Genome-Wide Identification of Banana Csl Gene Family and Their Different Responses to Low Temperature between Chilling-Sensitive and Tolerant Cultivars
Abstract
:1. Introduction
2. Results
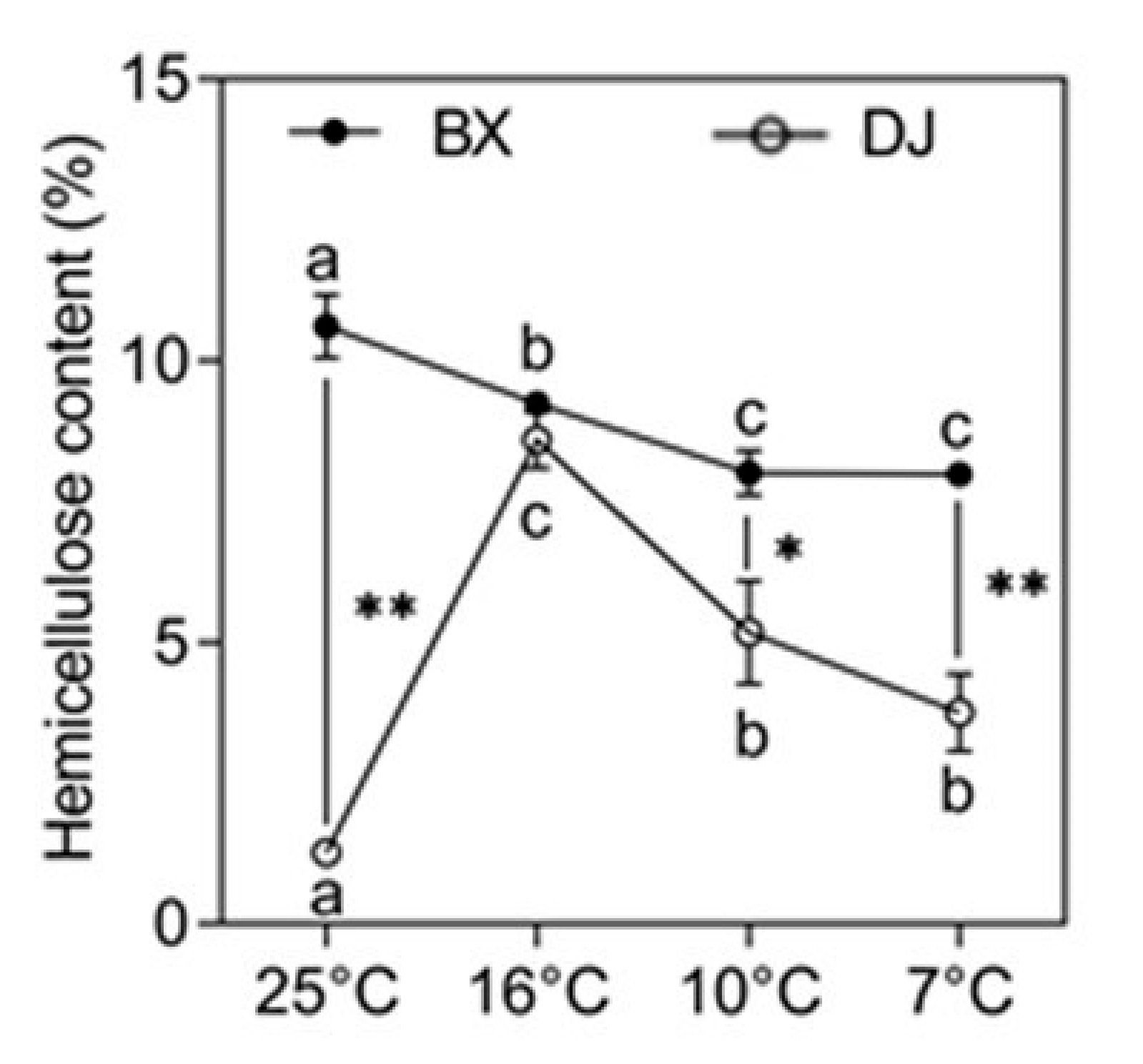
2.1. The Response of Hemicellulose in Banana to LT Stress
2.2. MaCsl and Their Molecular Structural Features
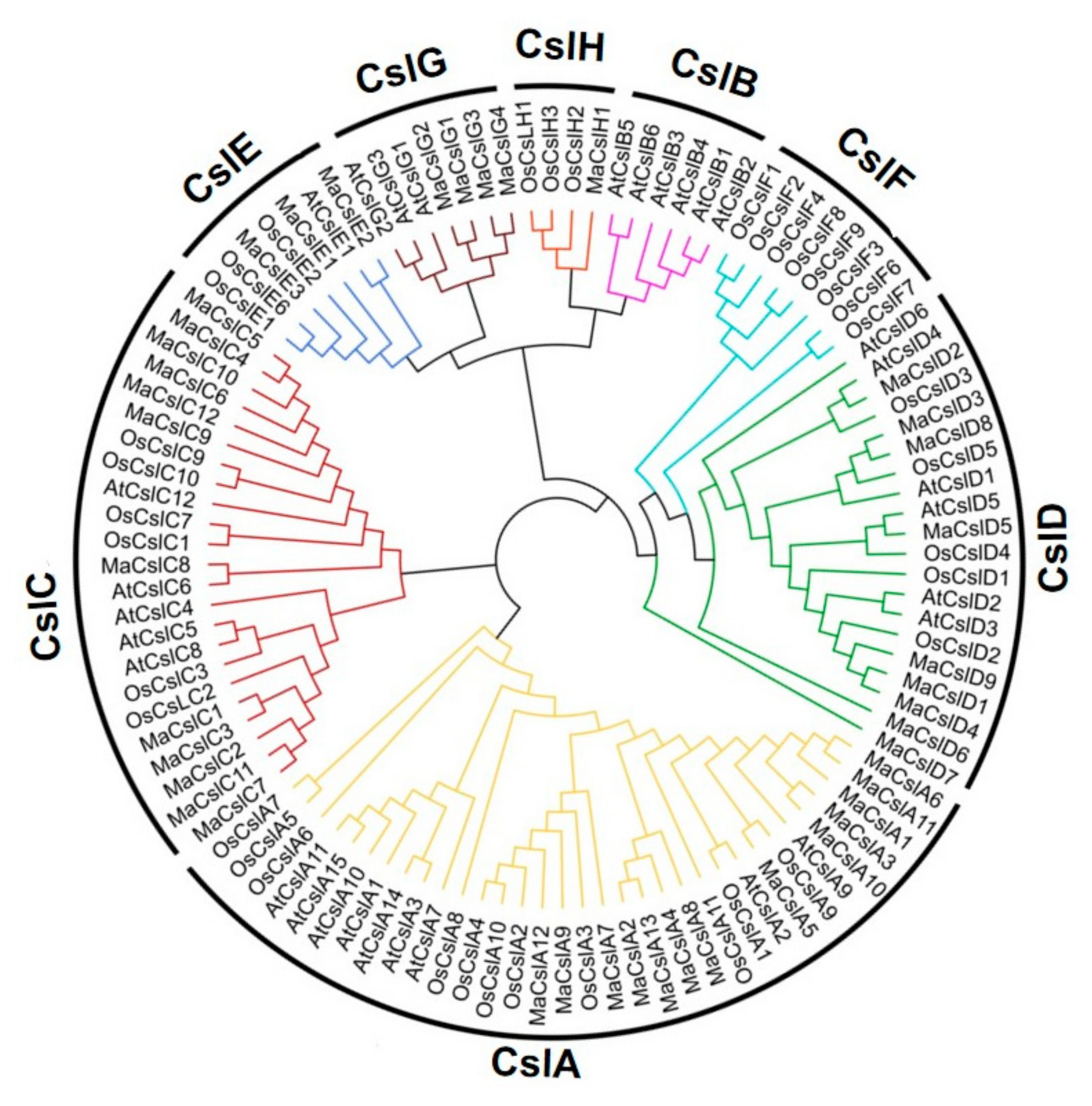
2.2.1. Phylogenetic Analysis of the MaCsls
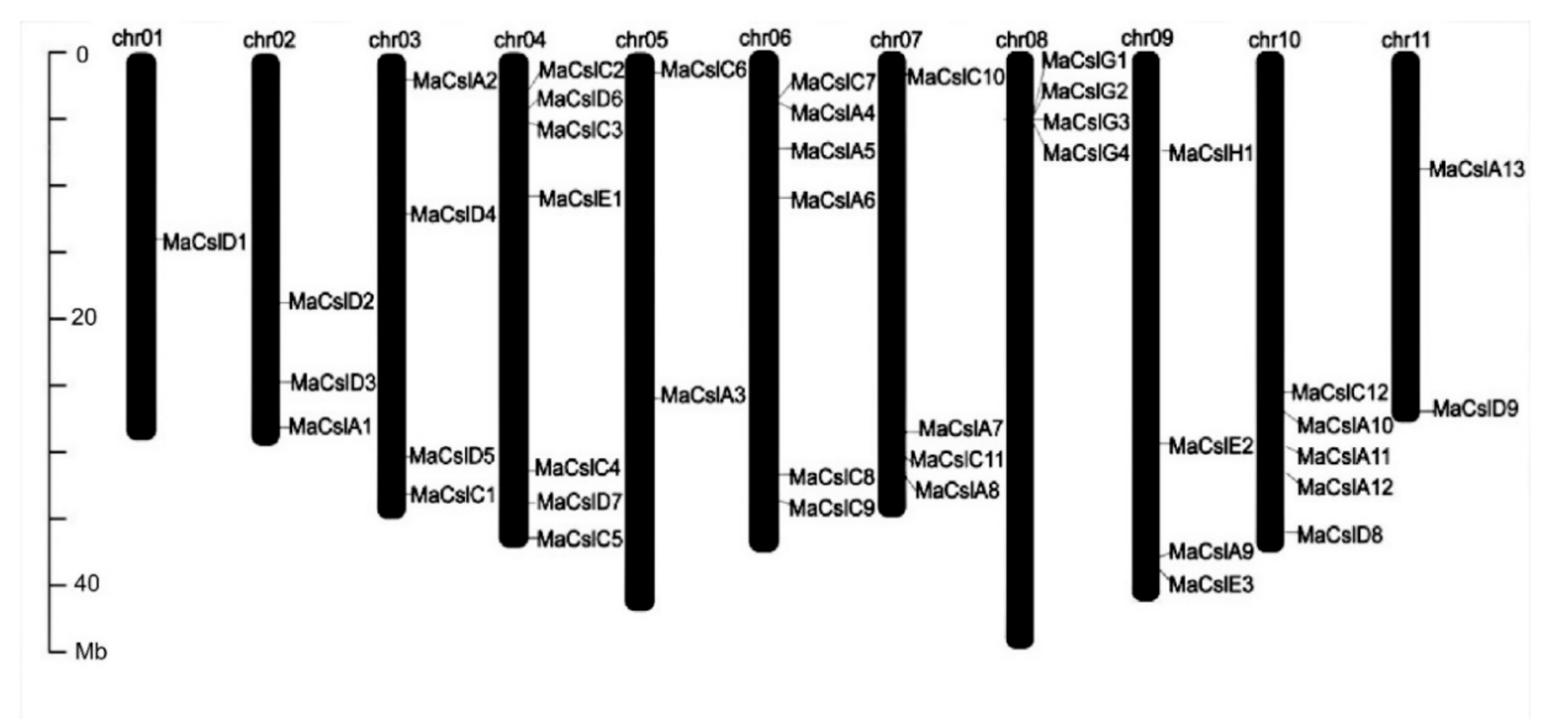
2.2.2. Identification of Csl Genes in Musa acuminata
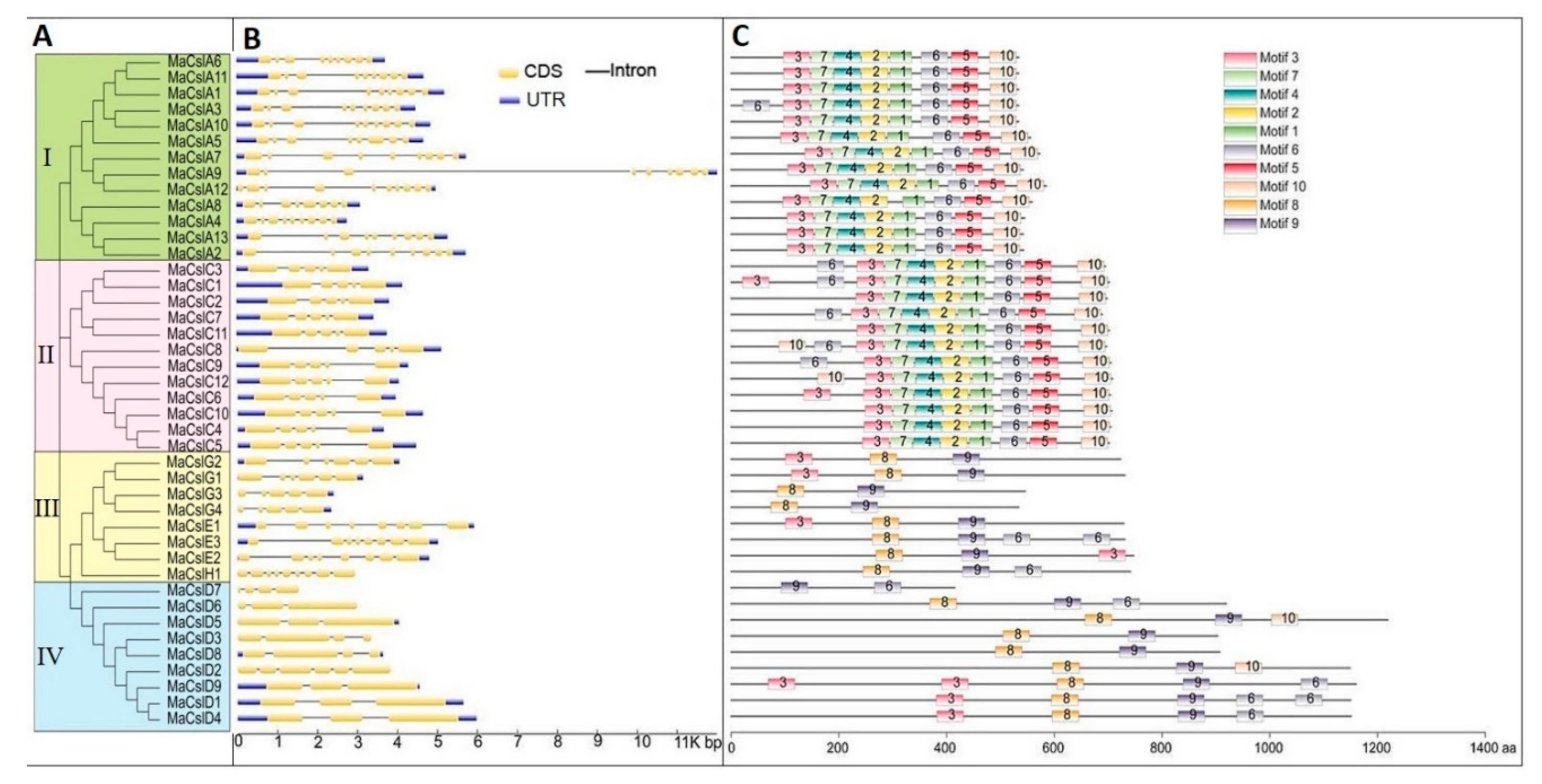
2.2.3. Phylogenetic Evolutionary Tree, Gene Structure, and Conserved Motifs
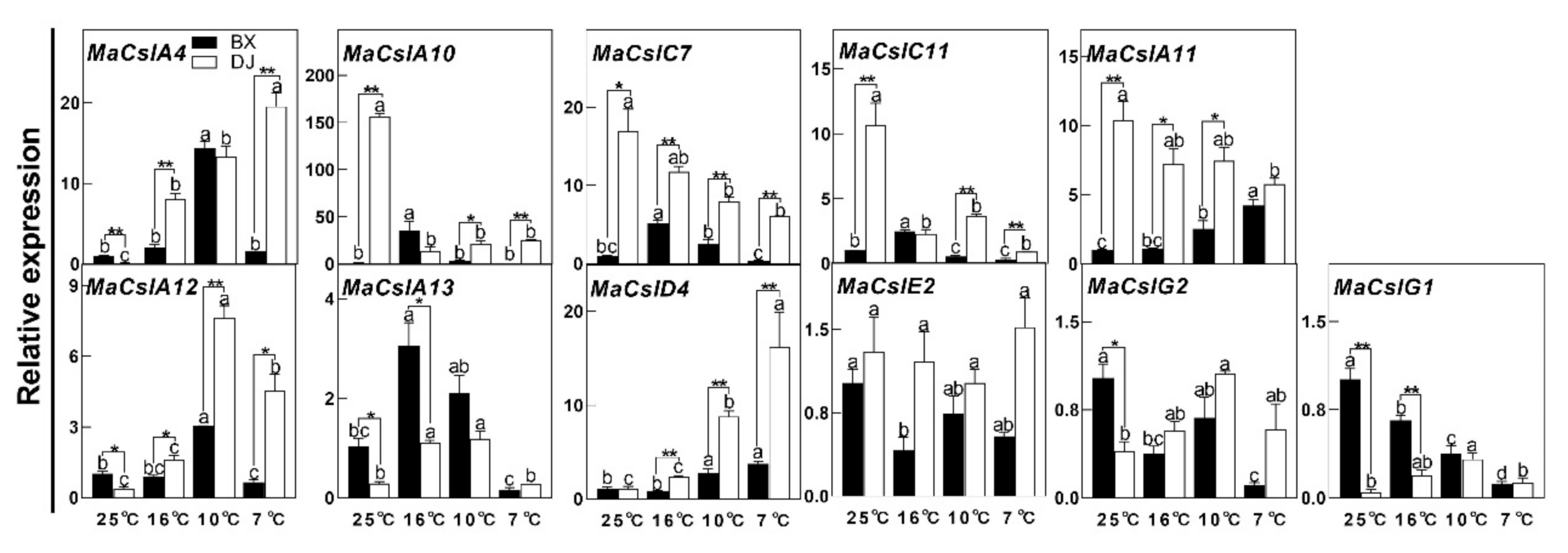
2.3. Differences in the Responses of MaCsls to LT Stress between CS and CT Cultivars
3. Discussion
3.1. The Features of MaCsls
3.2. The Involvement of MaCsls in Tolerance to LT Stress
4. Materials and Methods
4.1. Plant Materials and Natural LT Conditions
4.2. Measurement of Hemicellulose Content of Banana Leaves
4.3. RNA-Seq Analysis
4.4. Identification of Csls in Banana
4.5. Physicochemical Properties and Phylogenetic Analysis
4.6. Conserved Motif and Gene Structure Analysis
4.7. Quantification of the Expression Level of MaCsls Using qPCR
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
Abbreviations
| bp | Base pair |
| BX | Baxijiao |
| CK | The control |
| CS | Chilling-sensitive |
| Csl | Cellulose synthase-like gene |
| CT | Chilling-tolerant |
| DEG | Differentially expressed gene |
| DJ | Dongguandajiao |
| kD | Kilodalton |
| LT | Low temperature |
| LT16 | LT of 16 °C |
| LT10 | LT of 10 °C |
| LT7 | LT of 7 °C |
| MEME | Multiple Em for Motif Elucidation |
| MLG | Mixed-linkage glucan |
| MW | Molecular weight |
| pI | Isoelectric point |
| qPCR | Quantitative real-time PCR |
| RNA-Seq | RNA sequencing |
| UTR | Untranslated region |
References
- Hu, H.; Zhang, R.; Dong, S.; Li, Y.; Fan, C.; Wang, Y.; Xia, T.; Chen, P.; Wang, L.; Feng, S.; et al. AtCslD3 and GtCslD3 mediate root growth and cell elongation downstream of the ethylene response pathway in Arabidopsis. J. Exp. Bot. 2018, 69, 1065–1080. [Google Scholar] [CrossRef] [PubMed]
- Hunter, C.T.; Kirienko, D.H.; Sylvester, A.W.; Peter, G.F.; McCarty, D.R.; Koch, K.E. Cellulose synthase-like D1 is integral to normal cell division, expansion, and leaf development in maize. Plant Physiol. 2012, 158, 708–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hyles, J.; Vautrin, S.; Pettolino, F.; MacMillan, C.; Stachurski, Z.; Breen, J.; Berges, H.; Wicker, T.; Spielmeyer, W. Repeat-length variation in a wheat cellulose synthase-like gene is associated with altered tiller number and stem cell wall composition. J. Exp. Bot. 2017, 68, 1519–1529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Han, X.; Xu, S.; Li, C.; Wei, X.; Liu, Y.; Zhang, R.; Tang, X.; Zhou, J.; Huang, Z. Glycoside hydrolase family 39 #-xylosidase of sphingomonas showing salt/ethanol/trypsin tolerance, low-pH/low-temperature activity, and transxylosylation activity. J. Agric. Food Chem. 2018, 66, 9465–9472. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Shi, D.; Li, J.; Kong, Y.; Yu, Y.; Chai, G.; Hu, R.; Wang, J.; Hahn, M.G.; Zhou, G. Cellulose synthase-like A2, a glucomannan synthase, is involved in maintaining adherent mucilage structure in Arabidopsis seed. Plant Physiol. 2014, 164, 1842–1856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, X.; Pang, H.; Abbas, M.; Yan, X.; Dai, X.; Li, Y.; Li, Q. Characterization of cellulose synthase-like D (CslD) family revealed the involvement of PtrCslD5 in root hair formation in Populus trichocarpa. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Jiang, S.; Lin, G.; Cai, J.; Ye, X.; Chen, H.; Li, M.; Li, H.; Takáč, T.; Šamaj, J.; et al. Wound-induced pectin methylesterases enhance banana (Musa spp. AAA) susceptibility to Fusarium oxysporum f. sp. cubense. J. Exp. Bot. 2013, 64, 2219–2229. [Google Scholar] [CrossRef]
