Factors Influencing Genomic Prediction Accuracies of Tropical Maize Resistance to Fall Armyworm and Weevils
Abstract
1. Introduction
2. Material and Methods
2.1. Genetic Material and Field Experiments
2.2. Genotyping, Quality Control, and Imputation for Genomic Prediction Analyses
2.3. FAW and MW Resistance Phenotyping
2.4. Statistical Analyses of the Phenotypic Data
2.5. Strategies for TS and BS Determination
2.5.1. MW Resistance Traits
2.5.2. FAW Damage Resistance
2.5.3. Random-Based TS Determination
2.5.4. Pedigree-Based TS Determination
2.6. Genomic Prediction Algorithms
2.6.1. Bayesian Models
2.6.2. Mixed Models
2.6.3. Machine Learning Algorithms
2.7. Cross-Validations and PA Estimation
3. Results
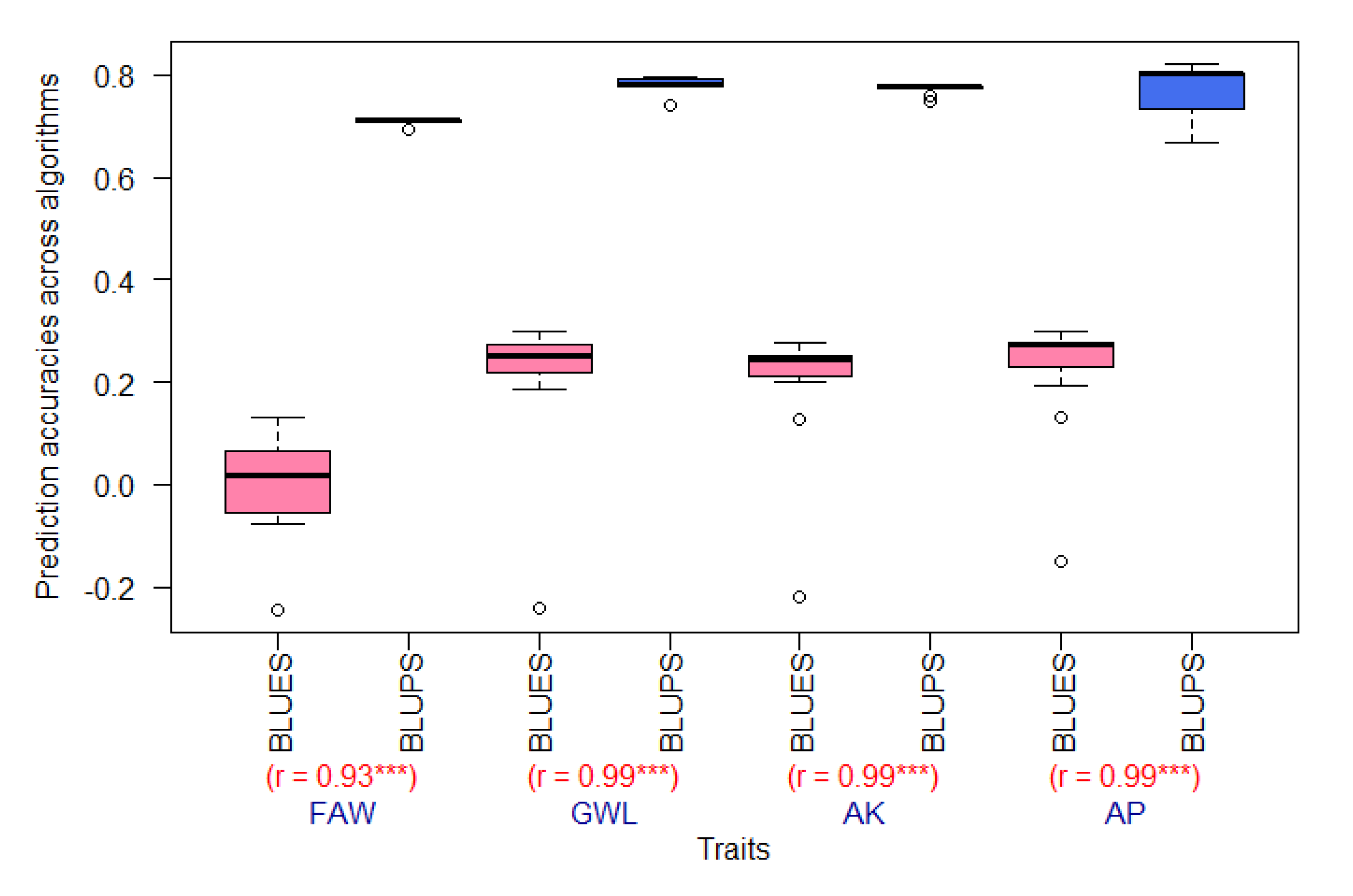
3.1. Higher PAs Achieved for FAW and MW-Resistance Traits with BLUPs when Compared to BLUEs across GP Algorithms
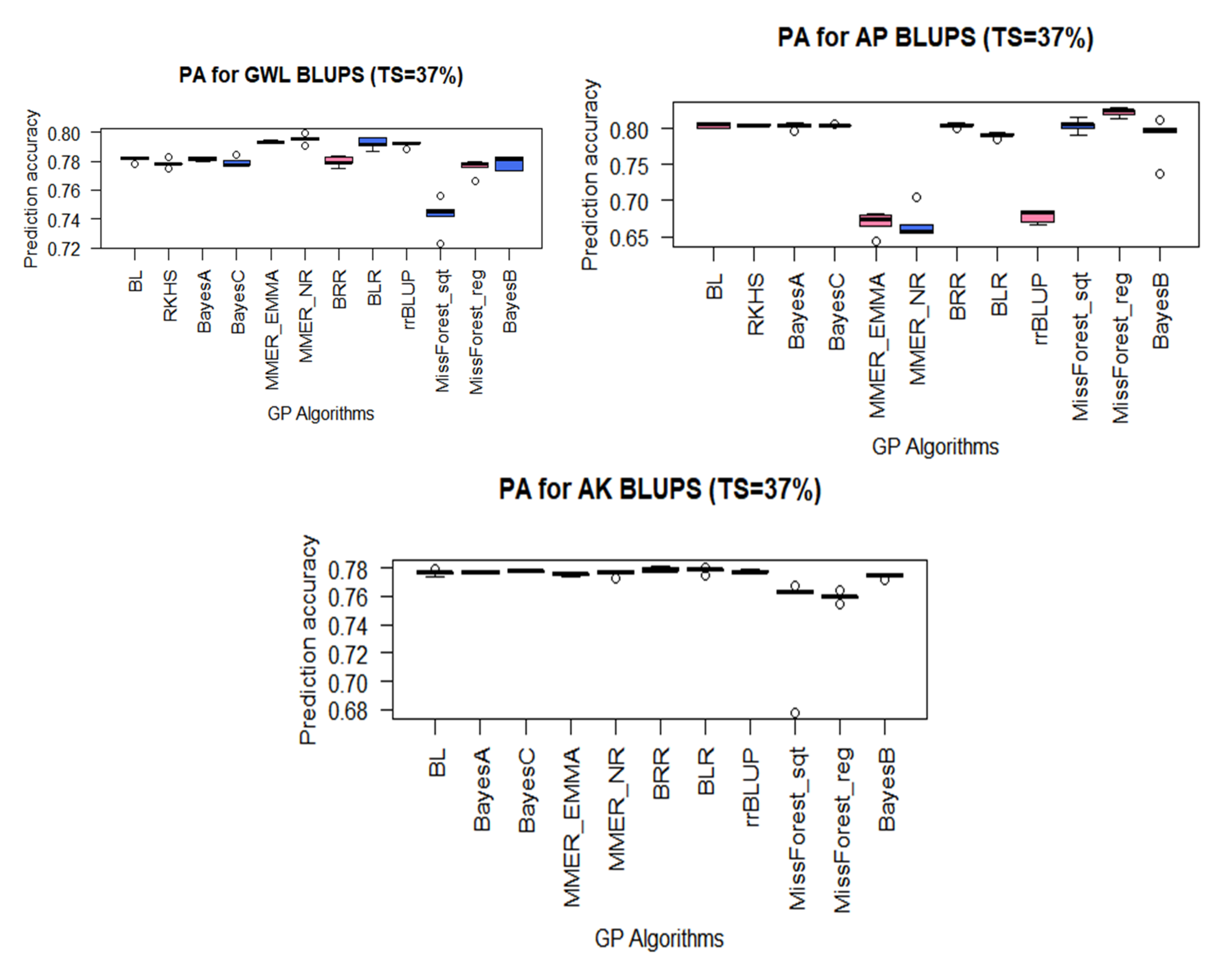
3.2. PAs for MW Resistance Traits Using BLUPs
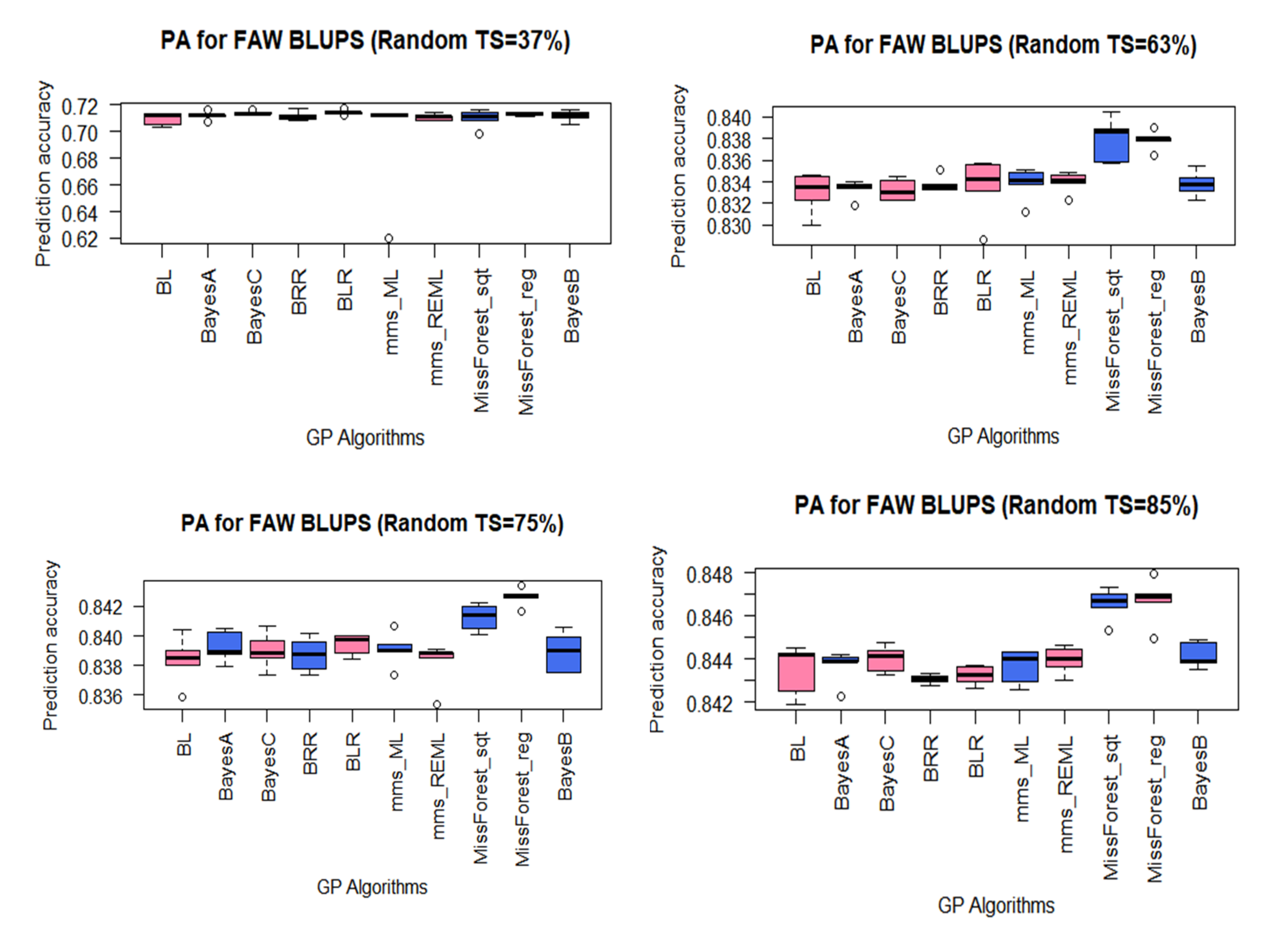
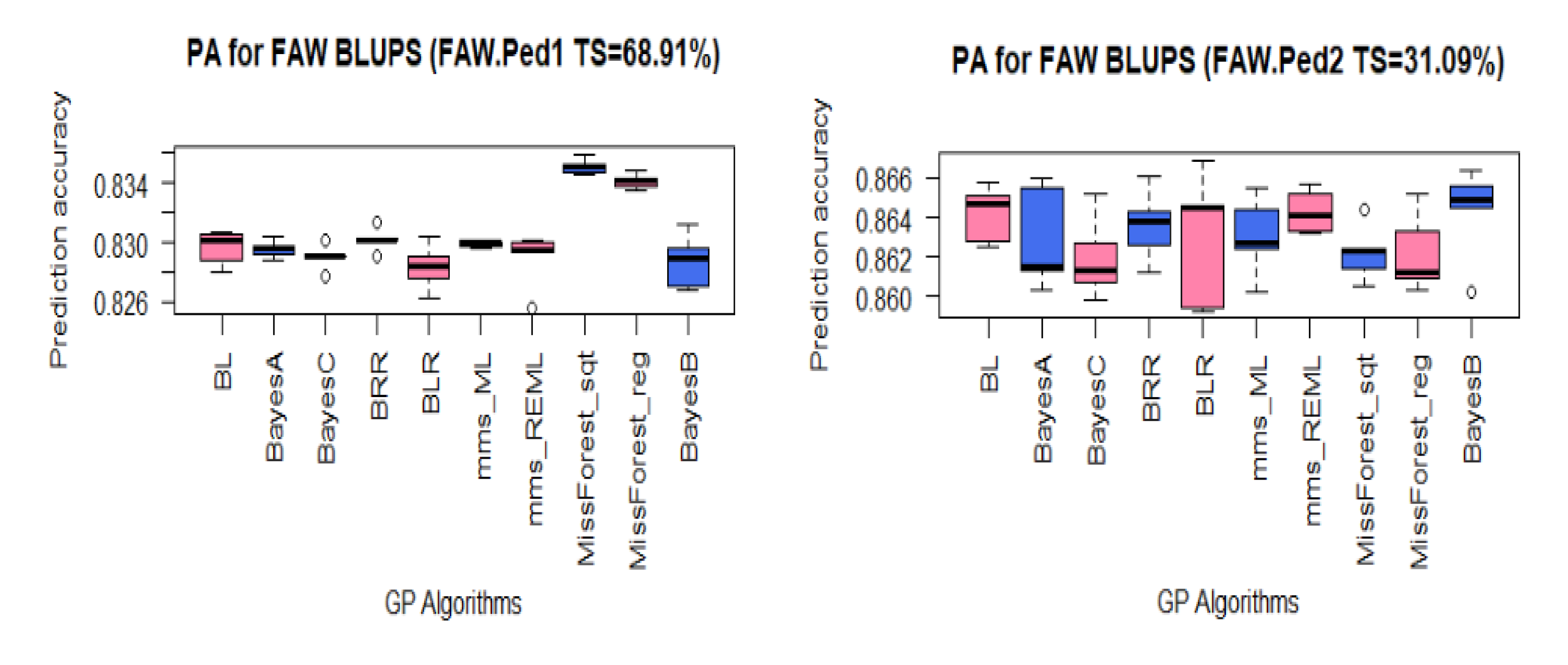
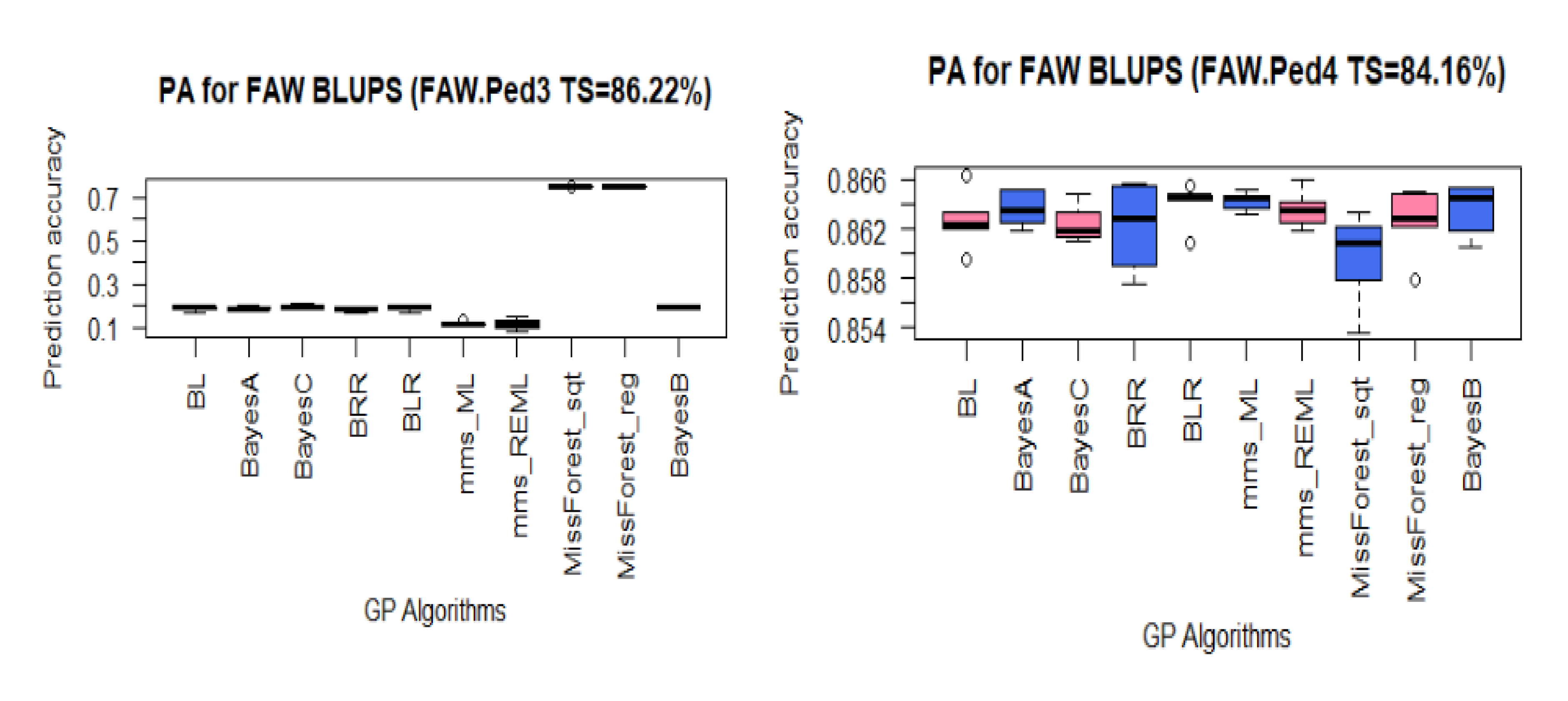
3.3. PA for FAW Resistance Using BLUPs
4. Discussion
4.1. Higher Pas Were Achieved for BLUPs Compared to BLUEs for Both FAW and MW-resistance Traits
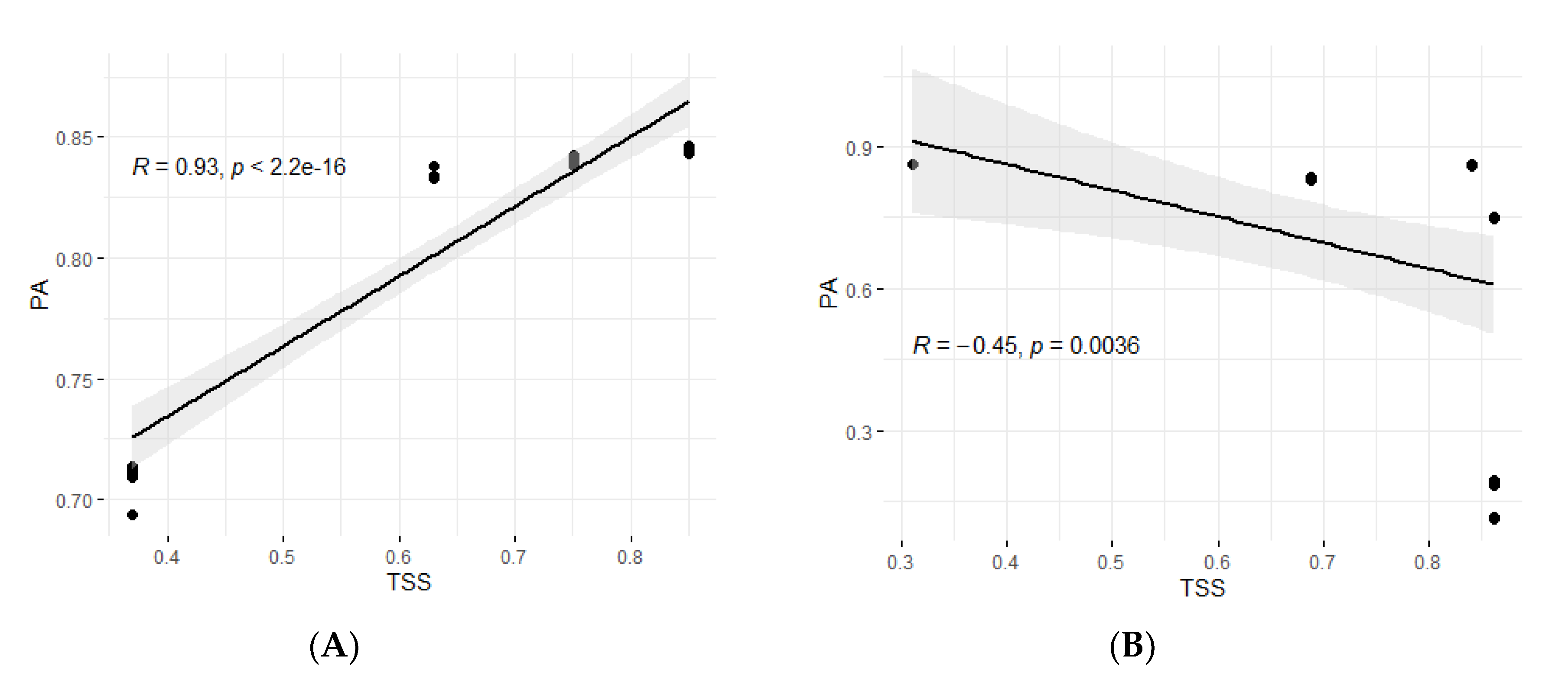
4.2. High PAs Were Achieved for FAW and MW-Resistance Traits Using Moderately Sized Training Sets
4.3. GP Algorithms Performed Differently on FAW and MW Maize Resistance Traits
4.4. Influences of the Sizes and the Compositions of TS and BS on PAs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Demissie, G.; Tefera, T.; Tadesse, A. Importance of husk covering on field infestation of maize by Sitophilus zeamais Motsch (Coleoptera: Curculionidea) at Bako, Western Ethiopia. Afr. J. Biotechnol. 2008, 7, 3777–3782. [Google Scholar]
- Shiferaw, B.; Prasanna, B.M.; Hellin, J.; Bänziger, M. Crops that feed the world 6. Past successes and future challenges to the role played by maize in global food security. Food Secur. 2011, 3, 307–327. [Google Scholar] [CrossRef]
- Awata, L.A.O.; Tongoona, P.; Danquah, E.; Ifie, B.E.; Suresh, L.M.; Jumbo, M.B.; Marchelo-D’ragga, P.W.; Sitonik, C. Understanding tropical maize (Zea mays L.): The major monocot in modernization and sustainability of agriculture in sub-Saharan Africa. Int. J. Adv. Agric. Res. 2019, 7, 32–77. [Google Scholar]
- Nyukuri, R.W.; Wanjala, F.M.; Kirui, S.C.; Cheramgoi, E.; Chirchir, E.; Mwale, R. Damage of stem borer species to Zea mays L.,Sorghum bicolor L. and three refugia graminae. Adv. Agric. Biol. 2014, 1, 37–45. [Google Scholar]
- Tefera, T.; Goftishu, M.; Ba, M.; Rangaswamy, M. A Guide to Biological Control of Fall Armyworm in Africa Using Egg Parasitoids, 1st ed.; ICIPE: Nairobi, Kenya, 2019. [Google Scholar]
- Munyiri, S.W.; Mugo, S.N.; Otim, M.; Mwololo, J.K.; Okori, P. Mechanisms and sources of resistance in tropical maize inbred lines to Chilo partellus stem borers. J. Agric. Sci. 2013, 5, 51–60. [Google Scholar] [CrossRef][Green Version]
- Mwololo, J.K.; Mugo, S.; Okori, P.; Tefera, T.; Otim, M.; Munyiri, S.W. Sources of resistance to the maize weevil Sitophilus zeamais in tropical maize. J. Agric. Sci. 2012, 4, 206–215. [Google Scholar]
- Mwololo, J.K. Resistance in Tropical Maize to the Maize Weevil and Larger Grain Borer. Ph.D. Thesis, Makerere University, Kampala, Uganda, 2013. [Google Scholar]
- Kasozi, L.C.; Derera, J.; Tongoona, P.; Tukamuhabwa, P.; Muwonge, A.; Asea, G. Genotypic variation for maize weevil resistance in eastern and southern Africa maize inbred lines. Uganda J. Agric. Sci. 2016, 17, 83–97. [Google Scholar] [CrossRef][Green Version]
- Tende, R.; Derera, J.; Mugo, S.; Oikeh, S. Estimation of genetic diversity of germplasm used to develop insect-pest resistant maize. Maydica 2016, 61, 1–8. [Google Scholar]
- Khakata, S.; Mbute, F.N.; Chemining’wa, G.N.; Mwimali, M.; Karanja, J.; Harvey, J.; Mwololo, J.K. Post-harvest evaluation of selected inbred lines to maize weevil Sitophilus zeamais resistance. J. Plant Breed. Crop Sci. 2018, 10, 105–114. [Google Scholar] [CrossRef][Green Version]
