Predominance and Diversity of GLRaV-3 in Native Vines of Mediterranean Croatia
Abstract
1. Introduction
2. Results
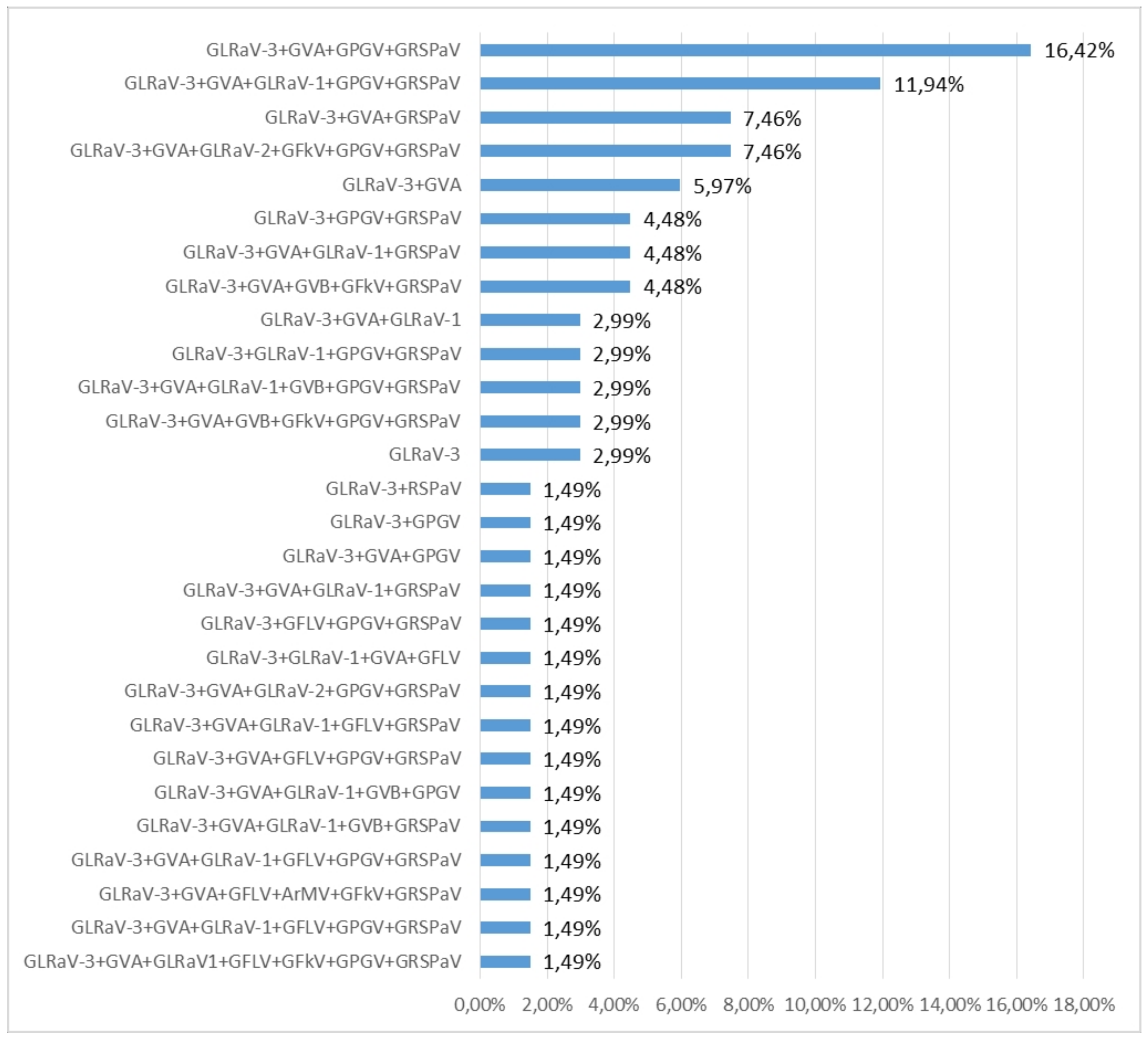
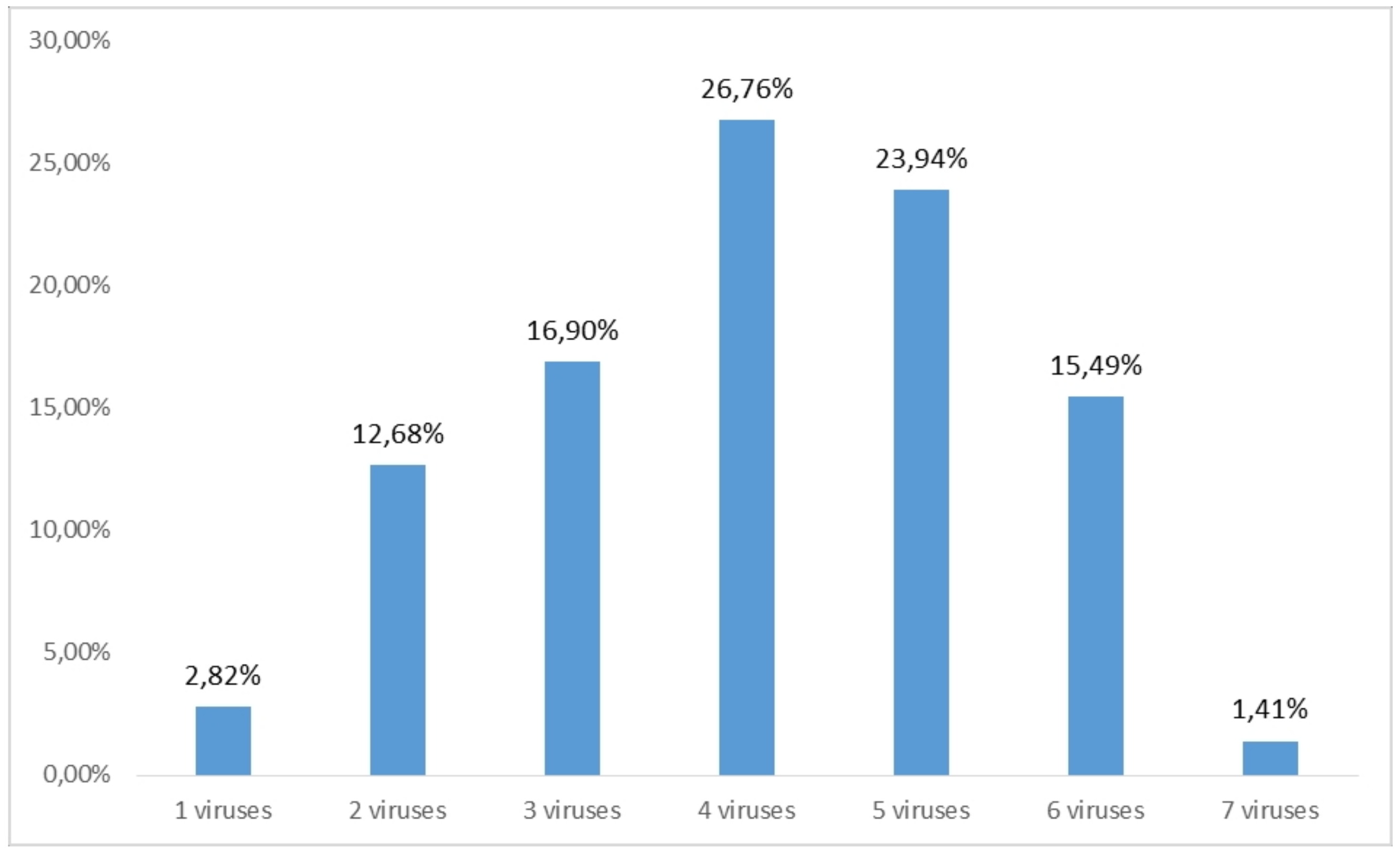
2.1. RT-PCR and ELISA Virus Detection
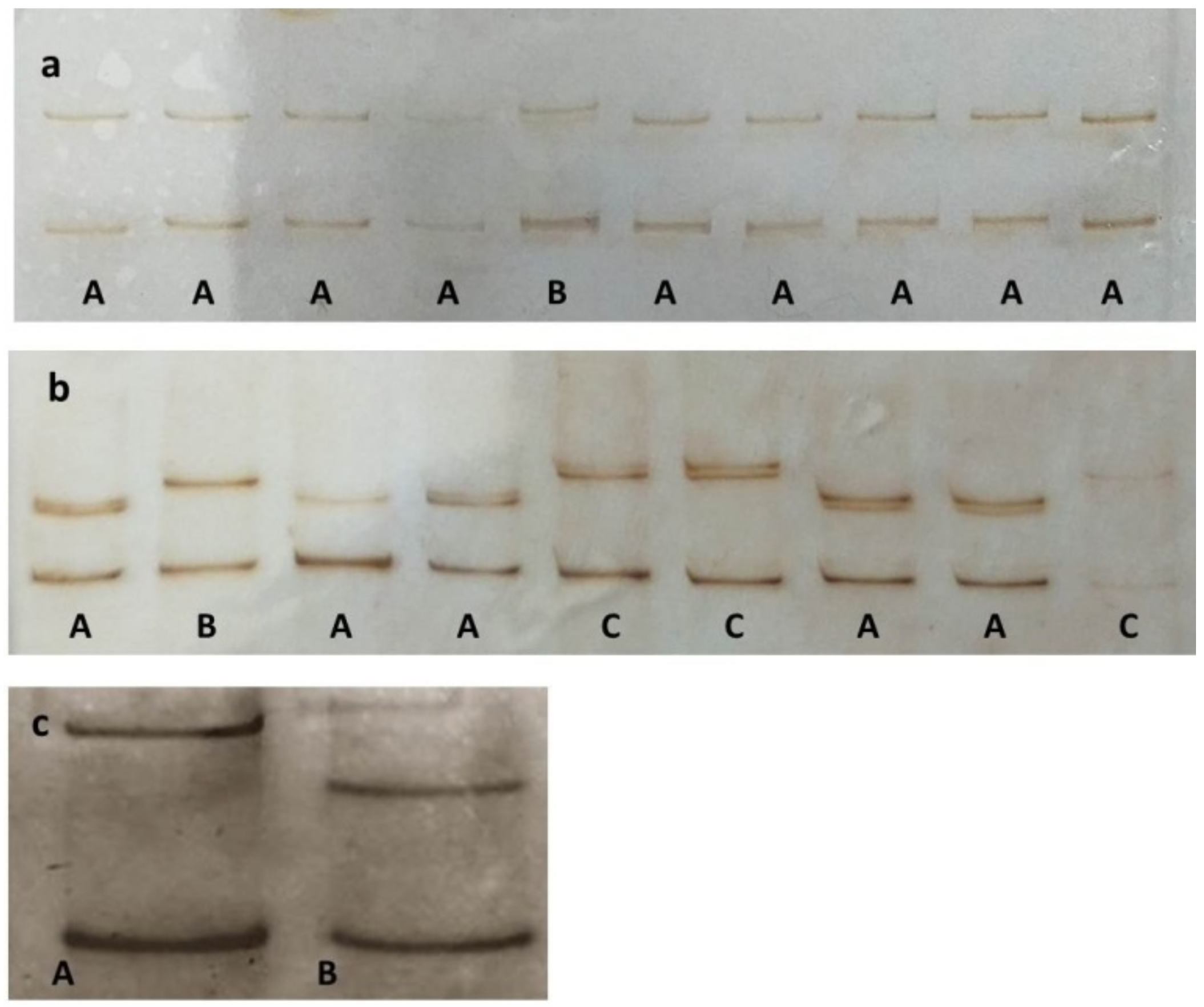
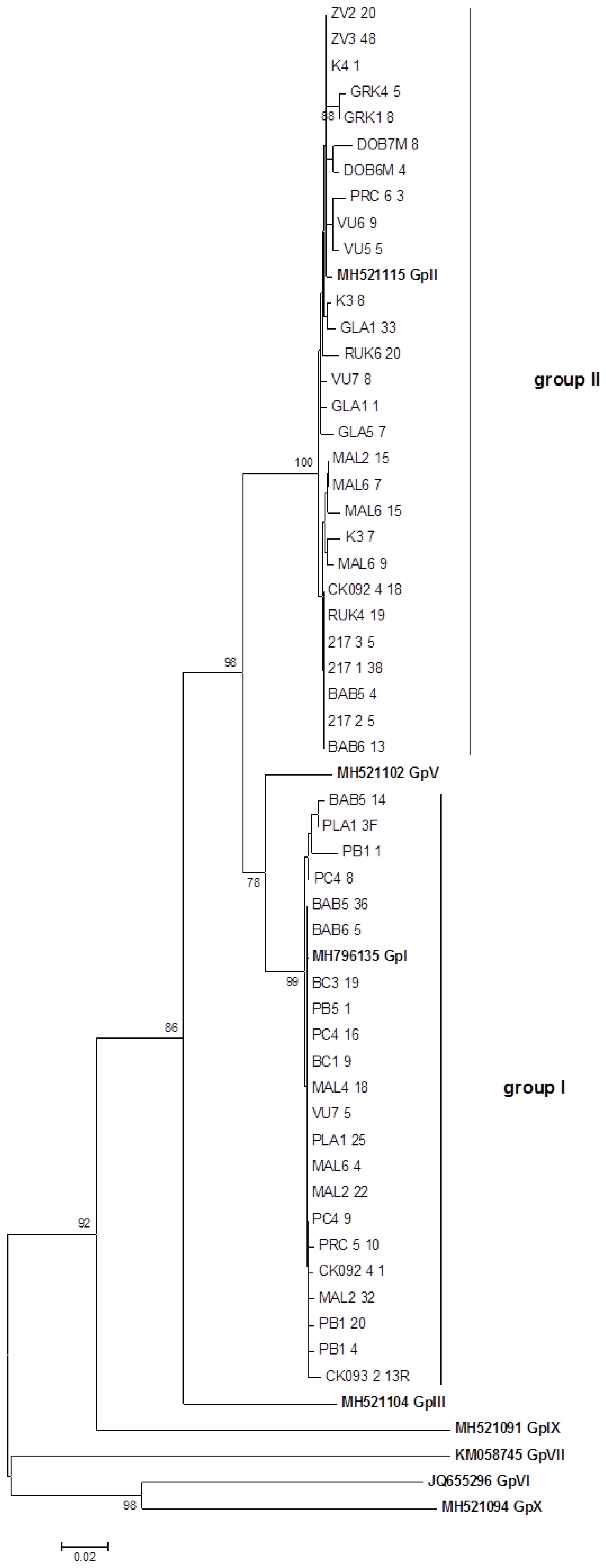
2.2. SSCP Analysis and Molecular Diversity of GLRaV-3
3. Discussion
3.1. Virus Distribution
3.2. Study of GLRaV-3 Natural Population
4. Materials and Methods
4.1. Viral Source
4.2. ELISA Virus Detection
4.3. RNA Extractions, cDNA Synthesis and PCR Virus Detection
4.4. Cloning and Single Stranded Conformational Polymorphism Analysis
4.5. Nucleotide Sequence Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Martelli, G.P. An Overview on Grapevine Viruses, Viroids and the Diseases They Cause. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Meng, B., Martelli, G.P., Golino, D.A., Fuchs, M., Eds.; Springer: Cham, Switzerland, 2017; pp. 31–46. ISBN 9783319577067. [Google Scholar]
- Martelli, G.P.; Sabanadzovic, S.; Ghanem-Sabanadzovic, N.A.; Saldarelli, P. Maculavirus, a new genus of plant viruses. Arch. Virol. 2002, 147, 1847–1853. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014. [Google Scholar]
- Naidu, R.; Rowhani, A.; Fuchs, M.; Golino, D.; Martelli, G.P. Grapevine Leafroll: A complex viral disease affecting a high-value fruit crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef] [PubMed]
- Gugerli, P. Grapevine leafroll and related viruses. In Proceedings of the Extended Abstracts 14th Meeting of ICVG, Locorotondo, Italy, 12–17 September 2003; pp. 25–31. [Google Scholar]
- Turturo, C.; Saldarelli, P.; Yafeng, D.; Digiaro, M.; Minafra, A.; Savino, V.; Martelli, G.P. Genetic variability and population structure of Grapevine leafroll-associated virus 3 isolates. J. Gen. Virol. 2005, 86, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013, 4, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Lehad, A.; Selmi, I.; Louanchi, M.; Aitouada, M.; Mahfoudhi, N. Genetic diversity of grapevine leafroll-associated virus 3 in Algeria. J. Plant Pathol. 2015, 97, 203–207. [Google Scholar] [CrossRef]
