Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data
Abstract
1. Introduction
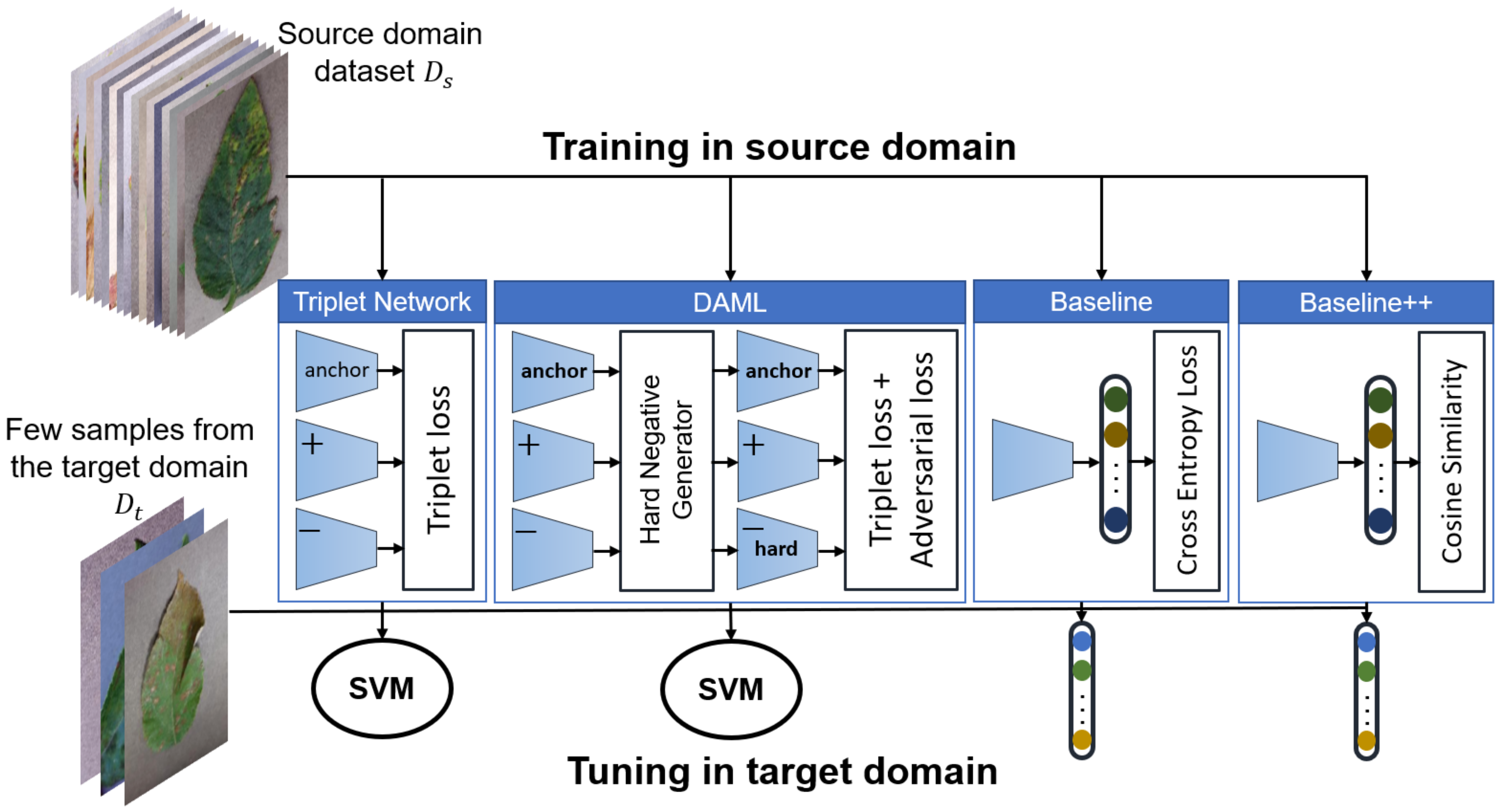
- Development and evaluation of several FSL approaches to classify plant diseases based on Triplet network, Deep Adversarial Metric Learning (DAML) [40] and transfer learning using both linear and cosine-similarity classifiers [23]. All approaches were evaluated using images captured under the same as well as different conditions. We have found that fine-tuning of a pretrained model outperforms all other approaches. Figure 1 shows an overview of the approaches developed in this study.