- Sodedji, F.K.A.; Kwemoi, D.B.; Kasozi, C.L.; Asea, G.; Kyamanywa, S. Genetic analysis for resistance to Sitophilus zeamais (Motschulsky) among provitamin-A maize germplasm. Maydica 2018, 63, 8. Available online: https://journals-crea.4science.it/index.php/maydica/article/view/1698 (accessed on 13 November 2019).
- Munyiri, W.S.; Mugo, N.S.; Otim, M.; Tefera, T.; Beyene, Y.; Mwololo, K.J.; Okori, P. Responses of tropical maize landraces to damage by Chilo partellus stem borer. Afr. J. Biotechnol. 2013, 12, 1229–1235. [Google Scholar]
- Munyiri, S.W.; Mugo, S.N.; Mwololo, J.K. Mechanisms and levels of resistance in hybrids, open pollinated varieties and landraces to Chilo partellus maize stem borers. Int. Res. J. Agric. Sci. Soil Sci. 2015, 5, 81–90. [Google Scholar]
- Mwololo, J.K.; Munyiri, S.W.; Semagn, K.; Mugo, S. Genetic diversity analysis in tropical maize germplasm for stem borer and storage pest resistance using molecular markers and phenotypic traits. Mol. Plant Breed. 2015, 6, 1–22. [Google Scholar]
- Goergen, G.; Kumar, P.L.; Sankung, S.B.; Togola, A.; Tamò, M. First report of outbreaks of the fall armyworm Spodoptera frugiperda (JE Smith) (Lepidoptera, Noctuidae), a new alien invasive pest in West and Central Africa. PLoS ONE 2016, 11, e0165632. [Google Scholar] [CrossRef]
- Padhee, A.K.; Prassanna, B.M. The emerging threat of Fall Armyworm in India. Indian Farming 2019, 69, 51–54. [Google Scholar]
- Prasanna, B.M.; Huesing, J.E.; Eddy, R.; Peschke, V.M.; Regina, E.; Virginia, M.P. Fall Armyworm in Africa: A Guide for Integrated Pest Management, 1st ed.; West Africa Regional Training of Trainers and Awareness Generation Workshop on Fall Armyworm Management, IITA, Cotonou, Bénin; Prasanna, B.M., Regina, E., Virginia, M.P., Eds.; CIMMYT: Queretaro, Mexico, 2018. [Google Scholar]
- Gedil, M.; Menkir, A. An integrated molecular and conventional breeding scheme for enhancing genetic gain in maize in Africa. Front. Plant Sci. 2019, 10, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Murenga, M.; Derera, J.; Mugo, S.; Tongoona, P. A review of genetic analysis and response to selection for resistance to Busseola fusca and Chilo partellus, stem borers in tropical maize germplasm: A Kenyan perspective. Maydica 2016, 61, 1–11. [Google Scholar]
- Crossa, J.; Pérez-Rodríguez, P.; Cuevas, J.; Montesinos-López, O.; Jarquín, D.; de los Campos, G.; Burgueno, J.; Gonzales-Camacho, J.M.; Perez-Elizade, S.; Beyene, Y.; et al. Genomic selection in plant breeding: Methods, models, and perspectives. Trends Plant Sci. 2017, 22, 1–15. [Google Scholar] [CrossRef]
- Roorkiwal, M.; Jarquin, D.; Singh, M.K.; Gaur, P.M.; Bharadwaj, C.; Rathore, A.; Howard, R.; Srinivasan, S.; Jain, A.; Garg, V.; et al. Genomic-enabled prediction models using multi-environment trials to estimate the effect of genotype × environment interaction on prediction accuracy in chickpea. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Robertsen, C.D.; Hjortshøj, R.L.; Janss, L.L. Genomic selection in cereal breeding. Agronomy 2019, 9, 95. [Google Scholar] [CrossRef]
- Munyiri, S.W.; Mugo, S.N. Quantitative trait loci for resistance to spotted and African maize stem borers (Chilo partellus and Busseola fusca) in a tropical maize (Zea mays L.) population. Afr. J. Biotechnol. 2017, 16, 1579–1589. [Google Scholar] [CrossRef]
- Badji, A.; Kwemoi, D.B.; Machida, L.; Okii, D.; Mwila, N.; Agbahoungba, S.; Kumi, F.; Ibanda, A.; Bararyenya, A.; Solemangey, M.; et al. Genetic basis of maize resistance to multiple-insect pests: Integrated genome-wide comparative mapping and candidate. Genes 2020, 11, 689. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, T.H.E.; Hayes, B.J.; Goddard, M.E. Prediction of total genetic value using genome-wide dense marker maps. Genetics 2001, 157, 1819–1829. [Google Scholar] [PubMed]