- Burger, J.T.; Maree, H.J.; Gouveia, P.; Naidu, R.A. Grapevine leafroll-associated virus 3. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer: Cham, Switzerland, 2017; pp. 167–194. ISBN 9783319577067. [Google Scholar]
- Diaz-Lara, A.; Klaassen, V.; Stevens, K.; Sudarshana, M.R.; Rowhani, A.; Maree, H.J.; Chooi, K.M.; Blouin, A.G.; Habili, N.; Song, Y.; et al. Characterization of grapevine leafroll-associated virus 3 genetic variants and application towards RT-qPCR assay design. PLoS ONE 2018, 13. [Google Scholar] [CrossRef]
- Rahali, M.; Migliaro, D.; Laiadi, Z.; Bertazzon, N.; Angelini, E.; Crespan, M. Genetic identification, origin and sanitary status of grapevine cultivars (Vitis vinifera L.) grown in Babar, Algeria. Vitis 2019, 58, 153–158. [Google Scholar]
- Regner, F.; Hack, R.; Gangl, H.; Leitner, G.; Mandl, K.; Tiefenbrunner, W. Genetic variability and incidence of systemic diseases in wild vines (Vitis vinifera ssp. silvestris) along the Danube. Vitis J. Grapevine Res. 2004, 43, 123–130. [Google Scholar]
- Gisbert, C.; Peiró, R.; San Pedro, T.; Olmos, A.; Jiménez, C.; García, J. Recovering ancient Grapevine varieties: from genetic variability to in vitro conservation, a case study. In Grapes Wines Advances in Production, Processing, Analysis and Valorization; InTech: Rijeka, Croatia, 2018; p. 1. [Google Scholar]
- Karoglan Kontić, J.; Preiner, D.; Šimon, S.; Zdunić, G.; Poljuha, D.; Maletić, E. Sanitary status of Croatian native grapevine varieties. Agric. Conspec. Sci. 2009, 74, 99–103. [Google Scholar]
- Poljuha, D.; Sladonja, B.; Bubola, M. Incidence of viruses infecting grapevine varieties in Istria (Croatia). J. Food, Agric. Environ. 2010, 8, 166–169. [Google Scholar]