- We investigate the impact of model complexity on the performance of the FSL approaches, ResNet18, ResNet34, and ResNet50 were used in this study. The results showed that fine-tuning using linear classifiers benefits from increased model complexity while this cannot be confirmed for other approaches.
- We examined two different formulations in this study. In the first, we classify plants and diseases together, while in the second we focus on diseases only. The second formulation helped the model to achieve better results in identifying new diseases and to better adapt to images captured under different conditions.
2. Materials and Methods
2.1. Datasets
2.2. Learning Approaches
2.3. Transfer Learning: Baseline and Baseline++
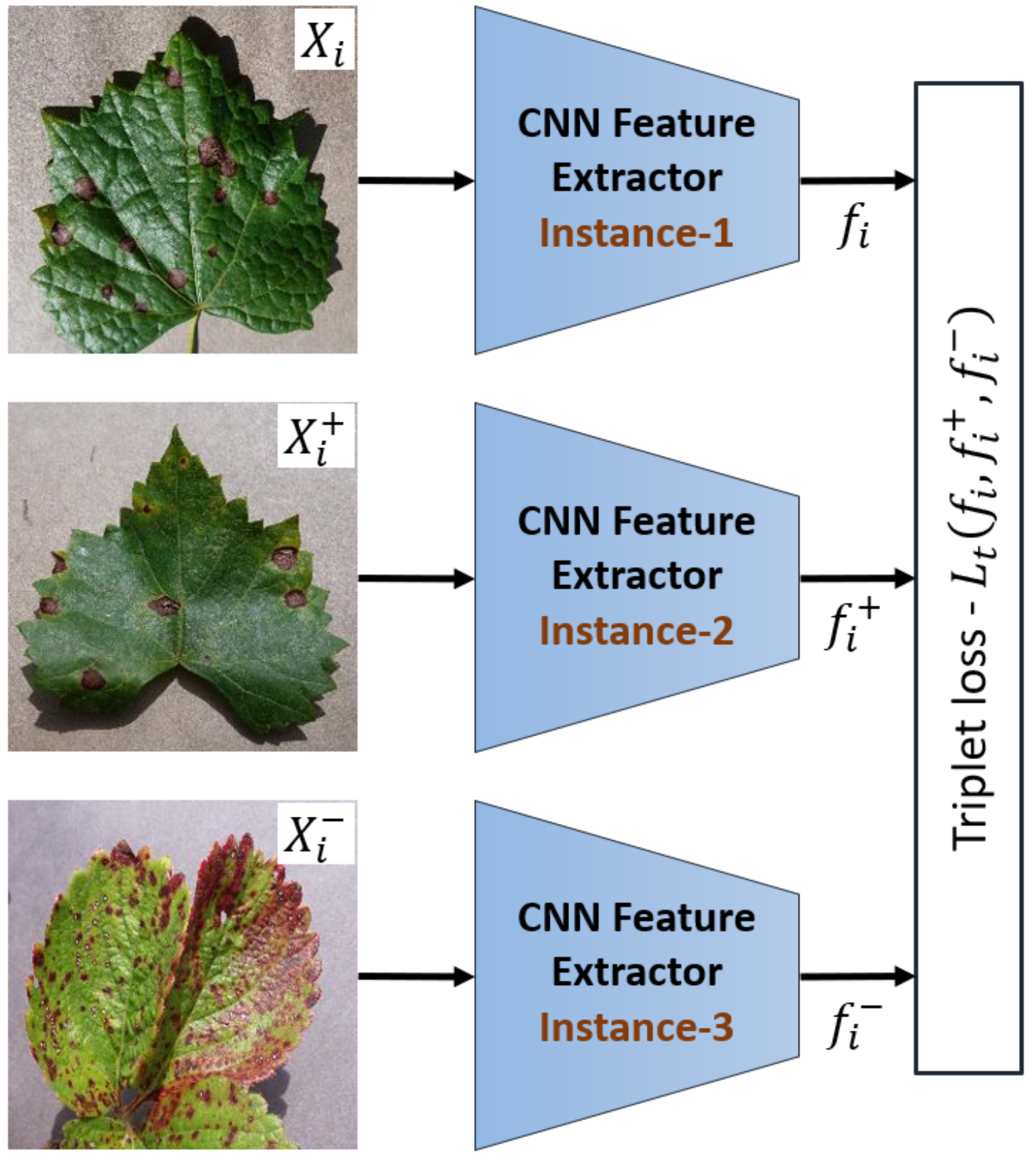
2.4. Metric Learning Using Triplet Network
2.5. Deep Adversarial Metric Learning (DAML)
2.6. Deep Architectures and Experimental Setup
3. Results and Discussion
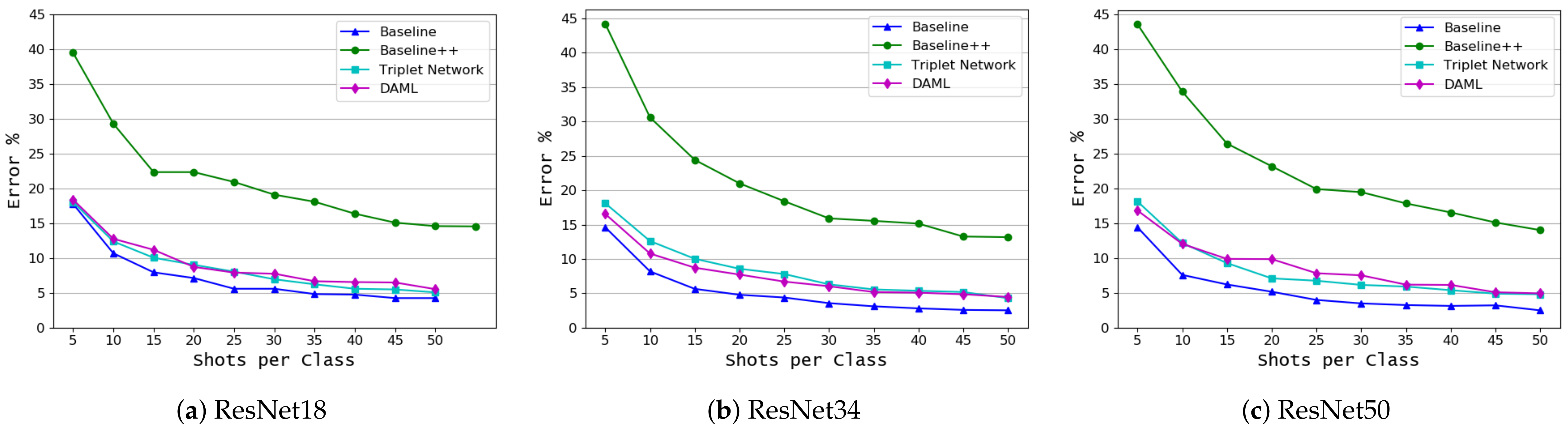
3.1. Crop and Disease Classification
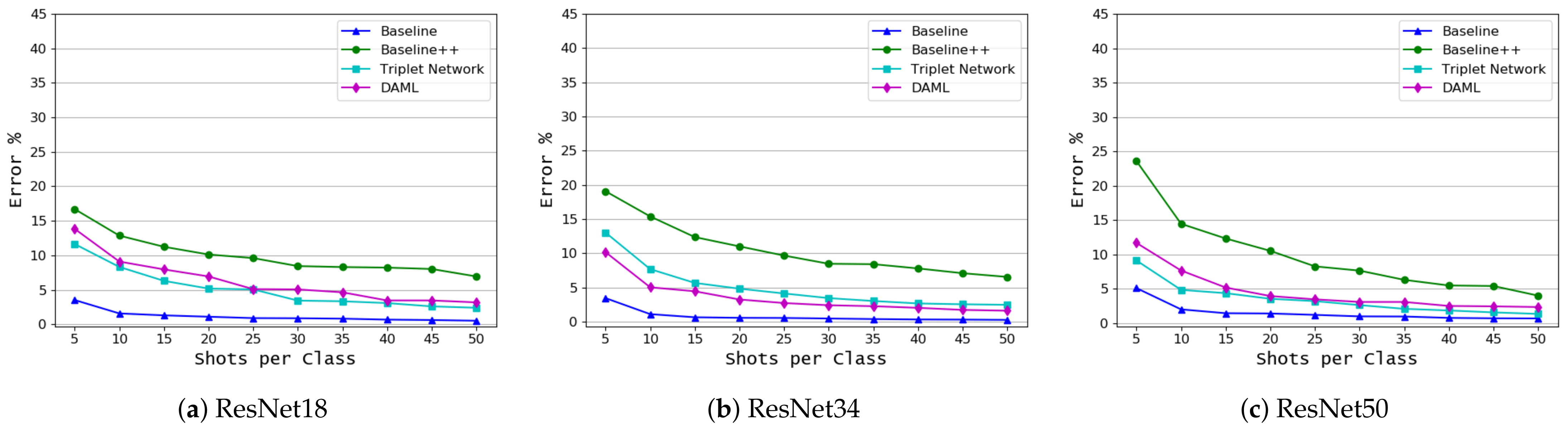
3.2. Disease Classification Based on Common Disease Name
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Johannes, A.; Picon, A.; Alvarez-Gila, A.; Echazarra, J.; Rodriguez-Vaamonde, S.; Navajas, A.D.; Ortiz-Barredo, A. Automatic plant disease diagnosis using mobile capture devices, applied on a wheat use case. Comput. Electron. Agric. 2017, 138, 200–209. [Google Scholar] [CrossRef]
- Esgario, J.G.; Krohling, R.A.; Ventura, J.A. Deep learning for classification and severity estimation of coffee leaf biotic stress. Comput. Electron. Agric. 2020, 169. [Google Scholar] [CrossRef]
- Ramcharan, A.; Baranowski, K.; McCloskey, P.; Ahmed, B.; Legg, J.; Hughes, D.P. Deep learning for image-based cassava disease detection. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Sambasivam, G.; Opiyo, G.D. A predictive machine learning application in agriculture: Cassava disease detection and classification with imbalanced dataset using convolutional neural networks. Egypt. Inform. J. 2020. [Google Scholar] [CrossRef]
