.E.; Espinoza, F.; Soliman, M.; Galdeano, F.; Ortiz, J.P.A. Hybridity and autopolyploidy increase the expressivity of apospory in diploid Paspalum rufum. Plant Syst. Evol. 2016, 302, 1471–1481. [Google Scholar] [CrossRef]
- Tanksley, S.D. Mapping polygenes. Annu. Rev. Genet. 1993, 27, 205–233. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Jahufer, M.Z.Z.; Brouwer, J.B.; Pang, E.C.K. An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: The basic concepts. Euphytica 2005, 142, 169–196. [Google Scholar] [CrossRef]
- Rios, E.F.; Blount, A.; Kenworthy, K.E.; Acuña, C.A.; Quesenberry, K.H. Seasonal expression of apospory in bahiagrass. Trop. Grassl. Forrajes Trop. 2013, 1, 116–118. [Google Scholar] [CrossRef]
- Klatt, S.; Hadacek, F.; Hodač, L.; Brinkmann, G.; Eilerts, M.; Hojsgaard, D.; Hörandl, E. Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 2016, 7, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunarathne, P.; Reutemann, A.V.; Schedler, M.; Glücksberg, A.; Martínez, E.J.; Honfi, A.I.; Hojsgaard, D.H. Sexual modulation in a polyploid grass: A reproductive contest between environmentally inducible sexual and genetically dominant apomictic pathways. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef]
- van der Staay, M. Solar Topo. Available online: http://www.solartopo.com/daylength.htm (accessed on 14 July 2021).
- Merkel, A. Climate-Data.org. Available online: https://es.climate-data.org/america-del-sur/argentina-11/ (accessed on 14 July 2021).
- Rodrigo, J.M.; Zappacosta, D.C.; Selva, J.P.; Garbus, I.; Albertini, E.; Echenique, V. Apomixis frequency under stress conditions in weeping lovegrass (Eragrostis curvula). PLoS ONE 2017, 12, e0175852. [Google Scholar] [CrossRef]
- Gounaris, E.K.; Sherwood, R.T.; Gounaris, I.; Hamilton, R.H.; Gustine, D.L. Inorganic salts modify embryo sac development in sexual and aposporous Cenchrus ciliaris. Sex. Plant Reprod. 1991, 4, 188–192. [Google Scholar] [CrossRef]
- Barke, B.H.; Daubert, M.; Hörandl, E. Establishment of apomixis in diploid F2 hybrids and inheritance of apospory from F1 to F2 hybrids of the Ranunculus auricomus complex. Front. Plant Sci. 2018, 9, 1111. [Google Scholar] [CrossRef] [PubMed]
- Soliman, M.; Podio, M.; Marconi, G.; Di Marsico, M.; Ortiz, J.P.A.; Albertini, E.; Delgado, L. Differential epigenetic marks are associated with apospory expressivity in diploid hybrids of Paspalum rufum. Plants 2021, 10, 793. [Google Scholar] [CrossRef]
- Paszkowski, J.; Grossniklaus, U. Selected aspects of transgenerational epigenetic inheritance and resetting in plants. Curr. Opin. Plant Biol. 2011, 14, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Quadrana, L.; Colot, V. Plant transgenerational epigenetics. Annu. Rev. Genet. 2016, 50, 467–491. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, J.P.A.; Pessino, S.C.; Bhat, V.; Hayward, M.D.; Quarin, C.L. A Genetic linkage map of diploid Paspalum notatum. Crop Sci. 2001, 41, 823–830. [Google Scholar] [CrossRef]
- Wang, Z.M.; Devos, K.M.; Liu, C.J.; Wang, R.Q.; Gale, M.D. Construction of RFLP-based maps of foxtail millet, Setaria italica (L.) P. Beauv. Theor. Appl. Genet. 1998, 96, 31–36. [Google Scholar] [CrossRef]