- Jannink, J.; Lorenz, A.J.; Iwata, H. Genomic selection in plant breeding: From theory to practice. Brief. Funct. Genom. 2010, 9, 166–177. [Google Scholar] [CrossRef]
- Massman, J.M.; Jung, H.J.G.; Bernardo, R. Genomewide selection versus marker-assisted recurrent selection to improve grain yield and stover-quality traits for cellulosic ethanol in maize. Crop Sci. 2013, 53, 58–66. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oukeh, S.; et al. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Vivek, B.S.; Krishna, G.K.; Vengadessan, V.; Babu, R.; Zaidi, P.H.; Kha, L.Q.; Mandal, S.S.; Grudloyma, P.; Takalkar, S.; Krothapalli, K.; et al. Use of genomic estimated breeding values results in rapid genetic gains for drought tolerance in maize. Plant Genome 2017, 10, 1–8. [Google Scholar] [CrossRef]
- Liu, X.; Wang, H.; Wang, H.; Guo, Z.; Xu, X.; Liu, J.; Wang, S.; Li, W.-X.; Zou, C.; Prasanna, B.M.; et al. Factors affecting genomic selection revealed by empirical evidence in maize. Crop J. 2018, 6, 341–352. [Google Scholar] [CrossRef]
- Yuan, Y.; Scheben, A.; Batley, J.; Edwards, D. Using genomics to adapt crops to climate change. In Sustainable Solutions for Food Security; Springer International Publishing: Cham, Switzerland, 2019; pp. 91–109. [Google Scholar]
- Zhang, X.; Pérez-Rodríguez, P.; Burgueño, J.; Olsen, M.; Buckler, E.; Atlin, G.; Prasanna, B.M.; Vargas, M.; San Vicente, F.; Crossa, J. Rapid cycling genomic selection in a multiparental tropical maize population. G3 Genes Genomes Genet. 2017, 7, 2315–2326. [Google Scholar] [CrossRef]
- Hassen, M.B.; Bartholomé, J.; Valè, G.; Cao, T.V.; Ahmadi, N. Genomic prediction accounting for genotype by environment interaction offers an effective framework for breeding simultaneously for adaptation to an abiotic stress and performance under normal cropping conditions in rice. G3 Genes Genomes Genet. 2018, 8, 2319–2332. [Google Scholar] [CrossRef]
- Muleta, K.T.; Pressoir, G.; Morris, G.P. Optimizing genomic selection for a sorghum breeding program in Haiti: A simulation study. G3 Genes Genomes Genet. 2019, 9, 391–401. [Google Scholar] [CrossRef]
- Suontama, M.; Klápště, J.; Telfer, E.; Graham, N.; Stovold, T.; Low, C.; McKinley, R.; Dungey, H. Efficiency of genomic prediction across two Eucalyptus nitens seed orchards with different selection histories. Heredity 2019, 122, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Voss-Fels, K.P.; Cooper, M.; Hayes, B.J. Accelerating crop genetic gains with genomic selection. Appl. Genet. 2019, 132, 669–686. [Google Scholar] [CrossRef] [PubMed]
- Kadam, D.C.; Lorenz, A.J. Evaluation of nonparametric models for genomic prediction of early-stage single crosses in maize. Crop Sci. 2019, 59, 1411–1423. [Google Scholar] [CrossRef]
- Whittaker, J.C.; Thompson, R.; Denham, M.C. Marker-assisted selection using ridge regression. Genet. Res. 2000, 75, 249–252. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.A.; van der Werf, J. Genomic Best Linear Unbiased Prediction (gBLUP) for the Estimation of Genomic Breeding Values; Springer: Berlin/Heidelberg, Germany, 2013; pp. 321–330. [Google Scholar]
- Zhang, H.; Yin, L.; Wang, M.; Yuan, X.; Liu, X. Factors affecting the accuracy of genomic selection for agricultural economic traits in maize, cattle, and pig populations. Front. Genet. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, T.; Iwata, H. EM algorithm for Bayesian estimation of genomic breeding values. BMC Genet. 2010, 11, 3. [Google Scholar] [CrossRef]
- Meuwissen, T.H.; Solberg, T.R.; Shepherd, R.; Woolliams, J.A. A fast algorithm for BayesB type of prediction of genome-wide estimates of genetic value. Genet. Sel. Evol. 2009, 41, 1–10. [Google Scholar] [CrossRef]
- Habier, D.; Fernando, R.L.; Kizilkaya, K.; Garrick, D.J. Extension of the bayesian alphabet for genomic selection. BMC Bioinform. 2011, 12, 186. [Google Scholar] [CrossRef]