- Vončina, D.; Preiner, D.; Šimon, S.; Cvjetković, B.; Maletić, E.; Pejić, I.; Kontić, J. Distribution of nine viruses in croatian autochthonous grapevine (Vitis vinifera L.) cultivars from dalmatian region included in clonal selection. J. Cent. Eur. Agric. 2019, 20, 262–273. [Google Scholar] [CrossRef]
- Vončina, D.; Al Rwahnih, M.; Rowhani, A.; Gouran, M.; Almeida, R.P.P. Viral diversity in autochthonous croatian grapevine cultivars. Plant Dis. 2017, 101, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Dimitrijevic, B. The occurrence of leafroll of grapevine in Yugoslavia. Zaštita Bilja 1970, 21, 373–378. [Google Scholar]
- Bertolini, E.; García, J.; Yuste, A.; Olmos, A. High prevalence of viruses in table grape from Spain detected by real-time RT-PCR. Eur. J. Plant Pathol. 2010, 128, 283–287. [Google Scholar] [CrossRef]
- Savino, V.; La Notte, P.; Bottalico, G.; Martelli, G.P. Situazione sanitaria della vite in Italia centro-meridionale. Quad. della Sc. di Spec. Sci. Vitic. ed Enol. 2001, 25, 67–76. [Google Scholar]
- Zindović, J.; Viršček Marn, M.; Mavrič Pleško, I. Phytosanitary status of grapevine in Montenegro. EPPO Bull. 2014, 44, 60–64. [Google Scholar] [CrossRef]
- Ahmed, H.M.H.; Digiaro, M.; Martelli, G.P. Viruses and virus diseases of grapevine in Egypt. EPPO Bull. 2004, 34, 395–398. [Google Scholar] [CrossRef]
- Digiaro, M.; Martelli, G.P.; Savino, V. Phloem-limited viruses of the grapevine in the Mediterranean and Near East: a synopsis. Opt. Méditerrranéennes Ser. B Stud. Res. 1999, 29, 83–92. [Google Scholar]
- Mahfoudhi, N.; Digiaro, M.; Savino, V.; Di Terlizzi, B.; Ahmed, H.M.H.; Digiaro, M.; Martelli, G.P. Viruses and virus diseases of grapevine in Tunisia. EPPO Bull. 1998, 28, 197–204. [Google Scholar] [CrossRef]
- Vončina, D.; Badurina, D.; Preiner, D.; Vjetkovic, B.; Maletic, E.; Kontic, J.K. Incidence of virus infections in grapevines from Croatian collection plantations. Phytopathol. Mediterr. 2011, 50, 316–326. [Google Scholar] [CrossRef]