- Dufour, P.; Deu, M.; Grivet, L.; D’Hont, A.; Paulet, F.; Bouet, A.; Lanaud, C.; Glaszmann, J.C.; Hamon, P. Construction of a composite sorghum genome map and comparison with sugarcane, a related complex polyploid. Theor. Appl. Genet. 1997, 94, 409–418. [Google Scholar] [CrossRef]
- Dolezel, J.; Bartos, J.; Voglmayr, H.; Greilhuber, J. Nuclear DNA content and genome size of trout and human. Cytometry 2003, 51A, 127–128. [Google Scholar] [CrossRef]
- Medianeira Machado, J.; Krycki, K.C.; Luis Weiler, R.; Simioni, C.; Dall´Agnol, M. Reproduction mode and apospory expressivity of selected hybrids of Paspalum notatum Flgge. J. Plant Breed. Crop Sci. 2021, 13, 58–63. [Google Scholar] [CrossRef]
- Mackay, I.J.; Cockram, J.; Howell, P.; Powell, W. Understanding the classics: The unifying concepts of transgressive segregation, inbreeding depression and heterosis and their central relevance for crop breeding. Plant Biotechnol. J. 2021, 19, 26–34. [Google Scholar] [CrossRef]
- Rieseberg, L.H.; Widmer, A.; Arntz, A.M.; Burke, J.M.; Carr, D.E.; Abbott, R.J.; Meagher, T.R. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Philos. Trans. R. Soc. B Biol. Sci. 2003, 358, 1141–1147. [Google Scholar] [CrossRef] [Green Version]
- Pellino, M.; Hojsgaard, D.; Hörandl, E.; Sharbel, T.F. Chasing the apomictic factors in the Ranunculus auricomus complex: Exploring gene expression patterns in microdissected sexual and apomictic ovules. Genes 2020, 11, 728. [Google Scholar] [CrossRef]
- Soliman, M.; Espinoza, F.; Ortiz, J.P.A.; Delgado, L. Heterochronic reproductive developmental processes between diploid and tetraploid cytotypes of Paspalum rufum. Ann. Bot. 2019, 123, 901–915. [Google Scholar] [CrossRef]
- Young, B.A.; Sherwood, R.T.; Bashaw, E.C. Cleared-pistil and thick-sectioning techniques for detecting aposporous apomixis in grasses. Can. J. Bot. 1979, 57, 1668–1672. [Google Scholar] [CrossRef]
- Vos, P.; Hogers, R.; Bleeker, M.; Reijans, M.; Van De Lee, T.; Hornes, M.; Friters, A.; Pot, J.; Paleman, J.; Kuiper, M.; et al. AFLP: A new technique for DNA fingerprinting. Nucleic Acids Res. 1995, 23, 4407–4414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cnops, G.; Den Boer, B.; Gerats, A.; Van Montagu, M.; Van Lijsebettens, M. Chromosome landing at the Arabidopsis TORNADO1 locus using an AFLP-based strategy. Mol. Gen. Genet. 1996, 253, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Ritter, E.; Gebhardt, C.; Salamini, F. Estimation of recombination frequencies and construction of RFLP linkage maps in plants from crosses between heterozygous parents. Genetics 1990, 125, 645–654. [Google Scholar] [CrossRef]
- Van Ooijen, J.W. JoinMap 4. Software for the Calculation of Genetic Likage Maps in Experimental Populations; Kyazma: Wageningen, The Netherlands, 2006. [Google Scholar]
- Kosambi, D.D. The estimation of map distances from recombination values. Ann. Eugen. 1943, 12, 172–175. [Google Scholar] [CrossRef]
- Al-Janabi, S.M.; Honeycutt, R.J.; McClelland, M.; Sobral, B.W.S. A genetic linkage map of Saccharum spontaneum L. “SES 208”. Genetics 1993, 134, 1249–1260. [Google Scholar] [CrossRef] [PubMed]
- Lowry, R. VassarStats: Website for Statistical Computation. Available online: http://vassarstats.net/ (accessed on 17 August 2021).