- Niu, Y.; Hu, B.; Li, X.; Chen, H.; Takáč, T.; Šamaj, J.; Xu, C. Comparative digital gene expression analysis of tissue-cultured plantlets of highly resistant and susceptible banana cultivars in response to Fusarium oxysporum. Int. J. Mol. Sci. 2018, 19, 350. [Google Scholar] [CrossRef] [Green Version]
- Yan, Y.; Takáč, T.; Li, X.; Chen, H.; Wang, Y.; Xu, E.; Xie, L.; Su, Z.; Šamaj, J.; Xu, C. Variable content and distribution of arabinogalactan proteins in banana (Musa spp.) under low temperature stress. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [Green Version]
- Meng, J.; Hu, B.; Yi, G.; Li, X.; Chen, H.; Wang, Y.; Yuan, W.; Xing, Y.; Sheng, Q.; Su, Z.; et al. Genome-wide analyses of banana fasciclin-like AGP genes and their differential expression under low-temperature stress in chilling sensitive and tolerant cultivars. Plant Cell Rep. 2020, 39, 693–708. [Google Scholar] [CrossRef]
- Richmond, T.A.; Somerville, C.R. The cellulose synthase superfamily. Plant Physiol. 2000, 124, 495–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy). Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazen, S.P.; Scott-Craig, J.S.; Walton, J.D. Cellulose synthase-like genes of rice. Plant Physiol. 2002, 128, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Farrokhi, N.; Burton, R.A.; Brownfield, L.; Hrmova, M.; Wilson, S.M.; Bacic, A.; Fincher, G.B. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 2006, 4, 145–167. [Google Scholar] [CrossRef] [PubMed]
- Little, A.; Schwerdt, J.G.; Shirley, N.J.; Khor, S.F.; Neumann, K.; O’ Donovan, L.A.; Lahnstein, J.; Collins, H.M.; Henderson, M.; Fincher, G.B.; et al. Revised phylogeny of the cellulose synthase gene superfamily: Insights into cell wall evolution. Plant Physiol. 2018, 177, 1124–1141. [Google Scholar] [CrossRef] [Green Version]
- Dhugga, K.S.; Barreiro, R.; Whitten, B.; Stecca, K.; Hazebroek, J.; Randhawa, G.S.; Dolan, M.; Kinney, A.J.; Tomes, D.; Nichols, S.; et al. Guar seed β-mannan synthase is a member of the cellulose synthase super gene family. Science 2004, 303, 363–366. [Google Scholar] [CrossRef]
- Goubet, F.; Barton, C.J.; Mortimer, J.C.; Yu, X.; Zhang, Z.; Miles, G.P.; Richens, J.; Liepman, A.H.; Seffen, K.; Dupree, P. Cell wall glucomannan in Arabidopsis is synthesised by CslA glycosyltransferases, and influences the progression of embryogenesis. Plant J. 2009, 60, 527–538. [Google Scholar] [CrossRef]
- Liepman, A.H.; Wilkerson, C.G.; Keegstra, K. Expression of cellulose synthase-like (Csl) genes in insect cells reveals that CslA family members encode mannan synthases. Proc. Natl. Acad. Sci. USA 2005, 102, 2221–2226. [Google Scholar] [CrossRef] [Green Version]
- He, C.; Zhang, J.; Liu, X.; Zeng, S.; Wu, K.; Yu, Z.; Wang, X.; Teixeira da Silva, J.A.; Lin, Z.; Duan, J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015, 88, 219–231. [Google Scholar] [CrossRef]
- Dwivany, F.M.; Yulia, D.; Burton, R.A.; Shirley, N.J.; Wilson, S.M.; Fincher, G.B.; Bacic, A.; Newbigin, E.; Doblin, M.S. The Cellulose-synthase like C (CslC) family of barley includes members that are integral membrane proteins targeted to the plasma membrane. Mol. Plant 2009, 2, 1025–1039. [Google Scholar] [CrossRef] [Green Version]