- Afechtal, M.; Mounir, M.; Minafra, A.; Saldarelli, P.; Kubaa, R.A. First report on the occurence of grapevine rupestris stem pitting-associated virus in Moroccan grapevines. J. Plant Pathol. 2019, 101, 405. [Google Scholar] [CrossRef]
- Montero, R.; El aou ouad, H.; Pacifico, D.; Marzachì, C.; Castillo, N.; García, E.; Del Saz, N.F.; Florez-Sarasa, I.; Flexas, J.; Bota, J. Effects of Grapevine leafroll-associated virus 3 on the physiology in asymptomatic plants of Vitis vinifera. Ann. Appl. Biol. 2017, 171, 155–171. [Google Scholar] [CrossRef]
- Xiao, H.; Shabanian, M.; Moore, C.; Li, C.; Meng, B. Survey for major viruses in commercial Vitis vinifera wine grapes in Ontario. Virol. J. 2018, 15, 127. [Google Scholar] [CrossRef]
- Meng, B.; Martelli, G.P.; Golino, D.A.; Fuchs, M. (Eds.) Grapevine viruses: Molecular biology, diagnostics and management. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer: Cham, Switzerland, 2017; pp. 1–698. [Google Scholar] [CrossRef]
- Gambino, G.; Cuozzo, D.; Fasoli, M.; Pagliarani, C.; Vitali, M.; Boccacci, P.; Pezzotti, M.; Mannini, F. Co-evolution between Grapevine rupestris stem pitting-associated virus and Vitis vinifera L. leads to decreased defence responses and increased transcription of genes related to photosynthesis. J. Exp. Bot. 2012, 63, 5919–5933. [Google Scholar] [CrossRef]
- Pleško, I.M.; Marn, M.V.; Seljak, G.; Žežlina, I. First report of Grapevine Pinot gris virus infecting grapevine in Slovenia. Plant Dis. 2014, 98, 1014. [Google Scholar] [CrossRef]
- Beuve, M.; Candresse, T.; Tannières, M.; Lemaire, O. First report of Grapevine Pinot gris virus (GPGV) in grapevine in France. Plant Dis. 2015, 99, 293. [Google Scholar] [CrossRef]
- Saldarelli, P.; Beber, R.; Covelli, L.; Bianchedi, P.; Credi, R.; Giampetruzzi, A.; Malossini, U.; Pirolo, C.; Poggi Pollini, C.; Ratti, C. Studies on a new grapevine disease in Trentino vineyards. J. Plant Pathol. 2013, 95, 60. [Google Scholar]
- Al Rwahnih, M.; Golino, D.; Rowhani, A. First report of Grapevine Pinot gris virus infecting grapevine in the United States. Plant Dis. 2016, 100, 1030. [Google Scholar] [CrossRef]