- Wilson, E.B. Probable inference, the law of succession, and statistical inference. J. Am. Stat. Assoc. 1927, 22, 209–212. [Google Scholar] [CrossRef]
- Upton, G.J.G. Fisher’s exact test. J. R. Stat. Soc. A Statist. Soc. 1992, 155, 395–402. [Google Scholar] [CrossRef]
- Sauro, J. MeasuringU. A/B Test Calculator. Available online: http://www.measuringu.com/ab-calc.php (accessed on 17 August 2021).
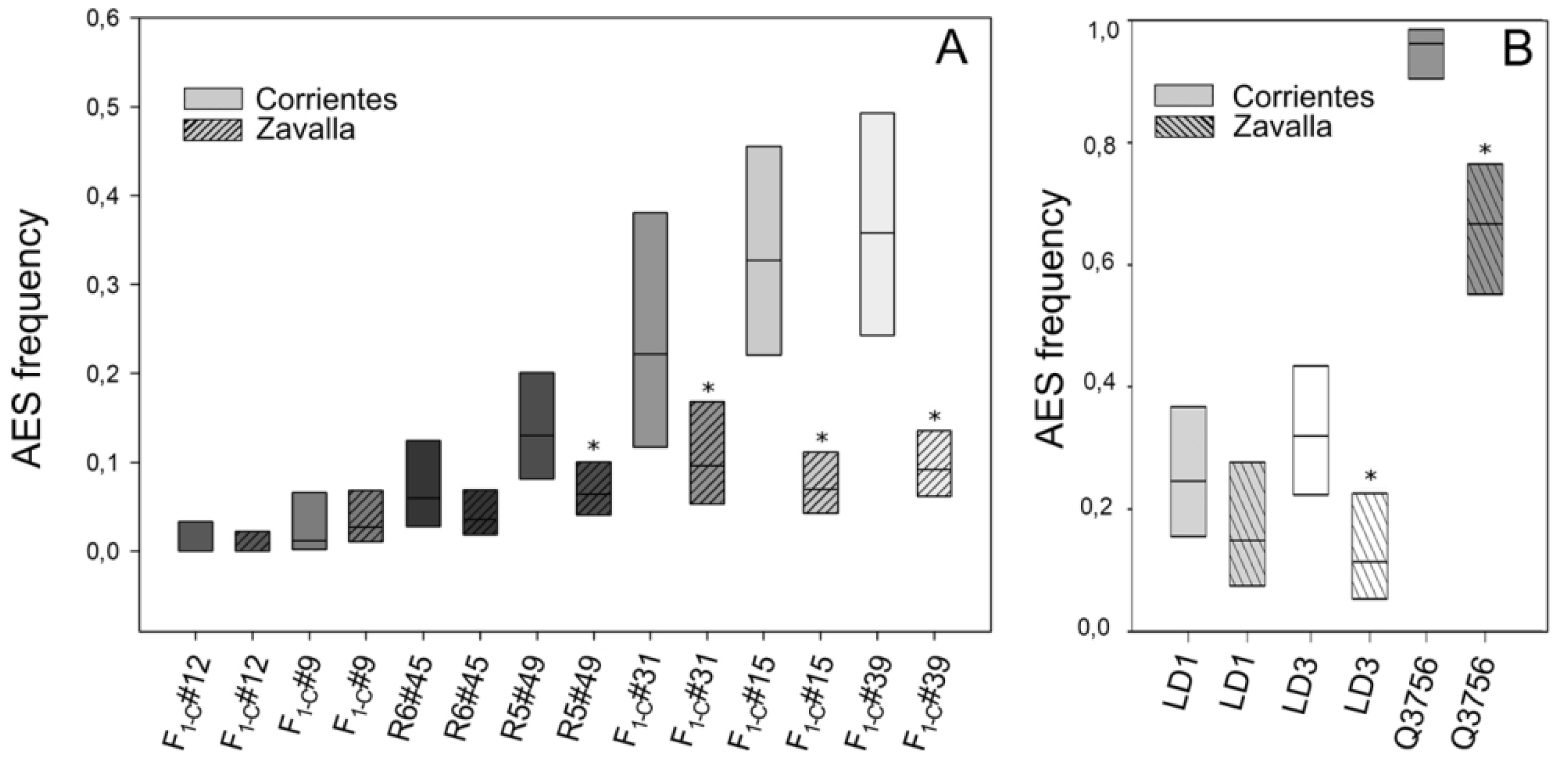
| Individual | Ploidy Level | %AES | Nº of Scored Ovules 1 | Origin | Reference |
|---|---|---|---|---|---|
| R6#45 | 2x | 5.77 | 104 | Paso Lucero, Corrientes, Argentina | Sartor et al., (2011), Delgado et al. (2014) |
| R5#49 | 2x | 13.3 | 123 | Saladas, Corrientes, Argentina | Sartor et al., (2011), Delgado et al., (2014) |
| F1-C#9 | 2x | 1.22 | 82 | R6#45 × R5#49 | Delgado et al. 2016 |
| F1-C#12 | 2x | 0.00 | 112 | R6#45 × R5#49 | Delgado et al. 2016 |
| F1-C#31 | 2x | 22.22 | 36 | R6#45 × R5#49 | Delgado et al. 2016 |
| F1-C#15 | 2x | 32.76 | 58 | R6#45 × R5#49 | Delgado et al. 2016 |
| F1-C#39 | 2x | 35.85 | 53 | R6#45 × R5#49 | Delgado et al. 2016 |
| LD1 | 4x | 31.94 | 72 | R6#49 2 | Delgado et al. 2016 |
| LD3 | 4x | 24.59 | 61 | R6#45 × R2#18 3 | Delgado et al. 2016 |
| Q3756 | 4x | 96.1 | 103 | Uruguay: unknown locality | Norrmann et al. 1989 |
| Individuals | Ovary Number | ||||||
|---|---|---|---|---|---|---|---|
| Total | AbES | SES | AES | SES + AES | NC | %AES 1 | |
| F1-Z#67 | 118 | 33 | 84 | 0 | 0 | 1 | 0.00 |
| F1-Z#74 | 132 | 81 | 43 | 0 | 0 | 8 | 0.00 |
| F1-Z#89 | 104 | 2 | 102 | 0 | 0 | 0 | 0.00 |
| F1-Z#91 | 117 | 17 | 97 | 0 | 1 | 2 | 0.85 |
| F1-Z#93 | 109 | 24 | 84 | 0 | 1 | 0 | 0.92 |
| F1-Z#55 | 105 | 4 | 100 | 0 | 1 | 0 | 0.95 |
| F1-Z#79 | 103 | 3 | 98 | 0 | 1 | 1 | 0.97 |
| F1-Z#83 | 99 | 1 | 97 | 0 | 1 | 0 | 1.01 |
| F1-Z#71 | 90 | 2 | 87 | 0 | 1 | 0 | 1.11 |