- Cocuron, J.C.; Lerouxel, O.; Drakakaki, G.; Alonso, A.P.; Liepman, A.H.; Keegstra, K.; Raikhel, N.; Wilkerson, C.G. A gene from the cellulose synthase-like C family encodes a beta-1,4 glucan synthase. Proc. Natl. Acad. Sci. USA 2007, 104, 8550–8555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Chandrasekar, B.; Rea, A.C.; Danhof, L.; Zemelis-Durfee, S.; Thrower, N.; Shepard, Z.S.; Pauly, M.; Brandizzi, F.; Keegstra, K. The synthesis of xyloglucan, an abundant plant cell wall polysaccharide, requires CslC function. Proc. Natl. Acad. Sci. USA 2020, 117, 20316–20324. [Google Scholar] [CrossRef] [PubMed]
- Verhertbruggen, Y.; Yin, L.; Oikawa, A.; Scheller, H.V. Mannan synthase activity in the CslD family. Plant Signal. Behav. 2015, 6, 1620–1623. [Google Scholar] [CrossRef] [Green Version]
- Dhugga, K.S. Biosynthesis of non-cellulosic polysaccharides of plant cell walls. Phytochemistry 2012, 74, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Szumlanski, A.L.; Gu, F.; Guo, F.; Nielsen, E. A role for CslD3 during cell-wall synthesis in apical plasma membranes of tip-growing root-hair cells. Nat. Cell Biol. 2011, 13, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Verhertbruggen, Y.; Oikawa, A.; Manisseri, C.; Knierim, B.; Prak, L.; Jensen, J.K.; Knox, J.P.; Auer, M.; Willats, W.G.T.; et al. The cooperative activities of CslD2, CslD3, and CslD5 are required for normal Arabidopsis development. Mol. Plant 2011, 4, 1024–1037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Bak, G.; Burgin, T.; Barnes, W.J.; Mayes, H.B.; Peña, M.J.; Urbanowicz, B.R.; Nielsen, E. Biochemical and genetic analysis identify CslD3 as a beta-1,4-glucan synthase that functions during plant cell wall synthesis. Plant Cell 2020, 32, 1749–1767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernal, A.J.; Jensen, J.K.; Harholt, J.; Sørensen, S.; Moller, I.; Blaukopf, C.; Johansen, B.; De Lotto, R.; Pauly, M.; Scheller, H.V.; et al. Disruption of AtCslD5 results in reduced growth, reduced xylan and homogalacturonan synthase activity and altered xylan occurrence in Arabidopsis. Plant J. 2007, 52, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Danilova, T.V.; Friebe, B.; Gill, B.S.; Poland, J.; Jackson, E. Development of a complete set of wheat–barley group-7 robertsonian translocation chromosomes conferring an increased content of β-glucan. Theor. Appl. Genet. 2017, 131, 377–388. [Google Scholar] [CrossRef]
- Burton, R.A.; Wilson, S.M.; Hrmova, M.; Harvey, A.J.; Shirley, N.J.; Medhurst, A.; Stone, B.A.; Newbigin, E.J.; Bacic, A.; Fincher, G.B. Cellulose synthase-like CslF genes mediate the synthesis of cell wall (1,3;1,4)-β-d-glucans. Science 2006, 311, 1940–1942. [Google Scholar] [CrossRef]
- Doblin, M.S.; Pettolino, F.A.; Wilson, S.M.; Campbell, R.; Burton, R.A.; Fincher, G.B.; Newbigin, E.; Bacic, A. A barley cellulose synthase-like CslH gene mediates (1,3;1,4)-beta-D-glucan synthesis in transgenic Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 5996–6001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vega-Sánchez, M.E.; Verhertbruggen, Y.; Christensen, U.; Chen, X.; Sharma, V.; Varanasi, P.; Jobling, S.A.; Talbot, M.; White, R.G.; Joo, M.; et al. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice. Plant Physiol. 2012, 159, 56–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Little, A.; Lahnstein, J.; Jeffery, D.W.; Khor, S.F.; Schwerdt, J.G.; Shirley, N.J.; Hooi, M.; Xing, X.; Burton, R.A.; Bulone, V. A novel (1,4)-β-linked glucoxylan is synthesized by members of the cellulose synthase-like F gene family in land plants. ACS Cent. Sci. 2019, 5, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheible, W.R.; Pauly, M. Glycosyltransferases and cell wall biosynthesis: Novel players and insights. Curr. Opin. Plant Biol. 2004, 7, 285–295. [Google Scholar] [CrossRef]
