Wheat, Rye, and Barley Genomes Can Associate during Meiosis in Newly Synthesized Trigeneric Hybrids
Abstract
1. Introduction
2. Results
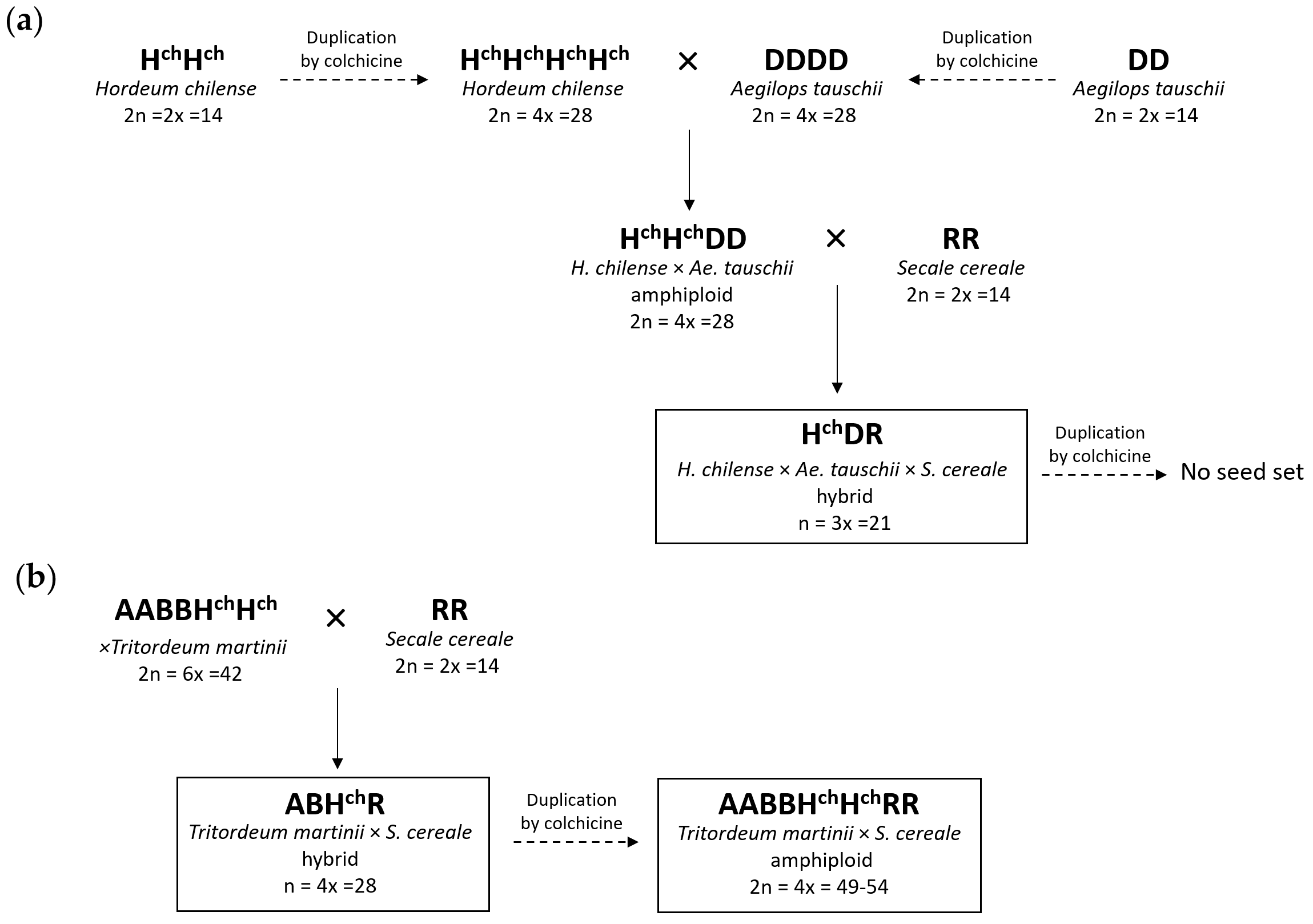
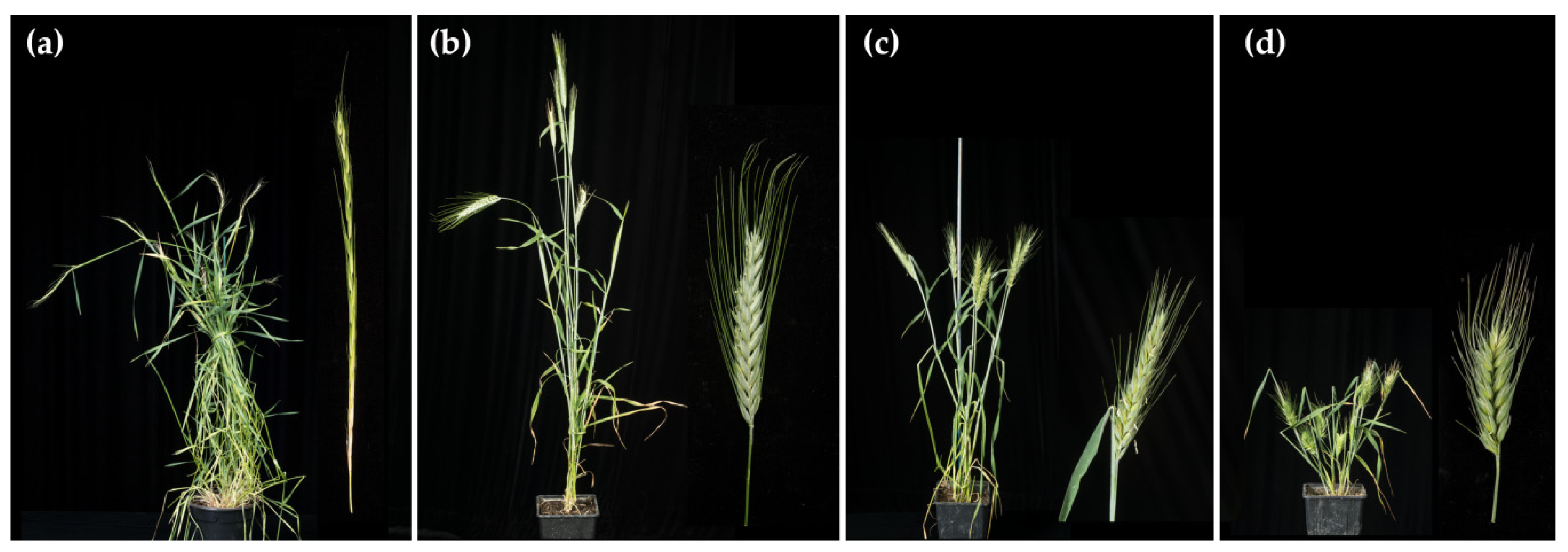
2.1. Production, Chromosome Constitution, and Morphology of the Trigeneric Hybrids and Amphiploids
2.1.1. Hybrid HchDR
2.1.2. Hybrid ABHchR
2.1.3. Amphiploid AABBHchHchRR
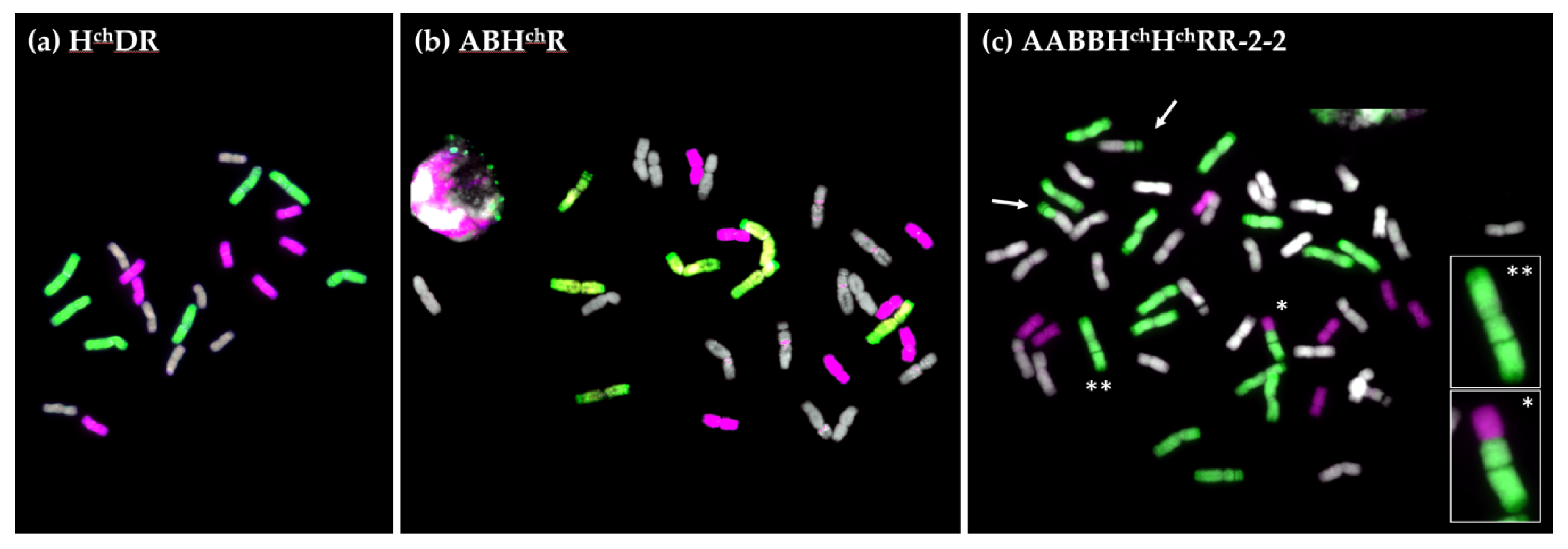
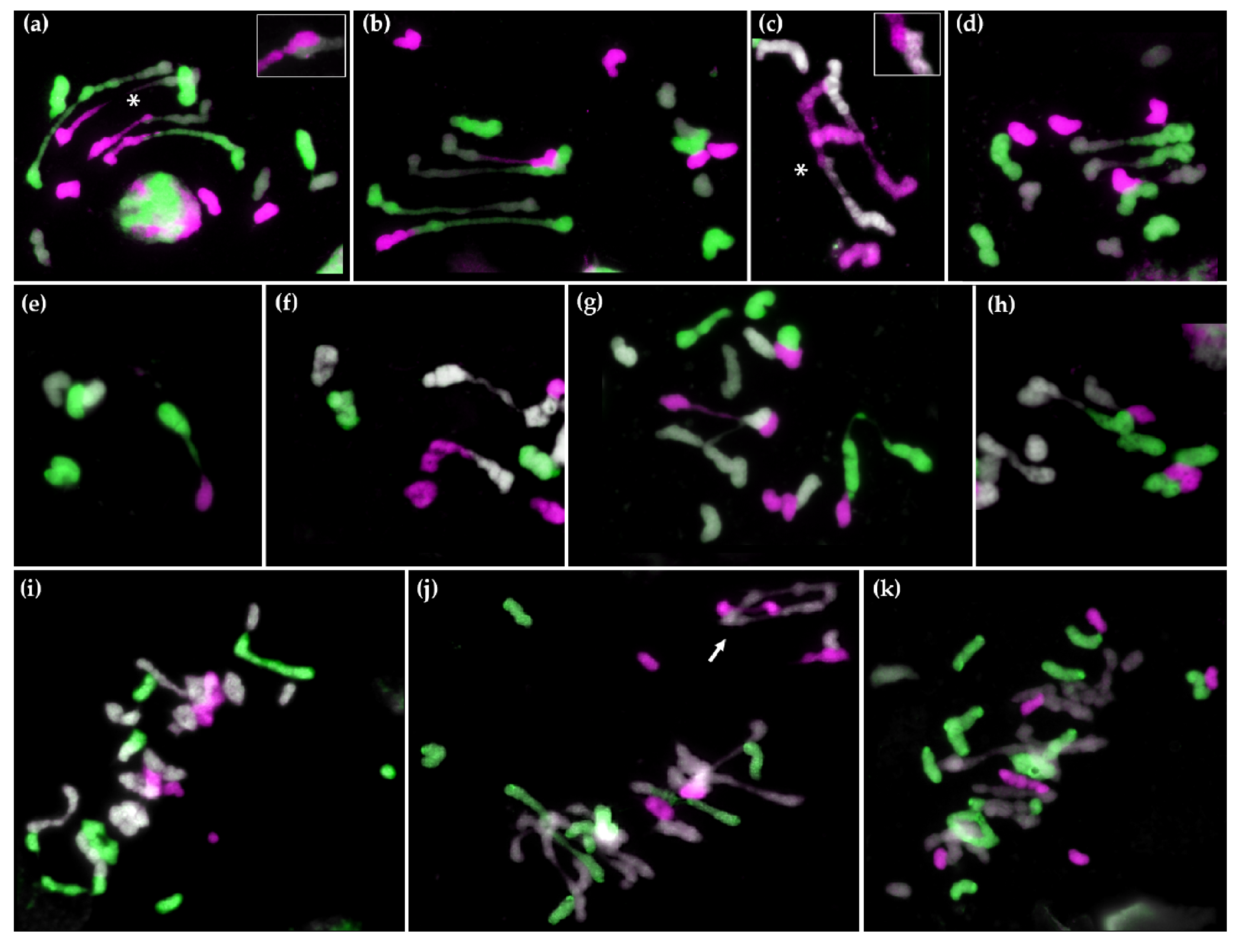
2.2. GISH Analysis of Meiotic Metaphase I Configuration in Hybrids and Amphiploids
2.2.1. Meiotic Metaphase I Configuration in the Trigeneric Hybrid HchDR
2.2.2. Meiotic Metaphase I Configuration in the Trigeneric Hybrid ABHchR and its Corresponding Amphiploid AABBHchHchRR
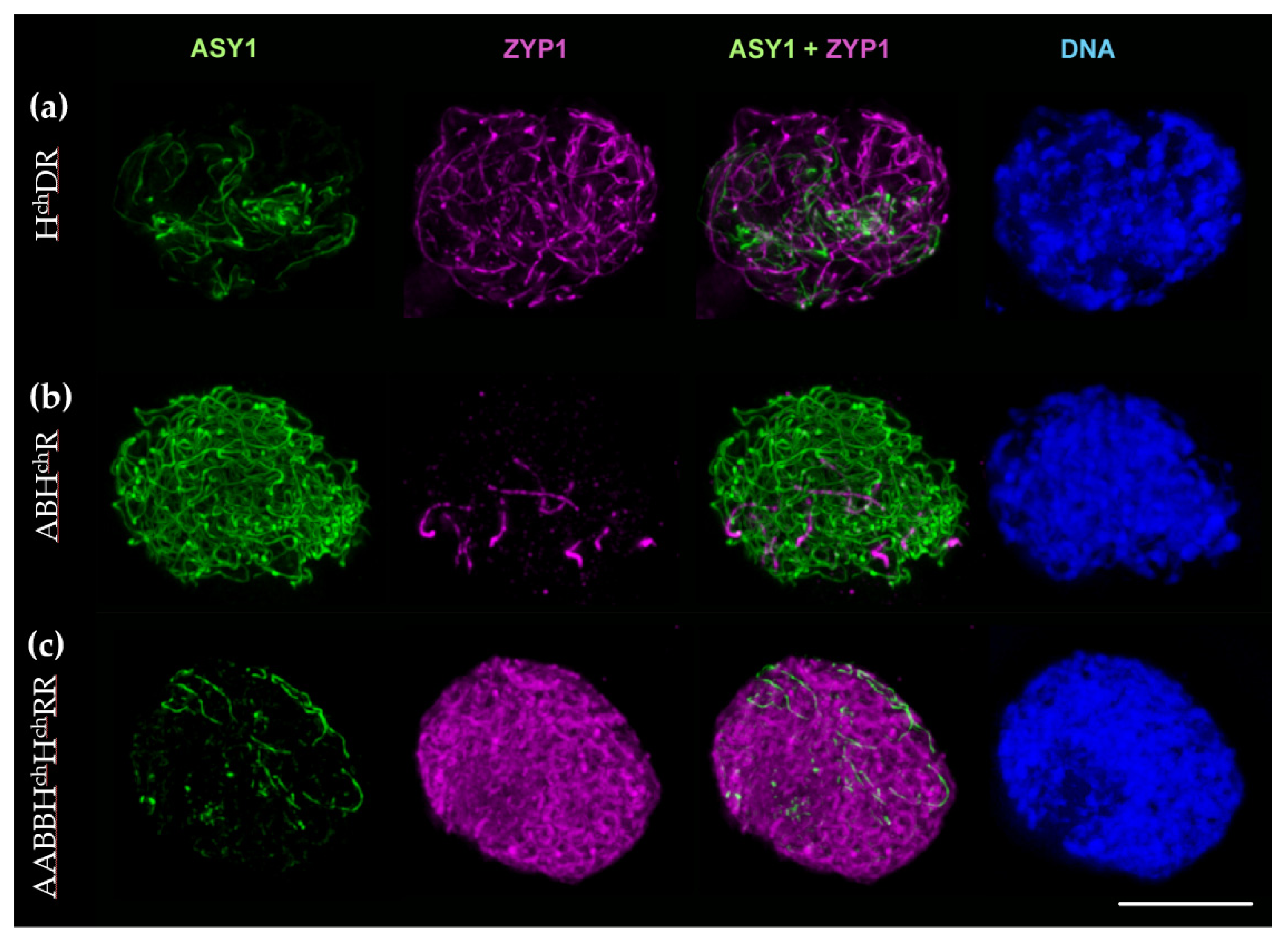
2.2.3. Synaptonemal Complex Formation in HchDR and ABHchR Hybrids, and the Amphiploid ABBHchHchRR
3. Discussion
3.1. CO Formation Can Take Place between Ae. tauschii, Barley and Rye in the HchDR hybrid
3.2. Most Associations between Wheat, Rye, and Barley in the ABHchR Hybrid Are Probably Achiasmatic
3.3. The Amphiploid AABBHchHchRR Is Highly Unstable and Mostly Sterile
4. Materials and Methods
4.1. Plant Material
4.2. Genomic In Situ Hybridization (GISH) of Mitotic and Meiotic Cells
4.3. Immunolocalisation of Meiotic Proteins
4.4. Image Acquisition and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Renny-Byfield, S.; Wendel, J.F. Doubling down on genomes: Polyploidy and crop plants. Am. J. Bot. 2014, 101, 1711–1725. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Naranjo, T. The effect of chromosome structure upon meiotic homologous and homoeologous recombinations in Triticeae. Agronomy 2019, 9, 552. [Google Scholar] [CrossRef]
- O’Mara, J.G. The cytogenetics of Triticale. Bot. Rev. 1953, 19, 587–605. [Google Scholar] [CrossRef]
- Martín, A.; Sánchez-Monge Laguna, E. Cytology and morphology of the amphiploid Hordeum chilense × Triticum turgidum conv. durum. Euphytica 1982, 31, 262–267. [Google Scholar]
- Fatih, A.M.B. Analysis of the breeding potential of wheat-Agropyron and wheat-Elymus derivatives: I. Agronomic and quality characteristics. Hereditas 1983, 98, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Forster, B.P.; Phillips, M.S.; Miller, T.E.; Baird, E.; Powell, W. Chromosome location of genes controlling tolerance to salt (NaCl) and vigour in Hordeum vulgare and H. chilense. Heredity 1990, 65, 99–107. [Google Scholar] [CrossRef]