- De Los Campos, G.; Naya, H.; Gianola, D.; Crossa, J.; Legarra, A.; Manfredi, E.; Weigel, K.; Cotes, J.M. Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 2009, 182, 375–385. [Google Scholar] [CrossRef]
- Zhou, X.; Carbonetto, P.; Stephens, M. Polygenic Modeling with Bayesian Sparse Linear Mixed Models. PLoS Genet. 2013, 9, e1003264. [Google Scholar] [CrossRef] [PubMed]
- Gianola, D.; Fernando, R.L.; Stella, A. Genomic-assisted prediction of genetic value with semiparametric procedures. Genetics 2006, 173, 1761–1776. [Google Scholar] [CrossRef] [PubMed]
- Gianola, D.; de los Campos, G.; González-Recio, O.; Long, N.; Okut, H.; Rosa, G.J.M.; Weigel, K.A.; Wu, X.-L. Statistical learning methods for genome-based analysis of quantitative traits. In Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany, 1–6 August 2010; Gesellschaft für Tierzuchtwissenschaften eV: Neustadt am Rübenberge, Germany, 2010; Volume 14, pp. 1–6. [Google Scholar]
- Howard, R.; Carriquiry, A.L.; Beavis, W.D. Parametric and nonparametric statistical methods for genomic selection of traits with additive and epistatic genetic architectures. G3 Genes Genomes Genet. 2014, 4, 1027–1046. [Google Scholar] [CrossRef] [PubMed]
- De los Campos, G.; Hickey, J.M.; Pong-Wong, R.; Daetwyler, H.D.; Calus, M.P.L. Whole-genome regression and prediction methods applied to plant and animal breeding. Genetics 2013, 193, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Heslot, N.; Yang, H.P.; Sorrells, M.E.; Jannink, J.L. Genomic selection in plant breeding: A comparison of models. Crop Sci. 2012, 52, 146–160. [Google Scholar] [CrossRef]
- Maltecca, C.; Parker, K.L.; Cassady, J.P.; Maltecca, C.; Parker, K.L.; Cassady, J.P. Application of multiple shrinkage methods to genomic predictions. J. Anim. Sci. 2014, 90, 1777–1787. [Google Scholar] [CrossRef]
- E Sousa, M.B.; Cuevas, J.; de Oliveira Couto, E.G.; Pérez-Rodríguez, P.; Jarquín, D.; Fritsche-Neto, R.; Burgueno, J.; Crossa, J. Genomic-enabled prediction in maize using Kernel models with genotype × environment interaction. G3 Genes Genomes Genet. 2017, 7, 1995–2014. [Google Scholar]
- Cuevas, J.; Granato, I.; Fritsche-Neto, R.; Montesinos-Lopez, O.A.; Burgueño, J.; e Sousa, M.B.; Crossa, J. Genomic-enabled prediction Kernel models with random intercepts for multi-environment trials. G3 Genes Genomes Genet. 2018, 8, 1347–1365. [Google Scholar] [CrossRef]
- Bellot, P.; de los Campos, G.; Pérez-Enciso, M. Can deep learning improve genomic prediction of complex human traits? Genetics 2018, 210, 809–819. [Google Scholar] [CrossRef]
- Montesinos-López, O.A.; Martín-Vallejo, J.; Crossa, J.; Gianola, D.; Hernández-Suárez, C.M.; Montesinos-López, A.; Juliana, P.; Singh, R. A benchmarking between deep learning, support vector machine and Bayesian threshold best linear unbiased prediction for predicting ordinal traits in plant breeding. G3 Genes Genomes Genet. 2019, 9, 601–618. [Google Scholar] [CrossRef]
- Crossa, J.; Beyene, Y.; Semagn, K.; Pérez, P.; Hickey, J.M.; Chen, C.; de los Campos, G.; Burgueno, J.; Windhausen, V.S.; Buckler, E.; et al. Genomic prediction in maize breeding populations with genotyping-by-sequencing. G3 Genes Genomes Genet. 2013, 3, 1903–1926. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Rodríguez, P.; Gianola, D.; González-Camacho, J.M.; Crossa, J.; Manès, Y.; Dreisigacker, S. Comparison between linear and non-parametric regression models for genome-enabled prediction in wheat. G3 Genes Genomes Genet. 2012, 2, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- De Andrade, L.R.B.; Bandeira e Sousa, M.; Oliveira, E.J.; de Resende, M.D.V.; Azevedo, C.F. Cassava yield traits predicted by genomic selection methods. PLoS ONE 2019, 14, e0224920. [Google Scholar] [CrossRef] [PubMed]
- Hickey, J.M.; Chiurugwi, T.; Mackay, I.; Powell, W. Genomic prediction unifies animal and plant breeding programs to form platforms for biological discovery. Nat. Genet. 2017, 49, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Akdemir, D.; Isidro-sánchez, J. Design of training populations for selective phenotyping in genomic prediction. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef]