- Zhu, J.; Lee, B.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.A.; Zhu, J. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bagheri, R.; Bashir, H.; Ahmad, J.; Iqbal, M.; Qureshi, M.I. Spinach (Spinacia oleracea L.) modulates its proteome differentially in response to salinity, cadmium and their combination stress. Plant Physiol. Bioch. 2015, 97, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Cristóbal, J.J.; Herrera-Rodríguez, M.B.; Beato, V.M.; Rexach, J.; Navarro-Gochicoa, M.T.; Maldonado, J.M.; González-Fontes, A. The expression of several cell wall-related genes in Arabidopsis roots is down-regulated under boron deficiency. Environ. Exp. Bot. 2008, 63, 351–358. [Google Scholar] [CrossRef]
- İşkil, R.; Surgun-Acar, Y. Expression analysis of cell wall assembly and remodelling-related genes in Arabidopsis roots subjected to boron stress and brassinosteroid at different developmental stages. Acta Bot. Bras. 2018, 32, 546–554. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Wu, X.; Liu, D.; Yao, J.; Liang, G.; Song, H.; Ismail, A.M.; Luo, J.; Zhang, Z. Cell wall polysaccharide-mediated cadmium tolerance between two Arabidopsis thaliana ecotypes. Front. Plant Sci. 2020, 11, 473. [Google Scholar] [CrossRef]
- Aditya, J.; Lewis, J.; Shirley, N.J.; Tan, H.; Fincher, M.H.B.; Burton, R.A.; Mather, D.E.; Tucker, M.R. The dynamics of cereal cyst nematode infection differ between susceptible and resistant barley cultivars and lead to changes in (1,3;1,4)-β-glucan levels and HvCslF gene transcript abundance. New Phytol. 2015, 207, 135–147. [Google Scholar] [CrossRef]
- Douchkov, D.; Lueck, S.; Hensel, G.; Kumlehn, J.; Rajaraman, J.; Johrde, A.; Doblin, M.S.; Beahan, C.T.; Kopischke, M.; Fuchs, R.; et al. The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016, 212, 421–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domon, J.; Baldwin, L.; Acket, S.; Caudeville, E.; Arnoult, S.; Zub, H.; Gillet, F.; Lejeune-Hénaut, I.; Brancourt-Hulmel, M.; Pelloux, J.; et al. Cell wall compositional modifications of Miscanthus ecotypes in response to cold acclimation. Phytochemistry 2013, 85, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Le Gall, H.; Philippe, F.; Domon, J.; Gillet, F.; Pelloux, J.; Rayon, C. Cell wall metabolism in response to abiotic stress. Plants 2015, 4, 112–166. [Google Scholar] [CrossRef] [PubMed]
- Le, M.Q.; Pagter, M.; Hincha, D.K. Global changes in gene expression, assayed by microarray hybridization and quantitative RT-PCR, during acclimation of three Arabidopsis thaliana accessions to sub-zero temperatures after cold acclimation. Plant Mol. Biol. 2015, 87, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Gao, J.; He, W.; Dou, T.; Ding, L.; Wu, J.; Li, C.; Peng, X.; Zhang, S.; Yi, G. Comparative transcriptomics analysis reveals difference of key gene expression between banana and plantain in response to cold stress. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, S.K.; Pandit, E.; Nayak, D.K.; Behera, L.; Mohapatra, T. Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol. 2019, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panter, P.E.; Kent, O.; Dale, M.; Smith, S.J.; Skipsey, M.; Thorlby, G.; Cummins, I.; Ramsay, N.; Begum, R.A.; Sanhueza, D.; et al. MUR1-mediated cell-wall fucosylation is required for freezing tolerance in Arabidopsis thaliana. New Phytol. 2019, 224, 1518–1531. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, D.; Johnson, K.L.; Hao, P.; Tuong, T.; Erban, A.; Sampathkumar, A.; Bacic, A.; Livingston, D.P.; Kopka, J.; Kuroha, T.; et al. Cell wall modification by the xyloglucan endotransglucosylase/hydrolase XTH19 influences freezing tolerance after cold and sub-zero acclimation. Plant Cell Environ. 2020. [Google Scholar] [CrossRef]