| F1-Z#45 | 73 | 13 | 57 | 0 | 1 | 2 | 1.37 |
| F1-Z#52 | 142 | 76 | 59 | 0 | 2 | 5 | 1.41 |
| F1-Z#73 | 115 | 6 | 107 | 0 | 2 | 0 | 1.74 |
| F1-Z#69 | 105 | 53 | 50 | 0 | 2 | 0 | 1.90 |
| F1-Z#63 | 97 | 2 | 92 | 0 | 2 | 1 | 2.06 |
| F1-Z#66 | 92 | 30 | 49 | 0 | 2 | 11 | 2.17 |
| F1-Z#47 | 106 | 30 | 71 | 0 | 3 | 2 | 2.83 |
| F1-Z#90 | 103 | 6 | 64 | 0 | 3 | 30 | 2.91 |
| F1-Z#87 | 100 | 4 | 91 | 0 | 3 | 2 | 3.00 |
| F1-Z#53 | 99 | 5 | 91 | 0 | 3 | 0 | 3.03 |
| F1-Z#68 | 98 | 5 | 89 | 0 | 3 | 1 | 3.06 |
| F1-Z#85 | 98 | 2 | 93 | 0 | 3 | 0 | 3.06 |
| F1-Z#81 | 130 | 21 | 102 | 0 | 4 | 3 | 3.08 |
| F1-Z#61 | 96 | 2 | 90 | 0 | 3 | 1 | 3.13 |
| F1-Z#58 | 94 | 6 | 83 | 0 | 3 | 2 | 3.19 |
| F1-Z#48 | 92 | 6 | 83 | 0 | 3 | 0 | 3.26 |
| F1-Z#84 | 89 | 3 | 83 | 0 | 3 | 0 | 3.37 |
| F1-Z#57 | 100 | 5 | 78 | 0 | 4 | 13 | 4.00 |
| F1-Z#44 | 98 | 52 | 35 | 2 | 2 | 7 | 4.08 |
| F1-Z#59 | 95 | 1 | 55 | 0 | 4 | 35 | 4.21 |
| F1-Z#77 | 117 | 44 | 68 | 0 | 5 | 0 | 4.27 |
| F1-Z#62 | 100 | 8 | 83 | 0 | 5 | 4 | 5.00 |
| F1-Z#49 | 76 | 4 | 67 | 0 | 4 | 1 | 5.26 |
| F1-Z#50 | 112 | 3 | 103 | 0 | 6 | 0 | 5.36 |
| F1-Z#70 | 110 | 16 | 88 | 0 | 6 | 0 | 5.45 |
| F1-Z#51 | 72 | 3 | 65 | 0 | 4 | 0 | 5.56 |
| F1-Z#56 | 114 | 4 | 103 | 0 | 7 | 0 | 6.14 |
| F1-Z#75 | 108 | 6 | 93 | 0 | 7 | 2 | 6.48 |
| F1-Z#82 | 108 | 10 | 90 | 0 | 7 | 1 | 6.48 |
| F1-Z#76 | 86 | 28 | 52 | 0 | 6 | 0 | 6.98 |
| F1-Z#78 | 78 | 0 | 72 | 0 | 6 | 0 | 7.69 |
| F1-Z#64 | 103 | 6 | 88 | 0 | 8 | 1 | 7.77 |
| F1-Z#92 | 112 | 1 | 101 | 0 | 9 | 1 | 8.04 |
| F1-Z#88 | 94 | 0 | 86 | 0 | 8 | 0 | 8.51 |
| F1-Z#65 | 105 | 3 | 93 | 0 | 9 | 0 | 8.57 |
| F1-Z#54 | 109 | 4 | 95 | 0 | 10 | 0 | 9.17 |
| F1-Z#60 | 82 | 9 | 64 | 0 | 9 | 0 | 10.98 |
| F1-Z#46 | 47 | 3 | 37 | 0 | 6 | 1 | 12.77 |
| F1-Z#72 | 79 | 1 | 67 | 1 | 10 | 0 | 13.92 |
| F1-Z#86 | 95 | 0 | 78 | 0 | 17 | 0 | 17.89 |
| R6#45 2 | 222 | 3 | 211 | 0 | 8 | 0 | 3.60 |