- Hohmann, U.; Busch, W.; Badaeva, K.; Friebe, B.; Gill, B.S. Molecular cytogenetic analysis of Agropyron chromatin specifying resistance to barley yellow dwarf virus in wheat. Genome 1996, 39, 336–347. [Google Scholar] [CrossRef]
- Koba, T.; Takumi, S.; Shimada, T. Isolation, identification and characterization of disomic and translocated barley chromosome addition lines of common wheat. Euphytica 1997, 96, 289–296. [Google Scholar] [CrossRef]
- Schlegel, R.; Cakmak, I.; Torun, B.; Eker, S.; Tolay, I.; Ekiz, H.; Kalayci, M.; Braun, H.J. Screening for zinc efficiency among wheat relatives and their utilisation for alien gene transfer. In Wheat: Prospects for Global Improvement; Springer: Dordrecht, The Netherlands, 1997; pp. 347–352. [Google Scholar]
- Reynolds, M.P.; Calderini, D.F.; Condon, A.G.; Rajaram, S. Physiological basis of yield gains in wheat associated with the LR19 translocation from Agropyron elongatum. In Wheat in a Global Environmen; Springer: Dordrecht, The Netherlands, 2001; pp. 345–351. [Google Scholar]
- Rakesh, V.; Sethi, G.S. Cytogenetic analysis and differential response of rye-introgressed bread wheat genotypes for cold tolerance. Indian J. Genet. Plant Breed. 2000, 60, 1–4. [Google Scholar]
- De Pace, C.; Snidaro, D.; Ciaffi, M.; Vittori, D.; Ciofo, A.; Cenci, A.; Tanzarella, O.A.; Qualset, C.O.; Mugnozza, G.S. Introgression of Dasypyrum villosum chromatin into common wheat improves grain protein quality. Euphytica 2001, 117, 67–75. [Google Scholar] [CrossRef]
- Chen, G.; Zheng, Q.; Bao, Y.; Liu, S.; Wang, H.; Li, X. Molecular cytogenetic identification of a novel dwarf wheat line with introgressed Thinopyrum ponticum chromatin. J. Biosci. 2011, 37, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Molnár-Láng, M.; Linc, G.; Szakács, É. Wheat-barley hybridization: The last 40 years. Euphytica 2014, 195, 315–329. [Google Scholar] [CrossRef]
- Wulff, B.B.; Moscou, M.J. Strategies for transferring resistance into wheat: From wide crosses to GM cassettes. Front. Plant Sci. 2014, 5, 692. [Google Scholar] [CrossRef]