- Djerbi, S.; Lindskog, M.; Arvestad, L.; Sterky, F.; Teeri, T.T. The genome sequence of black cottonwood (Populus trichocarpa) reveals 18 conserved cellulose synthase (CesA) genes. Planta 2005, 221, 739–746. [Google Scholar] [CrossRef]
- Suzuki, S.; Li, L.; Sun, Y.; Chiang, V.L. The cellulose synthase gene superfamily and biochemical functions of xylem-specific cellulose synthase-like genes in Populus trichocarpa. Plant Physiol. 2006, 142, 1233–1245. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.W.; Bushoven, J.T. The cellulose synthase (CesA) gene superfamily of the moss Physcomitrella patens. Plant Mol. Biol. 2007, 63, 207–219. [Google Scholar] [CrossRef] [PubMed]
- Appenzeller, L.; Doblin, M.; Barreiro, R.; Wang, H.; Niu, X.; Kollipara, K.; Carrigan, L.; Tomes, D.; Chapman, M.; Dhugga, K.S. Cellulose synthesis in maize: Isolation and expression analysis of the cellulose synthase (CesA) gene family. Cellulose 2004, 11, 287–299. [Google Scholar] [CrossRef]
- Li, Y.; Cheng, X.; Fu, Y.; Wu, Q.; Guo, Y.; Peng, J.; Zhang, W.; He, B. A genome-wide analysis of the cellulose synthase-like (Csl) gene family in maize. Biol. Plantarum. 2019, 63, 721–732. [Google Scholar] [CrossRef]
- Burton, R.A.; Shirley, N.J.; King, B.J.; Harvey, A.J.; Fincher, G.B. The CesA gene family of barley quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol. 2004, 134, 224–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nairn, C.J.; Haselkorn, T. Three loblolly pine CesA genes expressed in developing xylem are orthologous to secondary cell wall CesA genes of angiosperms. New Phytol. 2005, 166, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Xu, L.; Yu, J.; Tian, P.; Hu, X. Genome-wide characterization of the cellulose synthase gene superfamily in Solanum lycopersicum. Gene 2018, 688, 71–83. [Google Scholar] [CrossRef]
- Cao, S.; Cheng, H.; Zhang, J.; Aslam, M.; Yan, M.; Hu, A.; Lin, L.; Ojolo, S.P.; Zhao, H.; Priyadarshani, S.; et al. Genome-wide identification, expression pattern analysis and evolution of the Ces/Csl gene superfamily in pineapple (Ananas comosus). Plants 2019, 8, 275. [Google Scholar] [CrossRef] [Green Version]
- Kaur, S.; Dhugga, K.S.; Beech, R.; Singh, J. Genome-wide analysis of the cellulose synthase-like (Csl) gene family in bread wheat (Triticum aestivum L.). BMC Plant Biol. 2017, 17, 193. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Liu, X.; Liang, Y.; Zhang, Y.; Cheng, X.; Cai, Y. Genome-wide characterization of the cellulose synthase gene superfamily in Pyrus bretschneideri and reveal its potential role in stone cell formation. Funct. Integr. Genomic. 2020, 20, 723–738. [Google Scholar] [CrossRef]
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.P.; Jenny, C.; et al. Multidisciplinary perspectives on banana (Musa spp.) domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [Green Version]
- FAOSTAT. 2020. Available online: https://www.fao.org/faostat/en/#data/QC/visualize (accessed on 6 March 2020).
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; de Souza, A.; Schultink, A.; Xiong, G. Hemicellulose biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Huang, J.; Xu, Y. The cellulose synthase superfamily in fully sequenced plants and algae. BMC Plant Biol. 2009, 9, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, N.; Holland, D.; Helentjaris, T.; Dhugga, K.S.; Xoconostle-Cazares, B.; Delmer, D.P. A comparative analysis of the plant cellulose synthase (CesA) gene family. Plant Physiol. 2000, 123, 1313–1324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CesA/Csl superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, C.; Li, Q.; Duan, C.; Chen, D.; Liu, J. Bioinformatics and expression analysis on Csl gene family in Dendrobium catenatum [in Chinese with English abstract]. GAB 2019, 38, 2159–2166. [Google Scholar] [CrossRef]