- Fuentes, A.F.; Yoon, S.; Lee, J.; Park, D.S. High-performance deep neural network-based tomato plant diseases and pests diagnosis system with refinement filter bank. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Wspanialy, P.; Moussa, M. A detection and severity estimation system for generic diseases of tomato greenhouse plants. Comput. Electron. Agric. 2020, 178. [Google Scholar] [CrossRef]
- Fuentes, A.; Yoon, S.; Kim, S.C.; Park, D.S. A robust deep-learning-based detector for real-time tomato plant diseases and pests recognition. Sensors 2017, 17, 2022. [Google Scholar] [CrossRef]
- Sibiya, M.; Sumbwanyambe, M. A Computational Procedure for the Recognition and Classification of Maize Leaf Diseases Out of Healthy Leaves Using Convolutional Neural Networks. AgriEngineering 2019, 1, 119–131. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, Y.; He, D.J.; Li, Y. Identification of apple leaf diseases based on deep convolutional neural networks. Symmetry 2018, 10, 11. [Google Scholar] [CrossRef]
- Thapa, R.; Snavely, N.; Belongie, S.; Khan, A. The plant pathology 2020 challenge dataset to classify foliar disease of apples. arXiv 2020, arXiv:2004.11958. [Google Scholar]
- Liu, B.; Ding, Z.; Tian, L.; He, D.; Li, S.; Wang, H. Grape Leaf Disease Identification Using Improved Deep Convolutional Neural Networks. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef]
- Tiwari, D.; Ashish, M.; Gangwar, N.; Sharma, A.; Patel, S.; Bhardwaj, S. Potato Leaf Diseases Detection Using Deep Learning. In Proceedings of the International Conference on Intelligent Computing and Control Systems (ICICCS 2020), Madurai, India, 13–15 May 2020; pp. 461–466. [Google Scholar] [CrossRef]
- Too, E.C.; Yujian, L.; Njuki, S.; Yingchun, L. A comparative study of fine-tuning deep learning models for plant disease identification. Comput. Electron. Agric. 2019, 161, 272–279. [Google Scholar] [CrossRef]
- Hughes, D.P.; Salathe, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR 2017), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar] [CrossRef]
- Liang, Q.; Xiang, S.; Hu, Y.; Coppola, G.; Zhang, D.; Sun, W. PD 2 SE-Net: Computer-assisted plant disease diagnosis and severity estimation network. Comput. Electron. Agric. 2019, 157, 518–529. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef]
- Saleem, M.H.; Potgieter, J.; Arif, K.M. Plant disease detection and classification by deep learning. Plants 2019, 8, 468. [Google Scholar] [CrossRef]