- Feuillet, C.; Langridge, P.; Waugh, R. Cereal breeding takes a walk on the wild side. Trends Genet. 2008, 24, 24–32. [Google Scholar] [CrossRef]
- Kihara, H. Considerations on the evolution and distribution of Aegilops species based on the analyser-method. Cytologia 1954, 19, 336–357. [Google Scholar] [CrossRef]
- Bothmer, R.V.; Jacobsen, N.; Baden, C.; Jorgense, R.B.; Linde-Laurse, I. An Ecogeographical Study of the Genus Hordeum, 2nd ed.; Systematic and Ecogeographic Studies on Crop Genepools; IPGRI: Rome, Italy, 1995; Volume 7. [Google Scholar]
- Gallardo, M.; Fereres, E. Resistencia a la sequía del tritórdeo (Hordeum chilense × Triticum turgidum) en relacion a la del trigo, cebada y triticale. Investig. Agrar. Prod. Prot. Veg. 1989, 4, 361–375. [Google Scholar]
- Martín, A.; Martínez, C.; Rubiales, D.; Ballesteros, J. Tritordeum: triticale’s new brother cereal. In Triticale: Today and Tomorrow; Güedes-Pinto, H., Darvey, N., Carnide, V.P., Eds.; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1996; pp. 57–72. [Google Scholar]
- Atienza, S.G.; Ramirez, C.M.; Hernandez, P.; Martín, A. Chromosomal location of genes for carotenoid pigments in Hordeum chilense. Plant Breed. 2004, 123, 303–304. [Google Scholar] [CrossRef]
- Cabrera, A.; Friebe, B.; Jiang, J.; Gill, B.S. Characterization of Hordeum chilense chromosomes by C-banding and in situ hybridization using highly repeated DNA probes. Genome 1995, 38, 435–442. [Google Scholar] [CrossRef]
- Armstrong, S.J.; Caryl, A.P.; Jones, G.H.; Franklin, F.C.H. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 2002, 115, 3645–3655. [Google Scholar] [CrossRef]
- Higgins, J.D.; Sanchez-Moran, E.; Armstrong, S.J.; Jones, G.H.; Franklin, F.C.H. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005, 19, 2488–2500. [Google Scholar] [CrossRef] [PubMed]
- Martín, A.C.; Shaw, P.; Phillips, D.; Reader, S.; Moore, G. Licensing MLH1 sites for crossover during meiosis. Nat. Commun. 2014, 5, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Wang, X. Chromosome pairing analysis in haploid wheat by spreading of meiotic nuclei. Carlsberg Res. Commun. 1988, 53, 135. [Google Scholar] [CrossRef]
- Martín, A.C.; Rey, M.D.; Shaw, P.; Moore, G. Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 2017, 126, 669–680. [Google Scholar] [CrossRef]