| R5#49 2 | 265 | 67 | 172 | 3 | 14 | 9 | 6.42 |
| F1-Z Mean (S.D.) | 4.56 (3.82) * | ||||||
| F1-C Mean (S.D.) | 9.31 (8.03) 3 | ||||||
| Individual | % AES (n) | % AES (n) | Mean (S.D) | |||
|---|---|---|---|---|---|---|
| Parental genotypes | F1-C#15 | 2017 | 6.97 (215) | 2018 | 9.90 (101) | 8.79 (1.81) 1 |
| F1-C#39 | 9.20 (239) | 9.33 (75) | ||||
| F2L #35 × #39 | F2H#7 | 2016 | 2.7(73) | 2017 | 0.9 (111) ns | 1.82 (0.01) |
| F2H#5 | 6.3 (112) | 7.7 (78) ns | 6.97 (0.01) | |||
| F2H#6 | 6.8 (88) | 5.0 (100) ns | 5.91 (0.01) | |||
| F2H#4 | 7.6 (118) | 6.5 (107) ns | 7.08 (0.01) | |||
| F2H#3 | 10.8 (102) | 9.4 (96) ns | 10.08 (0.01) | |||
| F2H#1 | 12.4 (105) | 9.9 (81) ns | 11.13 (0.02) | |||
| F2H#9 | 13.5 (74) | 6.0 (100) ns | 9.76 (0.05) | |||
| F2H#8 | 13.6 (103) | 4.8 (124) * | 7.27 (0.03) | |||
| F2H#2 | 16.7 (96) | 14.5 (55) ns | 15.61 (0.01) | |||
| F2L Mean (S.D.) | 10.05 (4.47) | 7.19 (3.83) | 8.62 (4.29) | |||
| Individual | % AES (n) | % AES (n) | Mean (S.D) | |||
|---|---|---|---|---|---|---|
| Parental genotypes | F1-C#12 | 2017 | 0.00 (168) | 2018 | 0.00 (128) | 1.40 (1.98) 1 |
| F1-C#9 | N.D. | 2.76 (145) | ||||
| F2L #9 × #12 | F2L#1 | 2016 | 0 (95) | 2017 | 0 (111) ns | 0 (0) |
| F2L#6 | 1.1 (94) | 0 (81) ns | 0.53 (0.75) | |||
| F2L#9 | 2.2 (90) | 2.1 (94) ns | 2.17 (0.07) | |||
| F2L#2 | 0.9 (110) | 1.1 (92) ns | 1 (0.13) | |||
| F2L#8 | 1 (101) | 1.1 (92) ns | 1.04 (0.07) | |||
| F2L#5 | 0.9 (109) | 2.5 (79) ns | 1.72 (1.14) | |||
| F2L#7 | 0 (60) | 2.9 (102) ns | 1.47 (2.08) | |||
| F2L#3 | 7 (71) | 5.4 (111) ns | 6.22 (1.16) | |||
| F2L#10 | 16.9 (83) | 9.7 (113) ns | 13.3 (5.04) | |||
| F2L Mean (S.D.) | 3.33 (5.51) | 2.75 (3.09) | 3.05 (4.34) | |||
| Parental | Number of Primer Combination | Number of Markers | Marker Segregation | ||
|---|---|---|---|---|---|
| SDAF (1:1) | BSDF (3:1) | Distorted 1 | |||