- Hasan, R.I.; Yusuf, S.M.; Alzubaidi, L. Review of the state of the art of deep learning for plant diseases: A broad analysis and discussion. Plants 2020, 9, 1302. [Google Scholar] [CrossRef]
- Lee, S.H.; Goëau, H.; Bonnet, P.; Joly, A. New perspectives on plant disease characterization based on deep learning. Comput. Electron. Agric. 2020, 170. [Google Scholar] [CrossRef]
- Wang, Y.; Yao, Q.; Kwok, J.T.; Ni, L.M. Generalizing from a Few Examples: A Survey on Few-shot Learning. ACM Comput. Surv. 2020, 53. [Google Scholar] [CrossRef]
- Medela, A.; Picon, A.; Saratxaga, C.L.; Belar, O.; Cabezon, V.; Cicchi, R.; Bilbao, R.; Glover, B. Few shot learning in histopathological images: Reducing the need of labeled data on biological datasets. In Proceedings of the International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1860–1864. [Google Scholar] [CrossRef]
- Chen, W.Y.; Wang, Y.C.F.; Liu, Y.C.; Kira, Z.; Huang, J.B. A closer look at few-shot classification. In Proceedings of the 7th International Conference on Learning Representations (ICLR 2019), New Orleans, LA, USA, 6–9 May 2019. [Google Scholar]
- Finn, C.; Abbeel, P.; Levine, S. Model-agnostic meta-learning for fast adaptation of deep networks. In Proceedings of the 34th International Conference on Machine Learning (ICML 2017), Sydney, Australia, 6–11 August 2017; Volume 3, pp. 1856–1868. [Google Scholar]
- Nichol, A.; Achiam, J.; Schulman, J. On first-order meta-learning algorithms. arXiv 2018, arXiv:1803.02999. [Google Scholar]
- Mishra, N.; Rohaninejad, M.; Chen, X.; Abbeel, P. A simple neural attentive meta-learner. In Proceedings of the 6th International Conference on Learning Representations (ICLR 2018), Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Van der Spoel, E.; Rozing, M.P.; Houwing-Duistermaat, J.J.; Eline Slagboom, P.; Beekman, M.; de Craen, A.J.M.; Westendorp, R.G.J.; van Heemst, D. Siamese Neural Networks for One-Shot Image Recognition. In Proceedings of the ICML—Deep Learning Workshop, Lille Grande Palais, Lille, France, 10–11 July 2015; Volume 7, pp. 956–963. [Google Scholar]
- Hoffer, E.; Ailon, N. Deep metric learning using triplet network. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015)—Workshop Track Proceedings, Copenhagen, Denmark,, 12–14 October 2015. [Google Scholar]