- Gosal, S.S.; Wani, S.H. (Eds.) Accelerated Plant Breeding, Volume 1: Cereal Crops; Springer Nature; Springer International Publishing: Cham, Switzerland, 2020; Volume 1. [Google Scholar]
- Sarinelli, J.M.; Murphy, J.P.; Tyagi, P.; Holland, J.B.; Johnson, J.W.; Mergoum, M.; Mason, R.E.; Babar, A.; Harrison, S.; Sutton, R.; et al. Training population selection and use of fixed effects to optimize genomic predictions in a historical USA winter wheat panel. Theor. Appl. Genet. 2019, 132, 1247–1261. [Google Scholar] [CrossRef]
- Cericola, F.; Jahoor, A.; Orabi, J.; Andersen, J.R.; Janss, L.L.; Jensen, J. Optimizing training population size and genotyping strategy for genomic prediction using association study results and pedigree information. A case of study in advanced wheat breeding lines. PLoS ONE 2017, 12, e0169606. [Google Scholar] [CrossRef]
- Isidro, J.; Jannink, J.L.; Akdemir, D.; Poland, J.; Heslot, N.; Sorrells, M.E. Training set optimization under population structure in genomic selection. Theor. Appl. Genet. 2015, 128, 145–158. [Google Scholar] [CrossRef]
- Andres, R.J.; Dunne, J.C.; Samayoa, L.F.; Holland, J.B. Enhancing Crop Breeding Using Population Genomics Approaches; Springer: Berlin/Heidelberg, Germany, 2020. [Google Scholar]
- Sitonik, C.; Suresh, L.M.; Beyene, Y.; Olsen, M.S.; Makumbi, D.; Oliver, K.; Das, B.; Bright, J.M.; Mugo, S.; Crossa, J.; et al. Genetic architecture of maize chlorotic mottle virus and maize lethal necrosis through GWAS, linkage analysis and genomic prediction in tropical maize germplasm. Theor. Appl Genet. 2019, 132, 2381–2399. [Google Scholar] [CrossRef]
- Nyaga, C.; Gowda, M.; Beyene, Y.; Muriithi, W.T.; Makumbi, D.; Olsen, M.S.; Suresh, L.M.; Bright, J.M.; Das, B.; Prasanna, B.M. Genome-wide analyses and prediction of resistance to mln in large tropical maize germplasm. Genes 2020, 11, 16. [Google Scholar] [CrossRef]
- Gowda, M.; Das, B.; Makumbi, D.; Babu, R.; Semagn, K.; Mahuku, G.; Olsen, M.S.; Bright, J.M.; Beyene, Y.; Prasanna, B.M. Genome-wide association and genomic prediction of resistance to maize lethal necrosis disease in tropical maize germplasm. Theor. Appl Genet. 2015, 128, 1957–1968. [Google Scholar] [CrossRef] [PubMed]
- Galli, G.; Lyra, D.H.; Alves, F.C.; Granato, Í.S.C.; e Sousa, M.B.; Fritsche-Neto, R. Impact of phenotypic correction method and missing phenotypic data on genomic prediction of maize hybrids. Crop Sci. 2018, 58, 1481–1491. [Google Scholar] [CrossRef]
- Molenaar, H.; Boehm, R.; Piepho, H.P. Phenotypic selection in ornamental breeding: It’s better to have the BLUPs than to have the BLUEs. Front. Plant Sci. 2018, 871, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Piepho, H.P.; Möhring, J.; Melchinger, A.E.; Büchse, A. BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 2008, 161, 209–228. [Google Scholar] [CrossRef]
- Piepho, H.P.; Möhring, J. Selection in cultivar trials—Is it ignorable? Crop Sci. 2006, 46, 192–201. [Google Scholar] [CrossRef]
- Dramadri, I.O.; Nkalubo, S.T.; Kelly, J.D. Identification of QTL Associated with drought tolerance in Andean common bean. Crop Sci. 2019, 59, 1007–1020. [Google Scholar] [CrossRef]
- Williams, W.P.; Buckley, P.M.; Davis, F.M. Combining ability for resistance in corn to fall armyworm and southwestern corn borer. Crop Sci. 1989, 29, 913–915. [Google Scholar] [CrossRef]
- Sodedji, F.A.K.; Kwemoi, D.B.; Asea, G.; Kyamanywa, S. Response of provitamin—A maize germplasm to storage weevil Sitophilus zeamais (Motschulsky). Int. J. Agron. Agric. Res. 2016, 9, 1–13. [Google Scholar]