- Reynard, J.-S.; Schumacher, S.; Menzel, W.; Fuchs, J.; Bohnert, P.; Glasa, M.; Wetzel, T.; Fuchs, R. First report of Grapevine pinot gris virus in German vineyards. Plant Dis. 2016, 100, 2545. [Google Scholar] [CrossRef]
- Abou Kubaa, R.; Choueiri, E.; Jreijiri, F.; El Khoury, Y.; Saldarelli, P. First report of grapevine Pinot gris virus in Lebanon and the Middle East. J. Plant Pathol. 2019, 1. [Google Scholar] [CrossRef]
- Fan, X.D.; Dong, Y.F.; Zhang, Z.P.; Ren, F.; Hu, G.J.; Li, Z.N.; Zhou, J. First report of Grapevine Pinot gris virus in grapevines in China. Plant Dis. 2016, 100, 540. [Google Scholar] [CrossRef]
- Cho, I.S.; Jung, S.M.; Cho, J.D.; Choi, G.S.; Lim, H.S. First report of Grapevine pinot gris virus infecting grapevine in Korea. New Dis Rep 2013, 27, 588–2044. [Google Scholar] [CrossRef]
- Bertazzon, N.; Filippin, L.; Forte, V.; Angelini, E. Grapevine Pinot gris virus seems to have recently been introduced to vineyards in Veneto, Italy. Arch. Virol. 2016, 161, 711–714. [Google Scholar] [CrossRef]
- Malagnini, V.; de Lillo, E.; Saldarelli, P.; Beber, R.; Duso, C.; Raiola, A.; Zanotelli, L.; Valenzano, D.; Giampetruzzi, A.; Morelli, M.; et al. Transmission of grapevine Pinot gris virus by Colomerus vitis (Acari: Eriophyidae) to grapevine. Arch. Virol. 2016, 161, 2595–2599. [Google Scholar] [CrossRef]
- Croatian Bureau of Statistics: Basic Survey on Vineyard Structure. 2015. Available online: https://www.dzs.hr/Hrv_Eng/publication/2016/01-01-33_01_2016.htm (accessed on 11 September 2020).
- Gouveia, P.; Santos, M.T.; Eiras-Dias, J.E.; Nolasco, G. Five phylogenetic groups identified in the coat protein gene of grapevine leafroll-associated virus 3 obtained from Portuguese grapevine varieties. Arch. Virol. 2011, 156, 413–420. [Google Scholar] [CrossRef]
- Farooq, A.B.U.; Ma, Y.; Wang, Z.; Zhuo, N.; Wenxing, X.; Wang, G.; Hong, N. Genetic diversity analyses reveal novel recombination events in Grapevine leafroll-associated virus 3 in China. Virus Res. 2013, 171, 15–21. [Google Scholar] [CrossRef]