- Oriol, V.; Charles, B.; Tim, L.; Koray, K.; Daan, W. Matching Networks for One Shot Learning. In Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, Spain, 5–10 December 2016; pp. 3630–3638. [Google Scholar]
- Snell, J.; Swersky, K.; Zemel, R.S. Prototypical Networks for Few-shot Learning. Adv. Neural Inf. Process. Syst. 2017, 30, 4077–4087. [Google Scholar]
- Sung, F.; Yang, Y.; Zhang, L.; Xiang, T.; Torr, P.H.; Hospedales, T.M. Learning to Compare: Relation Network for Few-Shot Learning. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1199–1208. [Google Scholar] [CrossRef]
- Wang, Y.X.; Girshick, R.; Hebert, M.; Hariharan, B. Low-Shot Learning from Imaginary Data. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 7278–7286. [Google Scholar] [CrossRef]
- Antoniou, A.; Storkey, A.; Edwards, H. Data augmentation generative adversarial networks. arXiv 2017, arXiv:1711.04340. [Google Scholar]
- Guo, Y.; Codella, N.C.; Karlinsky, L.; Codella, J.V.; Smith, J.R.; Saenko, K.; Rosing, T.; Feris, R. A Broader Study of Cross-Domain Few-Shot Learning; Vedaldi, A., Bischof, H., Brox, T., Frahm, J.M., Eds.; Computer Vision—ECCV 2020. ECCV 2020. Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 12372, pp. 124–141. [Google Scholar] [CrossRef]
- A low shot learning method for tea leaf’s disease identification. Comput. Electron. Agric. 2019, 163, 104852. [CrossRef]
- Janarthan, S.; Thuseethan, S.; Rajasegarar, S.; Lyu, Q.; Zheng, Y.; Yearwood, J. Deep Metric Learning Based Citrus Disease Classification With Sparse Data. IEEE Access 2020, 8, 162588–162600. [Google Scholar] [CrossRef]
- Wang, B.; Wang, D. Plant leaves classification: A few-shot learning method based on siamese network. IEEE Access 2019, 7, 151754–151763. [Google Scholar] [CrossRef]
- Argüeso, D.; Picon, A.; Irusta, U.; Medela, A.; San-Emeterio, M.G.; Bereciartua, A.; Alvarez-Gila, A. Few-Shot Learning approach for plant disease classification using images taken in the field. Comput. Electron. Agric. 2020, 175. [Google Scholar] [CrossRef]
- Hsu, C.W.; Lin, C.J. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 2002, 13, 415–425. [Google Scholar]