- Kasozi, L.C.; Derera, J.; Tongoona, P.; Zziwa, S.; Foundation, M.G.; Box, P.O. Comparing the effectiveness of the “weevil warehouse” and “laboratory bioassay” as techniques for screening maize genotypes for weevil resistance. J. Food Secur. 2018, 6, 170–177. [Google Scholar]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2007, 23, 2633–2635. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Development Core Team, Ed.; R Foundation for Statistical Computing: Vienna, Austria, 2011; Volume 1, p. 409. [Google Scholar]
- Bates, D.M.; Maechler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- De los Campos, G.; Pérez, P.; Vazquez, A.I.; Crossa, J. Genome-enabled prediction using the BLR (Bayesian linear regression) R-package. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 299–320. [Google Scholar]
- Pérez, P.; de los Campos, G. BGLR: A statistical package for whole genome regression and prediction. Genetics 2014, 198, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; de los Campos, G.; Crossa, J.; Gianola, D. Genomic-enabled prediction based on molecular markers and pedigree using the bayesian linear regression package in R. Plant Genome J. 2010, 3, 106. [Google Scholar] [CrossRef]
- Park, T.; Casella, G. The Bayesian Lasso. J. Am. Stat. Assoc. 2008, 103, 681–686. [Google Scholar] [CrossRef]
- Covarrubias-Pazaran, G. Genome-assisted prediction of quantitative traits using the R package sommer. PLoS ONE 2016, 11, e0156744. [Google Scholar] [CrossRef]
- Henderson, C.R.; Henderson, A.C.R. Best linear unbiased estimation and prediction under a selection model published by: International biometric society stable. Biometrics 1975, 31, 423–447. [Google Scholar] [CrossRef]
- Kang, H.M.; Zaitlen, N.A.; Wade, C.M.; Kirby, A.; Heckerman, D.; Daly, M.J.; Eskin, E. Efficient control of population structure in model organism association mapping. Genetics 2008, 178, 1709–1723. [Google Scholar] [CrossRef]
- Gilmour, A.R.; Thompson, R.; Cullis, B.R. Average information REML: An efficient algorithm for variance parameter estimation in linear mixed models. Biometrics 1995, 51, 1440–1450. [Google Scholar] [CrossRef]
- Searle, S.R. Applying the EM algorithm to calculating ML and REML estimates of variance components. In Proceedings of the American Statistical Association Meetings, San Francisco, CA, USA, 8–12 August 1993; pp. 1–9. [Google Scholar]
- Tunnicliffe, G.W. On the use of marginal likelihood in time series model estimation. JRSS 1989, 51, 15–27. [Google Scholar]
- Vanraden, P.M. Efficient methods to compute genomic predictions. J. Dairy Sci. 2008, 91, 4414–4423. [Google Scholar] [CrossRef]
- Endelman, J.B. Ridge regression and other kernels for genomic selection with R package rrBLUP. Plant Genome J. 2011, 4, 250. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. Missforest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ishwaran, H. Genomics random forests for genomic data analysis. Genomics 2012, 99, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Romay, M.C.; Millard, M.J.; Glaubitz, J.C.; Peiffer, J.A.; Swarts, K.L.; Casstevens, T.M.; Elshire, R.J.; Acharya, C.B.; Mitchell, S.E.; Flint-Garcia, S.A.; et al. Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol. 2013, 14, R55. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Flint-Garcia, S.A.; De Leon, N.; McMullen, M.D.; Kaeppler, S.M.; Buckler, E.S. The genetic architecture of maize stalk strength. PLoS ONE 2013, 8, e67066. [Google Scholar] [CrossRef]
- Lorenz, A.J.; Smith, K.P.; Jannink, J.L. Potential and optimization of genomic selection for Fusarium head blight resistance in six-row barley. Crop Sci. 2012, 52, 1609–1621. [Google Scholar] [CrossRef]
- Arruda, M.P.; Brown, P.J.; Lipka, A.E.; Krill, A.M.; Thurber, C.; Kolb, F.L. Genomic selection for predicting fusarium head blight resistance in a wheat breeding program. Plant Genome 2015, 8, 1–12. [Google Scholar] [CrossRef]
- Foiada, F.; Westermeier, P.; Kessel, B.; Ouzunova, M.; Wimmer, V.; Mayerhofer, W.; Presterl, T.; Dilger, M.; Kreps, R.; Eder, J.; et al. Improving resistance to the European corn borer: A comprehensive study in elite maize using QTL mapping and genome-wide prediction. Theor. Appl Genet. 2015, 128, 875–891. [Google Scholar] [CrossRef]