| R6#45 | 33 | 278 | 266 | - | 12 |
| R5#49 | 33 | 239 | 221 | - | 18 |
| R6#45-R6#49 | 33 | 110 | - | 94 | 16 |
| Marker | Type of Marker | LG (M/P) | Position (cM) (M/P) | b Parameter 1 | R2 | p-Value |
|---|---|---|---|---|---|---|
| C25_263.9 | Maternal | 2-M | 14.5 | 0.29 | 0.21 | 0.004 |
| B4_490.5 | Paternal | 3-P | 104.7 | 0.24 | 0.13 | 0.027 |
| C5_411.5 | Maternal | 3-M | 16.5 | −0.22 | 0.12 | 0.032 |
| C35_334 | Maternal | 3-M | 48.9 | −0.23 | 0.13 | 0.025 |
| B5_360.1 | Maternal | 3-M | 52.2 | −0.25 | 0.15 | 0.018 |
| C4_227.4 | Maternal | 3-M | 54.4 | −0.22 | 0.11 | 0.043 |
| C32_150.4 | Maternal | 3-M | 56.7 | −0.23 | 0.13 | 0.025 |
| C33_187.4 | Maternal | 3-M | 61.4 | −0.23 | 0.13 | 0.026 |
| XC31_2201 | Biparental | 4-M/4-P | 0.0/36.67 | 0.42 | 0.20 | 0.005 |
| XB4_324.5 | Biparental | 4-M/4-P | 18.8/56.65 | 0.27 | 0.12 | 0.036 |
| C23_279.7 | Paternal | 4-P | 75.2 | 0.21 | 0.10 | 0.049 |
| B1_236.6 | Maternal | 4-M | 21.6 | −0.22 | 0.34 | 0.042 |
| C32_244.8 | Maternal | 4-M | 37.7 | 0.29 | 0.19 | 0.006 |
| C25_292.5 | Maternal | 4-M | 44.4 | −0.21 | 0.10 | 0.047 |
| C33_384.1 (B16_164.6) 2 | Maternal | 4-M | 48.1 | −0.22 | 0.12 | 0.036 |
| C17_402.3 | Maternal | 4-M | 66.6 | 0.24 | 0.13 | 0.025 |
| B1_72.8 | Maternal | 5-M | 47.2 | −0.23 | 0.13 | 0.030 |
| C5_221.1 | Paternal | 10-P | 0.0 | 0.22 | 0.12 | 0.035 |
| B3_217.8 | Paternal | 10-P | 4.6 | 0.22 | 0.12 | 0.035 |
| C29_141.3 | Paternal | 10-P | 10.8 | 0.22 | 0.11 | 0.038 |
| C43_342.72 (C29_332.2) 2 | Maternal | 4-M | 38.3 | −0.29 | 0.19 | 0.006 |
| C8_142.52 (C32_244.8) 2 | Maternal | 4-M | 37.7 | 0.25 | 0.15 | 0.016 |
| C29_151.42 | Paternal | 10-P | 10.8 | 0.22 | 0.11 | 0.038 |
| B48_256.83 | Paternal | Unmapped | - | 0.22 | 0.12 | 0.032 |
| C5_194.8 | Paternal | Unmapped | - | 0.22 | 0.12 | 0.035 |
| XC5_1093 | Biparental | Unmapped | - | 0.25 | 0.11 | 0.039 |