- Duan, Y.; Zheng, W.; Lin, X.; Lu, J.; Zhou, J. Deep Adversarial Metric Learning. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 2780–2789. [Google Scholar] [CrossRef]
- Weinberger, K.Q.; Blitzer, J.; Saul, L.K. Distance metric learning for large margin nearest neighbor classification. Adv. Neural Inf. Process. Syst. 2005, 1473–1480. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. In Proceedings of the 3rd International Conference on Learning Representations (ICLR 2015)—Conference Track Proceedings, Copenhagen, Denmark, 12–14 October 2015. [Google Scholar]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An imperative style, high-performance deep learning library. Adv. Neural Inf. Process. Syst. 2019, 32, 8026–8037. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar] [CrossRef]





| Class | Crop | Disease | Samples | |
|---|---|---|---|---|
| Train | Test | |||
| C1 | Apple | Apple cab | 504 | 126 |
| C2 | Apple | Black rot | 496 | 125 |
| C3 | Apple | Cedar apple rust | 220 | 55 |
| C4 | Apple | Healthy | 1316 | 329 |
| C5 | Blueberry | Healthy | 1202 | 300 |
| C6 | Cherry (including sour) | Healthy | 684 | 170 |
| C7 | Cherry (including sour) | Powdery mildew | 842 | 210 |
| C8 | Corn (maize) | Cercospora leaf spot Gray leaf spot | 410 | 103 |
| C9 | Corn (maize) | Common rust | 953 | 239 |
| C10 | Corn (maize) | Healthy | 929 | 233 |
| C11 | Corn (maize) | Northern Leaf Blight | 788 | 197 |
| C12 | Grape | Black rot | 944 | 236 |
| C13 | Grape | Esca (Black Measles) | 1107 | 276 |
| C14 | Grape | Healthy | 339 | 84 |
| C15 | Grape | Leaf blight (Isariopsis Leaf Spot) | 861 | 215 |
| C16 | Orange | Haunglongbing (Citrus greening) | 4405 | 1102 |
| C17 | Peach | Bacterial spot | 1838 | 459 |
| C18 | Peach | Healthy | 288 | 72 |
| C19 | Pepper bell | Bacterial spot | 797 | 200 |
| C20 | Pepper bell | Healthy | 1183 | 295 |
| C21 | Potato | Early blight | 800 | 200 |
| C22 | Potato | Healthy | 121 | 31 |
| C23 | Potato | Late blight | 800 | 200 |
| C24 | Raspberry | Healthy | 297 | 74 |
| C25 | Soybean | Healthy | 4072 | 1018 |
| C26 | Squash | Powdery mildew | 1468 | 367 |
| C27 | Strawberry | Healthy | 364 | 92 |
| C28 | Strawberry | Leaf scorch | 887 | 222 |
| C29 | Tomato | Bacterial spot | 1702 | 425 |
| C30 | Tomato | Early blight | 800 | 200 |
| C31 | Tomato | Healthy | 1273 | 318 |