- Sharma, A.M.; Wang, J.; Duffy, S.; Zhang, S.; Wong, M.K.; Rashed, A.; Cooper, M.L.; Daane, K.M.; Almeida, R.P.P. Occurrence of grapevine leafroll-associated virus complex in Napa Valley. PLoS ONE 2011, 6, 4–10. [Google Scholar] [CrossRef]
- Pesqueira, A.M.; Cabaleiro, C.; Velasco, L. Genetic analysis of Grapevine leafroll-associated virus 3 population from Galicia, Spain. Plant Pathol. 2016, 65, 310–321. [Google Scholar] [CrossRef]
- Jooste, A.E.C.; Pietersen, G.; Burger, J.T. Distribution of grapevine leafroll associated virus-3 variants in South African vineyards. Eur. J. Plant Pathol. 2011, 131, 371–381. [Google Scholar] [CrossRef]
- Clark, M.F.; Adams, A.N. Characteristics of the microplate method of enzyme linked immunosorbent assay for the detection of plant viruses. J. Gen. Virol. 1977, 34, 475–483. [Google Scholar] [CrossRef] [PubMed]
- MacKenzie, D.J.; McLean, M.A.; Mukerji, S.; Green, M. Improved RNA extraction from woody plants for the detection of viral pathogens by reverse transcription-polymerase chain reaction. Plant Dis. 1997, 81, 222–226. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Gribaudo, I. Simultaneous detection of nine grapevine viruses by multiplex reverse transcription-polymerase chain reaction with coamplification of a plant RNA as internal control. Phytopathology 2006, 96, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Johnson, R.; Peressini, S.; Forsline, P.L.; Gonsalves, D. Rupestris stem pitting associated virus-1 is consistently detected in grapevines that are infected with rupestris stem pitting. Eur. J. Plant Pathol. 1999, 105, 191–199. [Google Scholar] [CrossRef]