| Marker | Type of Marker | LG (M/P) | Position (cM) (M/P) | b Parameter 1 | R2 | p-Value |
|---|---|---|---|---|---|---|
| XC4_1399 | Biparental | 1-P | 52.1 | 0.24 | 0.12 | 0.02 |
| C4_173.5 | Maternal | 3-M | 25.9 | 0.22 | 0.12 | 0.02 |
| C17_260.6 | Paternal | 3-P | 30.7 | 0.20 | 0.10 | 0.02 |
| C17_261.7 | Paternal | 3-P | 27.0 | 0.21 | 0.11 | 0.02 |
| B5_193.1 | Maternal | 3-M | 101.2 | 0.24 | 0.15 | 0.01 |
| XC4_2241 | Biparental | 3-M/3-P | 111.89/37.5 | 0.27 | 0.16 | 0.00 |
| XC25_3915 | Biparental | 6-P | 2.9 | −0.27 | 0.11 | 0.02 |
| B54_74.7 | Paternal | 6-P | 66.0 | −0.19 | 0.10 | 0.03 |
| C4_120.6 | Maternal | 7-M | 55.6 | 0.22 | 0.12 | 0.02 |
| B54_309.9 | Maternal | 7-M | 71.3 | −0.26 | 0.17 | 0.00 |
| C23_372.6 | Maternal | 8-M | 71.8 | −0.21 | 0.11 | 0.02 |
| C31_336.52 (C12_83) 2 | Maternal | 10-M | 28.7 | 0.25 | 0.14 | 0.01 |
| C12_214.8 | Maternal | Unmapped | - | 0.23 | 0.13 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, M.; Bocchini, M.; Stein, J.; Ortiz, J.P.A.; Albertini, E.; Delgado, L. Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum. Plants 2021, 10, 2100. https://doi.org/10.3390/plants10102100
Soliman M, Bocchini M, Stein J, Ortiz JPA, Albertini E, Delgado L. Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum. Plants. 2021; 10(10):2100. https://doi.org/10.3390/plants10102100
Chicago/Turabian StyleSoliman, Mariano, Marika Bocchini, Juliana Stein, Juan Pablo A. Ortiz, Emidio Albertini, and Luciana Delgado. 2021. "Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum" Plants 10, no. 10: 2100. https://doi.org/10.3390/plants10102100
APA StyleSoliman, M., Bocchini, M., Stein, J., Ortiz, J. P. A., Albertini, E., & Delgado, L. (2021). Environmental and Genetic Factors Affecting Apospory Expressivity in Diploid Paspalum rufum. Plants, 10(10), 2100. https://doi.org/10.3390/plants10102100