| C32 | Tomato | Late blight | 1527 | 382 |
| C33 | Tomato | Leaf mold | 761 | 191 |
| C34 | Tomato | Septoria leaf spot | 1417 | 354 |
| C35 | Tomato | Spider mites Two-spotted spider mite | 1341 | 335 |
| C36 | Tomato | Target spot | 1123 | 281 |
| C37 | Tomato | Tomato mosaic virus | 299 | 74 |
| C38 | Tomato | Tomato Yellow Leaf Curl Virus | 4286 | 1071 |
| Class | Disease | Affected Plants | Samples | |
|---|---|---|---|---|
| Train | Test | |||
| CD1 | Apple scab | Apple | 504 | 126 |
| CD2 | Bacterial spot | Peach, Pepper bell, Tomato | 4337 | 1084 |
| CD3 | Black rot | Apple, Grape | 1140 | 361 |
| CD4 | Cedar apple rust | Apple | 220 | 55 |
| CD5 | Cercospora leaf spot Gray leaf spot | corn | 440 | 103 |
| CD6 | Common rust | corn | 953 | 239 |
| CD7 | Early blight | Potato, Tomato | 1600 | 400 |
| CD8 | Esca black measles | Grape | 1107 | 276 |
| CD9 | Haunglongbing Citrus greening | Orange | 4405 | 1102 |
| CD10 | Late blight | Potato, Tomato | 2327 | 582 |
| CD11 | Leaf blight Isariopsis Leaf Spot | Grape | 861 | 215 |
| CD12 | Leaf mold | Tomato | 761 | 191 |
| CD13 | Leaf scorch | Strawberry | 887 | 222 |
| CD14 | Northern Leaf blight | Corn | 817 | 197 |
| CD15 | Powdery mildew | Cherry, Squash | 2310 | 577 |
| CD16 | Septoria leaf spot | Tomato | 1417 | 354 |
| CD17 | Spider mites Two spotted spider mite | Tomato | 1341 | 335 |
| CD18 | Target spot | Tomato | 1123 | 281 |
| CD19 | Tomato mosaic virus | Tomato | 299 | 74 |
| CD20 | Tomato Yellow Leaf Curl Virus | Tomato | 4286 | 1071 |
| CD21 | Healthy | − | 4909 | 1200 |
| Class | Predominant Disease | Samples |
|---|---|---|
| CF1 | Leaf miner | 387 |
| CF2 | Rust | 531 |
| CF3 | Brown leaf spot | 348 |
| CF4 | Cercospora leaf spot | 147 |
| CF5 | (Several with same severity) | 62 |
| CF6 | Healthy | 272 |
| Approach | ResNet18/(Shots) | ResNet34/(Shots) | ResNet50/(Shots) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 25 | 50 | 5 | 25 | 50 | 5 | 25 | 50 | |
| Baseline | 17.78 | 5.58 | 4.24 | 14.6 | 4.38 | 2.52 | 14.42 | 3.98 | 2.48 |
| Baseline++ | 39.48 | 20.92 | 14.58 | 44.22 | 18.4 | 13.16 | 43.62 | 19.92 | 14.02 |
| Triplet Network | 18.06 | 8.04 | 5.06 | 18.1 | 7.8 | 4.28 | 18.14 | 6.74 | 4.8 |
| DAML | 18.38 | 7.9 | 5.52 | 16.54 | 6.7 | 4.46 | 16.84 | 7.82 | 4.92 |