- Riley, R.; Chapman, V. Genetic control of the cytologically diploid behavior of hexaploidy wheat. Nature 1958, 182, 713–715. [Google Scholar] [CrossRef]
- Sears, E.R.; Okamoto, M. Intergenomic chromosome relationship in hexaploidy wheat. In Proceedings of the 10th International Congress of Genetics, Montreal, QC, Canada, 20–27 August 1958; Volume 2, pp. 258–259. [Google Scholar]
- Rey, M.D.; Martín, A.C.; Higgins, J.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Exploiting the ZIP4 homologue within the wheat Ph1 locus has identified two lines exhibiting homoeologous crossover in wheat-wild relative hybrids. Mol. Breed. 2017, 37, 95. [Google Scholar] [CrossRef]
- Rey, M.D.; Martín, A.C.; Smedley, M.; Hayta, S.; Harwood, W.; Shaw, P.; Moore, G. Magnesium increases homoeologous crossover frequency during meiosis in ZIP4 (Ph1 Gene) mutant wheat-wild relative hybrids. Front. Plant Sci. 2018, 9, 509. [Google Scholar] [CrossRef]
- Cabrera, A.; Martín, A. A trigeneric hybrid between Hordeum, Aegilops, and Secale. Genome 1992, 35, 647–649. [Google Scholar] [CrossRef]
- Patto, M.V.; Niks, R.E. Leaf wax layer may prevent appressorium differentiation but does not influence orientation of the leaf rust fungus Puccinia hordei on Hordeum chilense leaves. Eur. J. Plant Pathol. 2001, 107, 795–803. [Google Scholar] [CrossRef]
- Martín, A.C.; Atienza, S.G.; Ramírez, M.C.; Barro, F.; Martín, A. Male fertility restoration of wheat in Hordeum chilense cytoplasm is associated with 6HchS chromosome addition. Aust. J. Agric. Res. 2008, 59, 206–213. [Google Scholar] [CrossRef]
- Martín, A.C.; Atienza, S.G.; Barro, F. Use of ccSSR markers for the determination of the purity of alloplasmic wheat in different Hordeum cytoplasms. Plant Breed. 2008, 127, 470–475. [Google Scholar] [CrossRef]
- Moore, G. Cereal genome evolution: Pastoral pursuits with ‘Lego’ genomes. Curr. Opin. Genet. Dev. 1995, 5, 717–724. [Google Scholar] [CrossRef]
- Orellana, J. Most of the homoeologous pairing at metaphase I in wheat-rye hybrids is not chiasmatic. Genetics 1985, 111, 917–931. [Google Scholar] [CrossRef]
- Martín, A. Cytology and morphology of the hybrid Hordeum chilense × Aegilops squarrosa. J. Hered. 1983, 74, 478. [Google Scholar] [CrossRef]