- Burton, R.A.; Collins, H.M.; Kibble, N.A.; Smith, J.A.; Shirley, N.J.; Jobling, S.A.; Henderson, M.; Singh, R.R.; Pettolino, F.; Wilson, S.M.; et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-beta-D-glucans and alters their fine structure. Plant Biotechnol. J. 2011, 9, 117–135. [Google Scholar] [CrossRef]
- Yin, Y.; Johns, M.A.; Cao, H.; Rupani, M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genom. 2014, 15, 260. [Google Scholar] [CrossRef] [Green Version]
- Carpita, N.C.; McCann, M.C. The maize mixed-linkage (1→3),(1→4)-β-D-glucan polysaccharide is synthesized at the golgi membrane. Plant Physiol. 2010, 153, 1362–1371. [Google Scholar] [CrossRef] [Green Version]
- Kubacka-Zębalska, M.; Kacperska, A. Low temperature-induced modifications of cell wall content and polysaccharide composition in leaves of winter oilseed rape (Brassica napus L. Var. Oleifera L.). Plant Sci. 1999, 148, 59–67. [Google Scholar] [CrossRef]
- Weiser, R.L.; Wallner, S.J.; Waddell, J.W. Cell wall and extensin mRNA changes during cold acclimation of pea seedlings. Plant Physiol. 1990, 93, 1021–1026. [Google Scholar] [CrossRef] [Green Version]
- Plancot, B.; Gügi, B.; Mollet, J.; Loutelier-Bourhis, C.; Govind, S.R.; Lerouge, P.; Follet-Gueye, M.; Vicré, M.; Alfonso, C.; Nguema-Ona, E.; et al. Desiccation tolerance in plants: Structural characterization of the cell wall hemicellulosic polysaccharides in three Selaginella species. Carbohyd. Polym. 2019, 208, 180–190. [Google Scholar] [CrossRef] [PubMed]
- Zang, D.; Wang, J.; Zhang, X.; Liu, Z.; Wang, Y. Arabidopsis heat shock transcription factor HSFA7b positively mediates salt stress tolerance by binding to an E-box-like motif to regulate gene expression. J. Exp. Bot. 2019, 70, 5355–5374. [Google Scholar] [CrossRef] [PubMed]
- Budot, B.O.; Encabo, J.R.; Ambita, I.D.V.; Atienza-Grande, G.A.; Satoh, K.; Kondoh, H.; Ulat, V.J.; Mauleon, R.; Kikuchi, S.; Choi, I. Suppression of cell wall-related genes associated with stunting of Oryza glaberrima infected with rice tungro spherical virus. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, J.; Schober, M.S.; Shirley, N.J.; Singh, R.R.; Jacobs, A.K.; Douchkov, D.; Schweizer, P.; Fincher, G.B.; Burton, R.A.; Little, A. Down-regulation of the glucan synthase-like 6 gene (HvGsl6) in barley leads to decreased callose accumulation and increased cell wall penetration by Blumeria graminis f. sp. hordei. New Phytol. 2016, 212, 434–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kesten, C.; Menna, A.; Sanchez-Rodriguez, C. Regulation of cellulose synthesis in response to stress. Curr. Opin. Plant Biol. 2017, 40, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; McFarlane, H.E.; Persson, S. The impact of abiotic factors on cellulose synthesis. J. Exp. Bot. 2016, 67, 543–552. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.P.; Nguema-Ona, E.E.; Vicré-Gibouin, M.; Sørensen, I.; Willats, W.G.T.; Driouich, A.; Farrant, J.M. Arabinose-rich polymers as an evolutionary strategy to plasticize resurrection plant cell walls against desiccation. Planta 2013, 237, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ji, Z.; Li, P. A study on the injury symptoms and physiological quota of banana and an effective measure for cold injury protection [in Chinese with English abstract]. J. South China Agri. Univ. 1982, 3, 1–12. [Google Scholar]
- Sluiter, A.D.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.W.; Crocker, D. Determination of Structural Carbohydrates and Lignin in Biomass; NREL: Golden, CO, USA, 2012. Available online: http://www.nrel.gov/biomass/analytical_procedures.html (accessed on 2 April 2020).