| Approach | ResNet18/(Shots) | ResNet34/(Shots) | ResNet50/(Shots) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 25 | 50 | 5 | 25 | 50 | 5 | 25 | 50 | |
| Baseline | 44.78 | 29.46 | 25.08 | 36.68 | 25.76 | 22.66 | 35.54 | 22.62 | 20.22 |
| Baseline++ | 54.96 | 43.84 | 38.38 | 56.7 | 38.5 | 33.32 | 60.12 | 38.32 | 32.7 |
| Triplet Network | 43.02 | 27.6 | 23.5 | 45.9 | 28.9 | 23.96 | 47.14 | 30.74 | 24.48 |
| DAML | 43 | 28.2 | 23.58 | 42.52 | 27.14 | 21.78 | 43.82 | 28.58 | 23.14 |
| Approach | ResNet18/(Shots) | ResNet34/(Shots) | ResNet50/(Shots) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 25 | 50 | 5 | 25 | 50 | 5 | 25 | 50 | |
| Baseline | 2.52 | 0.9 | 0.52 | 3.46 | 0.56 | 0.28 | 5.08 | 1.018 | 0.66 |
| Baseline++ | 16.7 | 9.6 | 6.94 | 19.08 | 9.68 | 6.56 | 23.58 | 8.24 | 3.98 |
| Triplet Network | 11.64 | 5.06 | 2.4 | 13.04 | 4.14 | 2.48 | 9.1 | 3.18 | 1.32 |
| DAML | 13.84 | 5.1 | 3.18 | 10.18 | 2.74 | 1.62 | 11.66 | 3.44 | 2.32 |
| Approach | ResNet18/(Shots) | ResNet34/(Shots) | ResNet50/(Shots) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 | 25 | 50 | 5 | 25 | 50 | 5 | 25 | 50 | |
| Baseline | 38.766 | 25.88 | 23.08 | 36.7 | 24.5 | 21.14 | 34.28 | 22.56 | 19.44 |
| Baseline++ | 47.38 | 36.38 | 33.34 | 45.5 | 36.24 | 33.46 | 49.42 | 35.28 | 30.76 |
| Triplet Network | 47.22 | 31.18 | 26.08 | 47.06 | 29.48 | 24.58 | 50.72 | 31.26 | 25.3 |
| DAML | 42.54 | 28.56 | 23.86 | 42.6 | 27.12 | 22.2 | 45.78 | 29.3 | 25.34 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Afifi, A.; Alhumam, A.; Abdelwahab, A. Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Plants 2021, 10, 28. https://doi.org/10.3390/plants10010028
Afifi A, Alhumam A, Abdelwahab A. Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Plants. 2021; 10(1):28. https://doi.org/10.3390/plants10010028
Chicago/Turabian StyleAfifi, Ahmed, Abdulaziz Alhumam, and Amira Abdelwahab. 2021. "Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data" Plants 10, no. 1: 28. https://doi.org/10.3390/plants10010028
APA StyleAfifi, A., Alhumam, A., & Abdelwahab, A. (2021). Convolutional Neural Network for Automatic Identification of Plant Diseases with Limited Data. Plants, 10(1), 28. https://doi.org/10.3390/plants10010028