- Thomas, H.M.; Pickering, R.A. Comparison of the hybrids Hordeum chilense × H. vulgare, H. chilense × H. bulbosum, H. chilense × Secale cereale and the amphiploid of H. chilense × H. vulgare. Theor. Appl. Genet. 1985, 69, 519–522. [Google Scholar] [CrossRef] [PubMed]
- Dvořák, J. Effect of rye on homoeologous chromosome pairing in wheat× rye hybrids. Can. J. Genet. Cytol. 1977, 19, 549–556. [Google Scholar] [CrossRef]
- Balyan, H.S.; Fedak, G. Meiotic study of hybrids between barley (Hordeum vulgare L.) and Triticale (× Triticosecale Wittmack). J. Hered. 1989, 80, 460–463. [Google Scholar] [CrossRef]
- Lambing, C.; Franklin, F.C.H.; Wang, C.J.R. Understanding and manipulating meiotic recombination in plants. Plant Physiol. 2017, 173, 1530–1542. [Google Scholar] [CrossRef]
- Fedak, G.; Armstrong, K.C. Production of trigeneric (barley× wheat)× rye hybrids. Theor. Appl. Genet. 1980, 56, 221–224. [Google Scholar] [CrossRef]
- Gupta, P.K.; Fedak, G. Meiosis in a hybrid between Hordeum procerum (6×) and Agropyron caninum (4×). Cereal Res. Commun. 1987, 15, 151–156. [Google Scholar]
- Holm, P.B.; Wang, X. The effect of chromosome 5B on synapsis and chiasma formation in wheat, Triticum aestivum cv. Chin. Spring. Carlsberg Res. Commun. 1988, 53, 191. [Google Scholar] [CrossRef]
- Kosina, R.; Heslop-Harrison, J.S. Molecular cytogenetics of an amphiploid trigeneric hybrid between Triticum durum, Thinopyrum distichum and Lophopyrum elongatum. Ann. Bot. 1996, 78, 583–589. [Google Scholar] [CrossRef]
- Jauhar, P.P.; Doğramaci, M.; Peterson, T.S. Synthesis and cytological characterization of trigeneric hybrids of durum wheat with and without Ph1. Genome 2004, 47, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Feliner, G.N.; Casacuberta, J.; Wendel, J.F. Genomics of evolutionary novelty in hybrids and polyploids. Front. Genet. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The significance of responses of the genome to challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef]
- Scoles, G.J.; Kaltsikes, P.J. The cytology and cytogenetics of triticale. Z. Pflanz. 1974, 73, 13–43. [Google Scholar]
- Müntzing, A. Triticale-Results and problems. In Fortschritte der Pflanzenzuechtung (Germany); VerlagPaul-Parey: Berlin, Germany, 1979. [Google Scholar]