- Hu, G.; Ellberg, S.; Burton, C.; Evans, C.; Satterfield, K.; Bockelman, H. Application of an orcinol-ferric chloride colorimetric assay in barley and wheat accessions for water-extractable and total arabinoxylan. J. Cereal Sci. 2020, 93, 102962. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Logacheva, M.D.; Dmitriev, S.E.; Penin, A.A. RNA-seq analysis of an apical meristem time series reveals a critical point in Arabidopsis thaliana flower initiation. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Xia, R.; Chen, H.; He, Y. TBtools, a toolkit for biologists integrating various biological data handling tools with a user-friendly interface. BioRxiv 2018, 13, 1194–1202. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. Clustal W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, B.; Jin, J.; Guo, A.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
| Gene Name | Gene ID | Chr | Start | End | Length (bp) | Strand | Size (aa) | pI | MW (kD) |
|---|---|---|---|---|---|---|---|---|---|
| MaCslA1 | Ma02_t22150 | chr02 | 27,565.079 | 27,570.275 | 1602 | − | 534 | 8.9 | 60,834.3 |
| MaCslA2 | Ma03_t01730 | chr03 | 1207.899 | 1213.633 | 1629 | + | 543 | 9.0 | 61,799.5 |
| MaCslA3 | Ma05_t18900 | chr05 | 25,534.488 | 25,538.961 | 1602 | − | 534 | 9.0 | 60,875.2 |
| MaCslA4 | Ma06_t04300 | chr06 | 3100.141 | 3102.901 | 1635 | − | 545 | 8.7 | 62,234.1 |
| MaCslA5 | Ma06_t09180 | chr06 | 6479.551 | 6484.223 | 1668 | + | 556 | 9.0 | 63,946.1 |
| MaCslA6 | Ma06_t14920 | chr06 | 10,164.997 | 10,168.70 | 1602 | + | 534 | 8.9 | 60,781.3 |
| MaCslA7 | Ma07_t19410 | chr07 | 27,422.775 | 27,428.514 | 1719 | + | 573 | 9.1 | 65,060.1 |
| MaCslA8 | Ma07_t22600 | chr07 | 30,481.370 | 30,484.454 | 1677 | + | 559 | 9.0 | 64,175.8 |
| MaCslA9 | Ma09_t27610 | chr09 | 38,562.778 | 38,574.772 | 1626 | + | 542 | 8.8 | 61,639.2 |
| MaCslA10 | Ma10_t11450 | chr10 | 24,986.818 | 24,991.666 | 1602 | + | 534 | 8.9 | 61,025.5 |
| MaCslA11 | Ma10_t15510 | chr10 | 27,582.419 | 27,587.095 | 1602 | − | 534 | 9.1 | 60,994.7 |
| MaCslA12 | Ma10_t18740 | chr10 | 29,549.996 | 29,554.980 | 1755 | − | 585 | 8.9 | 65,936.2 |
| MaCslA13 | Ma11_t08690 | chr11 | 6915.612 | 6920.893 | 1626 | + | 542 | 9.3 | 62,165.2 |
| MaCslC1 | Ma03_t29290 | chr03 | 32,234.549 | 32,238.695 | 2106 | + | 702 | 8.9 | 79,991.8 |
| MaCslC2 | Ma04_t02130 | chr04 | 1873.146 | 1876.963 | 2094 | − | 698 | 8.7 | 79,922.4 |
| MaCslC3 | Ma04_t05930 | chr04 | 4437.079 | 4440.382 | 2085 | + | 695 | 9.1 | 79,398.2 |
| MaCslC4 | Ma04_t29650 | chr04 | 30,511.868 | 30,515.518 | 2115 | + | 705 | 8.5 | 79,553.3 |
| MaCslC5 | Ma04_t38760 | chr04 | 36,164.056 | 36,168.517 | 2106 | + | 702 | 7.8 | 79,537.2 |
| MaCslC6 | Ma05_t01870 | chr05 | 1142.347 | 1146.294 | 2112 | − | 704 | 8.5 | 79,580.2 |
| MaCslC7 | Ma06_t03600 | chr06 | 2622.347 | 2625.771 | 2067 | + | 689 | 8.8 | 79,142.5 |
| MaCslC8 | Ma06_t29550 | chr06 | 30,901.418 | 30,906.541 | 2091 | + | 697 | 9.1 | 78,791.9 |
| MaCslC9 | Ma06_t31890 | chr06 | 32,901.867 | 32,906.161 | 2115 | + | 705 | 8.1 | 79,750.6 |
| MaCslC10 | Ma07_t00740 | chr07 | 619,565 | 624,194 | 2121 | − | 707 | 8.0 | 79,935.6 |
| MaCslC11 | Ma07_t20970 | chr07 | 28,958.080 | 28,961.838 | 2106 | − | 702 | 8.8 | 80,597.2 |
| MaCslC12 | Ma10_t09350 | chr10 | 23,545.265 | 23,549.303 | 2124 | − | 708 | 7.5 | 79,820.2 |
| MaCslD1 | Ma01_t18500 | chr01 | 13,756.260 | 13,761.905 | 3450 | + | 1150 | 6.9 | 128,482.2 |
| MaCslD2 | Ma02_t07580 | chr02 | 18,211.558 | 18,215.380 | 3447 | − | 1149 | 6.4 | 128,432.9 |
| MaCslD3 | Ma02_t17080 | chr02 | 24,143.640 | 24,146.984 | 2709 | − | 903 | 8.9 | 100,568.6 |
| MaCslD4 | Ma03_t14070 | chr03 | 11,229.456 | 11,235.431 | 3453 | + | 1151 | 7.5 | 128,361.1 |
| MaCslD5 | Ma03_t25420 | chr03 | 29,461.721 | 29,465.754 | 3657 | − | 1219 | 8.1 | 134,887.3 |
| MaCslD6 | Ma04_t04560 | chr04 | 3486.824 | 3489.813 | 2757 | + | 919 | 8.8 | 103,194.6 |
| MaCslD7 | Ma04_t33100 | chr04 | 32,908.316 | 32,909.842 | 1245 | − | 415 | 9.5 | 46,143.7 |
| MaCslD8 | Ma10_t26210 | chr10 | 34,009.560 | 34,013.193 | 2721 | − | 907 | 8.9 | 100,850.9 |
| MaCslD9 | Ma11_t21750 | chr11 | 25,761.358 | 25,765.90 | 3480 | + | 1160 | 6.8 | 128,916.7 |