- Riedelsheimer, C.; Technow, F.; Melchinger, A.E. Comparison of whole-genome prediction models for traits with contrasting genetic architecture in a diversity panel of maize inbred lines. BMC Genom. 2012, 13, 452. [Google Scholar] [CrossRef]
- Azodi, C.B.; Bolger, E.; McCarren, A.; Roantree, M.; de Los Campos, G.; Shiu, S.H. Benchmarking parametric and Machine Learning models for genomic prediction of complex traits. G3 Genes Genomes Genet. 2019, 9, 3691–3702. [Google Scholar] [CrossRef]
- Drouaillet, B.E.; Mendez, C.A.R. Combinatorial aptitude and resistance to leaf damage of Spodoptera frugiperda (J.E. Smith) in maize germplasm native to Tamaulipas. Rev. Mex. Cienc. Agríc. 2018, 9, 81–93. [Google Scholar]
- Alvarez, P.; Branco, J.; Filho, D.M. Diallel crossing among miaze populations for resistance to fall armyworm. Sci. Agric. 2002, 59, 731–741. [Google Scholar] [CrossRef]
- Viana, P.A.; Guimarães, P.E.O. Maize resistance to the lesser cornstalk borer and fall armyworm in Brazil. In Embrapa Milho e Sorgo-Artigo em Anais de Congresso (ALICE), Proceedings of the International Symposium on Insect Resistant Maize: Recent Advances and Utilization, Mexico City, Mexico, 27 November–3 December 1994; Centro Internacional de Mejoramiento de Maiz y Trigo (CIMMYT): Mexico City, Mexico, 1997. [Google Scholar]
- Musundire, L.; Dari, S.; Derera, J.; Co-Zimbabwe, S.; Wgt, P.O.B. Genetic analysis of grain yield performance and weevil [Sitophilus zeamais (Motschulsky)] resistance in southern African maize hybrids. Maydica 2015, 60, M35. [Google Scholar]
- Dhliwayo, T.; Pixley, K.V.; Kazembe, V. Combining ability for resistance to maize weevil among 14 southern African maize inbred lines. Crop Sci. 2005, 45, 662–667. [Google Scholar] [CrossRef]
- Asoro, F.G.; Newell, M.A.; Beavis, W.D.; Scott, M.P.; Jannink, J.-L. Accuracy and training population design for genomic selection on quantitative traits in elite North American oats. Plant Genome J. 2011, 4, 132. [Google Scholar] [CrossRef]
- Lenz, P.R.N.; Beaulieu, J.; Mansfield, S.D.; Clément, S.; Desponts, M.; Bousquet, J. Factors affecting the accuracy of genomic selection for growth and wood quality traits in an advanced-breeding population of black spruce (Picea mariana). BMC Genom. 2017, 18, 1–17. [Google Scholar] [CrossRef]
- Edwards, S.M.K.; Buntjer, J.B.; Jackson, R.; Bentley, A.R.; Lage, J.; Byrne, E.; Burt, C.; Jack, P.; Berry, S.; Flatman, E.; et al. The effects of training population design on genomic prediction accuracy in wheat. Theor. Appl Genet. 2019, 132, 1943–1952. [Google Scholar] [CrossRef]
- Wang, W.; Cao, X.H.; Miclǎu, M.; Xu, J.; Xiong, W. The promise of agriculture genomics. Int. J. Genom. 2017, 2017. [Google Scholar] [CrossRef]
- Spindel, J.; Iwata, H. Genomic selection in rice breeding. In Rice Genomics, Genet Breed; Springer: Singapore, 2018; pp. 473–496. [Google Scholar]
- Crossa, J.; Pérez, P.; Hickey, J.; Burgueño, J.; Ornella, L.; Cerón-Rojas, J.; Zhang, X.; Dreisigacker, S.; Babu, R.; Li, Y.; et al. Genomic prediction in CIMMYT maize and wheat breeding programs. Heredity 2014, 112, 48–60. [Google Scholar] [CrossRef]
- Ou, J.H.; Liao, C.T. Training set determination for genomic selection. Theor. Appl. Genet. 2019, 132, 2781–2792. [Google Scholar] [CrossRef]
- Mangin, B.; Rincent, R.; Rabier, C.E.; Moreau, L.; Goudemand-Dugue, E. Training set optimization of genomic prediction by means of EthAcc. PLoS ONE 2019, 14, e0205629. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, T.; Wimmer, V.; Auinger, H.J.; Erbe, M.; Knaak, C.; Ouzunova, M.; Simianer, H.; Schon, C.-C. Genome-based prediction of testcross values in maize. Theor. Appl. Genet. 2011, 123, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Hickey, J.M.; Dreisigacker, S.; Crossa, J.; Hearne, S.; Babu, R.; Prasanna, B.M.; Grondona, M.; Zambelli, A.; Windhausen, V.S.; Mathews, K.; et al. Evaluation of genomic selection training population designs and genotyping strategies in plant breeding programs using simulation. Crop Sci. 2014, 54, 1476–1488. [Google Scholar] [CrossRef]