- Gustafson, G.D.; Shaner, G. Influence of plant age on the expression of slow-mildewing resistance in wheat. Phytopathology 1982, 72, 746–749. [Google Scholar] [CrossRef]
- Fominaya, A.; Orellana, J. Does differential C-heterochromatin content affect chromosome pairing in octoploid triticale? Heredity 1988, 61, 167–173. [Google Scholar] [CrossRef]
- Lukaszewski, A.J.; Gustafson, J.P. Cytogenetics of triticale. Plant Breed. Rev. 1987, 5, 41–93. [Google Scholar]
- Riley, R.; Miller, T.E. Meiotic chromosome pairing in Triticale. Nature 1970, 227, 82–83. [Google Scholar] [CrossRef]
- Martín, A.C.; Borrill, P.; Higgins, J.; Alabdullah, A.; Ramírez-González, R.H.; Swarbreck, D.; Uauy, C.; Shaw, P.; Moore, G. Genome-wide transcription during early wheat meiosis is independent of synapsis, ploidy level, and the Ph1 Locus. Front. Plant Sci. 2018, 9, 1791. [Google Scholar] [CrossRef] [PubMed]
- Kalinka, A.; Achrem, M. Reorganization of wheat and rye genomes in octoploid triticale (× Triticosecale). Planta 2018, 247, 807–829. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.W.; Tanaka, H.; Nakata, N.; Tsujimoto, H. Molecular cytogenetic analyses of hexaploid lines spontaneously appearing in octoploid Triticale. Theor. Appl. Genet. 2006, 114, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.B.; Kaltsikes, P.J. A possible effect of heterochromatin on chromosome pairing. Proc. Natl. Acad. Sci. USA 1974, 71, 2787–2790. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.B.; Kaltsikes, P.J. A bouquet-like attachment plate for telomeres in leptotene of rye revealed by heterochromatin staining. Heredity 1976, 36, 155–162. [Google Scholar] [CrossRef]
- Merker, A. Chromosome substitutions, genetic recombination and the breeding of hexaploid triticale. Wheat Inf. Serv. 1976, 41, 44–48. [Google Scholar]
- Fu, S.; Yang, M.; Fei, Y.; Tan, F.; Ren, Z.; Yan, B.; Zhang, H.; Tang, Z. Alterations and abnormal mitosis of wheat chromosomes induced by wheat-rye monosomic addition lines. PLoS ONE 2013, 8, e70483. [Google Scholar] [CrossRef]
- Delgado, A.; Carvalho, A.; Martín, A.C.; Martín, A.; Lima-Brito, J.E. Genomic restructuring in F1 Hordeum chilense × durum wheat hybrids and corresponding hexaploid tritordeum lines revealed by DNA fingerprinting analyses fingerprinting analyses. J. Genet. 2017, 96, 13–23. [Google Scholar] [CrossRef]