| MaCslE1 | Ma04_t13090 | chr04 | 9902.007 | 9907.918 | 2187 | − | 729 | 8.3 | 83,277.4 |
| MaCslE2 | Ma09_t20060 | chr09 | 27,488.282 | 27,493.065 | 2241 | + | 747 | 8.2 | 84,590.1 |
| MaCslE3 | Ma09_t28670 | chr09 | 39,320.301 | 39,325.310 | 2193 | + | 731 | 8.5 | 82,679.5 |
| MaCslG1 | Ma08_t05160 | chr08 | 3537.407 | 3540.538 | 2193 | − | 731 | 8.2 | 81,197.7 |
| MaCslG2 | Ma08_t05170 | chr08 | 3544.521 | 3548.562 | 2169 | − | 723 | 6.9 | 80,645.9 |
| MaCslG3 | Ma08_t05180 | chr08 | 3548.894 | 3551.285 | 1638 | − | 546 | 6.7 | 60,810.7 |
| MaCslG4 | Ma08_t05190 | chr08 | 3561.425 | 3563.941 | 1602 | − | 534 | 6.5 | 59,472.2 |
| MaCslH1 | Ma09_t08420 | chr09 | 5566.954 | 5569.888 | 2223 | + | 741 | 7.2 | 83,277.7 |
| Gene Name | log2 Fold Change | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CKDJ vs. CKBX | LT16DJ vs. LT16BX | LT10DJ vs. LT10BX | LT7DJ vs. LT7BX | LT16BX vs. CKBX | LT10BX vs. CKBX | LT7BX vs. CKBX. | LT16DJ vs. CKDJ | LT10DJ vs. CKDJ | LT7DJ vs. CKDJ | |
| MaCslA1 | −2.52 | |||||||||
| MaCslA2 | ||||||||||
| MaCslA3 | −1.73 | −2.44 | ||||||||
| MaCslA4 | −2.10 | 1.51 | 2.00 | 2.31 | ||||||
| MaCslA5 | −1.70 | −1.99 | −2.20 | −2.46 | −2.88 | −2.81 | ||||
| MaCslA6 | ||||||||||
| MaCslA7 | −2.63 | −1.18 | −2.20 | |||||||
| MaCslA8 | 2.31 | −1.82 | 3.98 | |||||||
| MaCslA9 | −1.35 | −1.42 | −1.41 | −1.44 | ||||||
| MaCslA10 | 3.11 | 3.94 | −3.60 | −4.16 | −3.94 | |||||
| MaCslA11 | 1.96 | 2.12 | −1.76 | −2.17 | −2.20 | |||||
| MaCslA12 | −1.59 | −1.73 | 1.14 | |||||||
| MaCslA13 | 1.85 | −3.81 | −1.71 | |||||||
| MaCslC1 | ||||||||||
| MaCslC2 | −3.54 | −6.28 | −4.44 | −5.34 | −5.66 | |||||
| MaCslC3 | 4.90 | 3.93 | ||||||||
| MaCslC4 | ||||||||||
| MaCslC5 | 2.96 | −3.23 | 3.17 | 3.75 | −2.28 | −3.07 | −1.95 | |||
| MaCslC6 | −1.90 | −1.65 | ||||||||
| MaCslC7 | 3.86 | 2.59 | 3.37 | −2.97 | −3.22 | −3.20 | ||||
| MaCslC8 | −1.05 | −1.22 | −1.10 | |||||||
| MaCslC9 | −1.61 | |||||||||
| MaCslC10 | −3.45 | −3.96 | −4.22 | −3.94 | −3.53 | |||||
| MaCslC11 | 1.68 | 1.94 | −3.00 | −3.78 | −2.85 | −3.27 | −4.51 | |||
| MaCslC12 | −1.26 | |||||||||
| MaCslD1 | 1.58 | 1.34 | ||||||||
| MaCslD2 | 4.04 | |||||||||
| MaCslD3 | ||||||||||
| MaCslD4 | 1.27 | 1.34 | 1.52 | 2.16 | ||||||
| MaCslD5 | 2.46 | 2.70 | ||||||||
| MaCslD6 | ||||||||||
| MaCslD7 | ||||||||||
| MaCslD8 | ||||||||||
| MaCslD9 | −1.28 | 3.71 | 1.89 | 4.41 | ||||||
| MaCslE1 | −1.27 | |||||||||
| MaCslE2 | 1.50 | 1.98 | 1.37 | 2.58 | −1.55 | −1.70 | −2.11 | −1.16 | −1.89 | |
| MaCslE3 | −1.01 | −1.97 | −1.16 | |||||||
| MaCslG1 | 1.33 | 1.30 | 2.74 | −2.31 | ||||||
| MaCslG2 | −1.17 | 1.79 | −1.03 | −4.03 | ||||||
| MaCslG3 | −1.54 | −1.41 | −3.18 | |||||||
| MaCslG4 | ||||||||||
| MaCslH1 | −3.09 | −1.38 | −2.41 | −4.62 | −3.46 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, W.; Liu, J.; Takáč, T.; Chen, H.; Li, X.; Meng, J.; Tan, Y.; Ning, T.; He, Z.; Yi, G.; et al. Genome-Wide Identification of Banana Csl Gene Family and Their Different Responses to Low Temperature between Chilling-Sensitive and Tolerant Cultivars. Plants 2021, 10, 122. https://doi.org/10.3390/plants10010122
Yuan W, Liu J, Takáč T, Chen H, Li X, Meng J, Tan Y, Ning T, He Z, Yi G, et al. Genome-Wide Identification of Banana Csl Gene Family and Their Different Responses to Low Temperature between Chilling-Sensitive and Tolerant Cultivars. Plants. 2021; 10(1):122. https://doi.org/10.3390/plants10010122
Chicago/Turabian StyleYuan, Weina, Jing Liu, Tomáš Takáč, Houbin Chen, Xiaoquan Li, Jian Meng, Yehuan Tan, Tong Ning, Zhenting He, Ganjun Yi, and et al. 2021. "Genome-Wide Identification of Banana Csl Gene Family and Their Different Responses to Low Temperature between Chilling-Sensitive and Tolerant Cultivars" Plants 10, no. 1: 122. https://doi.org/10.3390/plants10010122
APA StyleYuan, W., Liu, J., Takáč, T., Chen, H., Li, X., Meng, J., Tan, Y., Ning, T., He, Z., Yi, G., & Xu, C. (2021). Genome-Wide Identification of Banana Csl Gene Family and Their Different Responses to Low Temperature between Chilling-Sensitive and Tolerant Cultivars. Plants, 10(1), 122. https://doi.org/10.3390/plants10010122