- Krchov, L.-M.; Bernardo, R. Relative efficiency of genomewide selection for testcross performance of doubled haploid lines in a maize breeding program. Crop Sci. 2015, 55, 2091–2099. [Google Scholar] [CrossRef]
- Mayor, P.J.; Bernardo, R. Genomewide selection and marker-assisted recurrent selection in doubled haploid versus F2 populations. Crop Sci. 2009, 49, 1719–1725. [Google Scholar] [CrossRef]

| Locations | Geographical Coordinates | Altitude above Sea Level | Minimum Rainfall | Soil Characteristics |
|---|---|---|---|---|
| Kasese | 0°16′10″ N 30°6′9″ E | 1330 m asl | 1000 mm | Sandy loam soils with a pH of 5.68 |
| Namulonge | 0°31′30″ N 32°36′54″ E | 1160 m asl | 1300 mm | Oxisols with a pH of 5.8 |
| FAW Datasets | FAW.Ped1 | FAW.Ped2 | FAW.Ped3 | FAW.Ped4 |
|---|---|---|---|---|
| TS composition | 235 DH CIMMYT lines | 106 Non-DH lines | 294 Non-CIMMYT SB and SP resistant lines | 287 DH and CIMMYT and IITA SB and SP lines |
| TS/Panel (%) | 68.91 | 31.09 | 86.22 | 84.16 |
| GP Algorithms | Abbreviations | Method Type | |
|---|---|---|---|
| 1 | Sommer with Average Information (AI) | mmer_AI | Parametric/Mixed model |
| 2 | Sommer with Expectation Maximization (EM) | mmer_EM | Parametric/Mixed model |
| 3 | Sommer with Efficient Mixed Model Association (EMMA) | mmer_EMMA | Parametric/Mixed model |
| 4 | Sommer with default Newton-Raphson (NR) | mmer-NR | Parametric/Mixed model |
| 5 | Ridge-regression Best linear unbiased Predictor | rrBLUP | Parametric/Mixed model |
| 6 | Mixed Model solution with Maximum Likelihood (ML) | mms_ML | Parametric/Mixed model |
| 7 | Mixed Model solution with Restricted Maximum Likelihood (REML) | mms_REML | Parametric/Mixed model |
| 8 | BayesB | BayesB | Parametric/Bayesian |
| 9 | BayesA | BayesA | Parametric/Bayesian |
| 10 | BayesC | BayesC | Parametric/Bayesian |
| 11 | Bayesian least absolute shrinkage and selection operator (LASSO) | BL | Parametric/Bayesian |
| 12 | Bayesian Ridge Regression | BRR | Parametric/Bayesian |
| 13 | Bayesian Linear Regression | BLR | Parametric/Bayesian |
| 14 | Reproducible kernel Hilbert space | RKHS | Semi-parametric/Bayesian |
| 15 | Random Forest with Square root | missForest_Sqt | Nonparametric/Machine Learning |
| 16 | Random Forest with Regression | missForest_Reg | Nonparametric/Machine Learning |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Badji, A.; Machida, L.; Kwemoi, D.B.; Kumi, F.; Okii, D.; Mwila, N.; Agbahoungba, S.; Ibanda, A.; Bararyenya, A.; Nghituwamhata, S.N.; et al. Factors Influencing Genomic Prediction Accuracies of Tropical Maize Resistance to Fall Armyworm and Weevils. Plants 2021, 10, 29. https://doi.org/10.3390/plants10010029
Badji A, Machida L, Kwemoi DB, Kumi F, Okii D, Mwila N, Agbahoungba S, Ibanda A, Bararyenya A, Nghituwamhata SN, et al. Factors Influencing Genomic Prediction Accuracies of Tropical Maize Resistance to Fall Armyworm and Weevils. Plants. 2021; 10(1):29. https://doi.org/10.3390/plants10010029
Chicago/Turabian StyleBadji, Arfang, Lewis Machida, Daniel Bomet Kwemoi, Frank Kumi, Dennis Okii, Natasha Mwila, Symphorien Agbahoungba, Angele Ibanda, Astere Bararyenya, Selma Ndapewa Nghituwamhata, and et al. 2021. "Factors Influencing Genomic Prediction Accuracies of Tropical Maize Resistance to Fall Armyworm and Weevils" Plants 10, no. 1: 29. https://doi.org/10.3390/plants10010029
APA StyleBadji, A., Machida, L., Kwemoi, D. B., Kumi, F., Okii, D., Mwila, N., Agbahoungba, S., Ibanda, A., Bararyenya, A., Nghituwamhata, S. N., Odong, T., Wasswa, P., Otim, M., Ochwo-Ssemakula, M., Talwana, H., Asea, G., Kyamanywa, S., & Rubaihayo, P. (2021). Factors Influencing Genomic Prediction Accuracies of Tropical Maize Resistance to Fall Armyworm and Weevils. Plants, 10(1), 29. https://doi.org/10.3390/plants10010029