- Delgado, A.; Carvalho, A.; Martín, A.C.; Martín, A.; Lima-Brito, J. Genomic reshuffling in advanced lines of hexaploid tritordeum. Genet. Resour. Crop. Evol. 2017, 64, 1331–1353. [Google Scholar] [CrossRef]
- Martín, A.C.; Atienza, S.G.; Ramírez, M.C.; Barro, F.; Martín, A. Molecular and cytological characterization of an extra acrocentric chromosome that restores male fertility of wheat in the msH1 CMS system. Theor. Appl. Genet. 2010, 121, 1093–1101. [Google Scholar] [CrossRef]
- Castillo, A.; Atienza, S.G.; Martín, A.C. Fertility of CMS wheat is restored by two Rf loci located on a recombined acrocentric chromosome. J. Exp. Bot. 2014, 65, 6667–6677. [Google Scholar] [CrossRef]
- Cabrera, A.; Martín, A. Cytology of the amphiploid Hordeum chilense (4×) × Aegilops squarrosa (4×). Theor. Appl. Genet. 1991, 81, 758–760. [Google Scholar] [CrossRef]
- Bell, G.D.H. Investigations in the Triticinae I. Colchicine techniques for chromosome doubling in interspecific and intergeneric hybridization. J. Agric. Sci. 1950, 40, 9–18. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Kato, A.; Lamb, J.C.; Birchler, J.A. Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 2004, 101, 13554–13559. [Google Scholar] [CrossRef] [PubMed]
- Rey, M.D.; Moore, G.; Martín, A.C. Identification and comparison of individual chromosomes of three accessions of Hordeum chilense, Hordeum vulgare, and Triticum aestivum by FISH. Genome 2018, 61, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Boden, S.A.; Langridge, P.; Spangenberg, G.; Able, J.A. TaASY1 promotes homologous chromosome interactions and is affected by deletion of Ph1. Plant J. 2009, 57, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Colas, I.; Macaulay, M.; Higgins, J.D.; Phillips, D.; Barakate, A.; Posch, M.; Armstrong, S.J.; Franklin, F.C.H.; Halpin, C.; Waugh, R.; et al. A spontaneous mutation in MutL-Homolog 3 (Hv MLH 3) affects synapsis and crossover resolution in the barley desynaptic mutant des10. New Phytol. 2016, 212, 693–707. [Google Scholar] [CrossRef]
| Line | Chromosome Number | Chromosome Configuration | Centromeric Translocation | ||
|---|---|---|---|---|---|
| AB Genome | Hch Genome | R Genome | |||
| AABBHchHchRR-1-1 | 46 + 2 t | 27 + 2 arms | 8 + 1 t | 9 + 2 arms + 1 t | 2 TR·AB |