- Saldarelli, P.; Giampetruzzi, A.; Morelli, M.; Malossini, U.; Pirolo, C.; Bianchedi, P.; Gualandri, V. Genetic variability of Grapevine Pinot gris virus and its association with Grapevine leaf mottling and deformation. Phytopathology 2015, 105, 555–563. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; de Catarino, A.M.; Susca, L.; Saldarelli, P.; Gualandri, V.; Martelli, G.P. First report of Grapevine pinot gris virus from table grapes in southern Italy. J. Plant Pathol. 2014, 96. [Google Scholar]
- Černi, S.; Ruščić, J.; Nolasco, G.; Gatin, Ž.; Krajačić, M.; Škorić, D. Stem pitting and seedling yellows symptoms of Citrus tristeza virus infection may be determined by minor sequence variants. Virus Genes 2008, 36, 241–249. [Google Scholar] [CrossRef]
- Beidler, J.L.; Hilliard, P.R.; Rill, R.L. Ultrasensitive staining of nucleic acids with silver. Anal. Biochem. 1982, 126, 374–380. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]

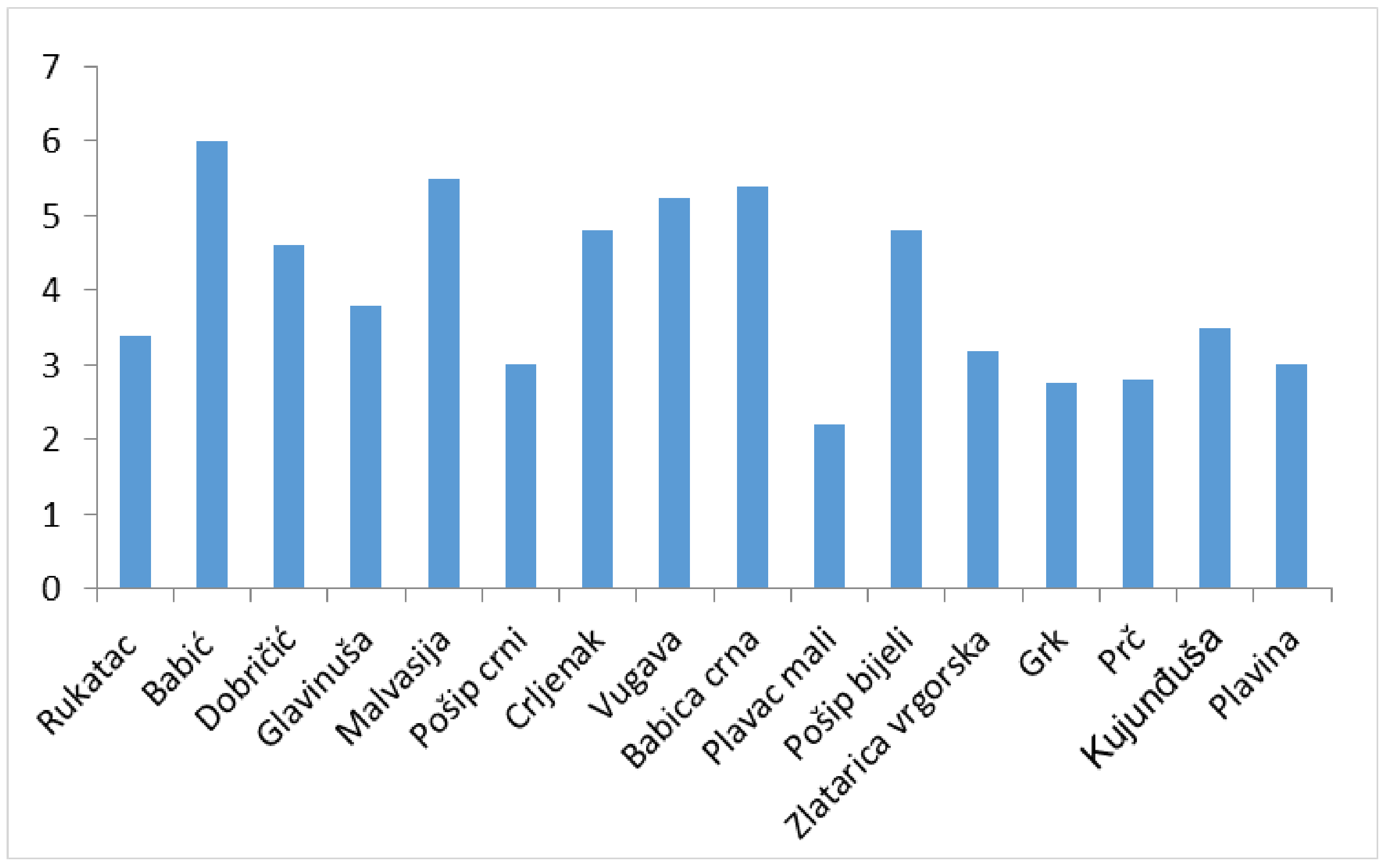
| Cultivar | Vine-Growing Area | Isolate Name | GLRaV-3 Phylogenetic Group | Co-Occurrence of Other Viruses | Leafroll Symptoms Presence | |
|---|---|---|---|---|---|---|
| Babica crna | Kaštela | BC1 | I | GVA,GVB,GFkV,GRSPaV | − | |
| Babica crna | Kaštela | BC3 | I | GVA,GVB,GFkV,GPGV,GRSPaV | − | |
| Babić | Primošten | Bab5 | I+II | GLRaV-2,GVA,GFkV,GPGV,GRSPaV | + | |
| Babić | Primošten | Bab6 | I+II | GLRaV-2,GVA,GFkV,GPGV,GRSPaV | + | |
| Crljenak kaštelanski | Kaštela | CK_092/4 | CK_093/2 | I+II | GVA,GFLV,ArMV,GFkV,GRSPaV | + |
| Crljenak kaštelanski | Kaštela | I | GVA,GPGV,GRSPaV | + | ||
| Dobričić | Šolta | Dob6 | II | GLRaV-1,GVA,GRSPaV | + | |
| Dobričić | Šolta | Dob7 | II | GLRaV-1,GVA,GRSPaV | + | |
| Glavinuša | Kaštela | Gla1 | II | GVA,GPGV,GRSPaV | + | |
| Glavinuša | Kaštela | Gla5 | II | GVA,GPGV,GRSPaV | + | |
| Grk | Korčula | Grk1 | II | GRSPaV | + | |
| Grk | Korčula | Grk4 | II | GPGV,GRSPaV | + | |
| Kujunduša | Imotski | K3 | II | GLRaV-2,GVA,GRSPaV | + | |
| Kujunđuša | Imotski | K4 | II | GVA,GRSPaV | + | |
| Malvasia dubr. | Konavle | Mal2 | I+II | GLRaV-1,GVA,GVB,GPGV,GRSPaV | + | |
| Malvasia dubr. | Konavle | Mal4 | I | GLRaV-1,GVA,GVB,GRSPaV | + | |