| AABBHchHchRR-1-2 | 51 | 28 + 2 arms | 10 | 11 + 2 arms | 2 TR·AB |
| AABBHchHchRR-1-3 | 49 | 28 + 1 arm | 9 | 11 + 1 arm | 1 TR·AB |
| AABBHchHchRR-2-1 | 47 + 1 t | 25 + 2 arms + 1 t | 7 | 13 + 2 arms | 2 TR·AB |
| AABBHchHchRR-2-2 | 51 | 27 + 2 arms | 7 + 1 arm | 14 + 3 arms | 2 TR·AB + 1 THch·R |
| No. and Type of Chromosome Associations | Total % of Associations | Crossover | ||||
|---|---|---|---|---|---|---|
| Rod Bivalent | Ring Bivalent | Trivalent | No. | % | ||
| Hordeum-Hordeum | 13 | - | - | 6.2 | 2 | 2.8 |
| Aegilops-Aegilops | 4 | - | - | 1.9 | - | - |
| Secale-Secale | 12 | - | - | 5.7 | 3 | 4.2 |
| Hordeum-Aegilops | 84 | 5 | - | 44.8 | 33 | 46.5 |
| Hordeum-Secale | 15 | - | - | 7.1 | 7 | 9.9 |
| Aegilops-Secale | 60 | - | - | 28.6 | 23 | 32.4 |
| Hordeum-Aegilops-Secale | - | - | 3 | 2.9 | 3 | 4.2 |
| Hordeum-Aegilops-Aegilops | - | - | 1 | 1.0 | - | - |
| Hordeum-Secale-Secale | - | - | 1 | 1.0 | - | - |
| Hordeum-Hordeum-Aegilops | - | - | 1 | 1.0 | - | - |
| 188 | 10 | 12 | 71 | |||
| Rod Bivalent | Total % of Associations | No. COs | |
|---|---|---|---|
| Triticum-Triticum | 34 | 41.0 | 1 |
| Hordeum-Hordeum | 6 | 7.2 | 0 |
| Secale-Secale | 9 | 10.8 | 2 |
| Triticum-Hordeum | 17 | 20.5 | 0 |
| Triticum-Secale | 13 | 15.7 | 2 |
| Hordeum-Secale | 4 | 4.8 | 1 |
| Total No. Associations | 83 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rey, M.-D.; Ramírez, C.; Martín, A.C. Wheat, Rye, and Barley Genomes Can Associate during Meiosis in Newly Synthesized Trigeneric Hybrids. Plants 2021, 10, 113. https://doi.org/10.3390/plants10010113
Rey M-D, Ramírez C, Martín AC. Wheat, Rye, and Barley Genomes Can Associate during Meiosis in Newly Synthesized Trigeneric Hybrids. Plants. 2021; 10(1):113. https://doi.org/10.3390/plants10010113
Chicago/Turabian StyleRey, María-Dolores, Carmen Ramírez, and Azahara C. Martín. 2021. "Wheat, Rye, and Barley Genomes Can Associate during Meiosis in Newly Synthesized Trigeneric Hybrids" Plants 10, no. 1: 113. https://doi.org/10.3390/plants10010113
APA StyleRey, M.-D., Ramírez, C., & Martín, A. C. (2021). Wheat, Rye, and Barley Genomes Can Associate during Meiosis in Newly Synthesized Trigeneric Hybrids. Plants, 10(1), 113. https://doi.org/10.3390/plants10010113