| Malvasia dubr. | Konavle | Mal6 | I+II | GLRaV-1,GVA,GVB,GPGV,GRSPaV | + | |
| Plavac mali | Vis | 217/1 | II | − | ||
| Plavac mali | Vis | 217/2 | II | − | ||
| Plavac mali | Vis | 217/3 | II | GPGV | − | |
| Plavina | Drniš | Pla1 | I | GVA,GPGV,GRSPaV | + | |
| Pošip bijeli | Korčula | PB1 | I | GLRaV-1,GVA,GPGV,GRSPaV | − | |
| Pošip bijeli | Korčula | PB5 | I | GLRaV-1,GVA,GPGV,GRSPaV | − | |
| Pošip crni | Korčula | PC4 | I | GLRaV-1,GPGV,GRSPaV | − | |
| Prč | Hvar | Prc5 | I | GLRaV-1,GVA | − | |
| Prč | Hvar | Prc6 | II | GVA,GRSPaV | − | |
| Rukatac | Korčula | Ruk4 | II | GVA | + | |
| Rukatac | Korčula | Ruk6 | II | GLRaV-1,GVA,GRSPaV | + | |
| Vugava | Vis | Vu5 | II | GLRaV-1,GVA,GFLV | + | |
| Vugava | Vis | Vu6 | II | GLRaV-1,GVA,GFLV,GRSPaV | + | |
| Vugava | Vis | Vu7 | I+II | GLRaV1,GVA,GFLV,GFkV,GPGV,GRSPaV | + | |
| Zlatarica vrgorska | Vrgorac | ZV2 | II | GVA,GPGV,GRSPaV | − | |
| Zlatarica vrgorska | Vrgorac | ZV3 | II | GVA,GRSPaV | − | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hančević, K.; Saldarelli, P.; Čarija, M.; Černi, S.; Zdunić, G.; Mucalo, A.; Radić, T. Predominance and Diversity of GLRaV-3 in Native Vines of Mediterranean Croatia. Plants 2021, 10, 17. https://doi.org/10.3390/plants10010017
Hančević K, Saldarelli P, Čarija M, Černi S, Zdunić G, Mucalo A, Radić T. Predominance and Diversity of GLRaV-3 in Native Vines of Mediterranean Croatia. Plants. 2021; 10(1):17. https://doi.org/10.3390/plants10010017
Chicago/Turabian StyleHančević, Katarina, Pasquale Saldarelli, Mate Čarija, Silvija Černi, Goran Zdunić, Ana Mucalo, and Tomislav Radić. 2021. "Predominance and Diversity of GLRaV-3 in Native Vines of Mediterranean Croatia" Plants 10, no. 1: 17. https://doi.org/10.3390/plants10010017
APA StyleHančević, K., Saldarelli, P., Čarija, M., Černi, S., Zdunić, G., Mucalo, A., & Radić, T. (2021). Predominance and Diversity of GLRaV-3 in Native Vines of Mediterranean Croatia. Plants, 10(1), 17. https://doi.org/10.3390/plants10010017








