There and Back Again: Hox Clusters Use Both DNA Strands
Abstract
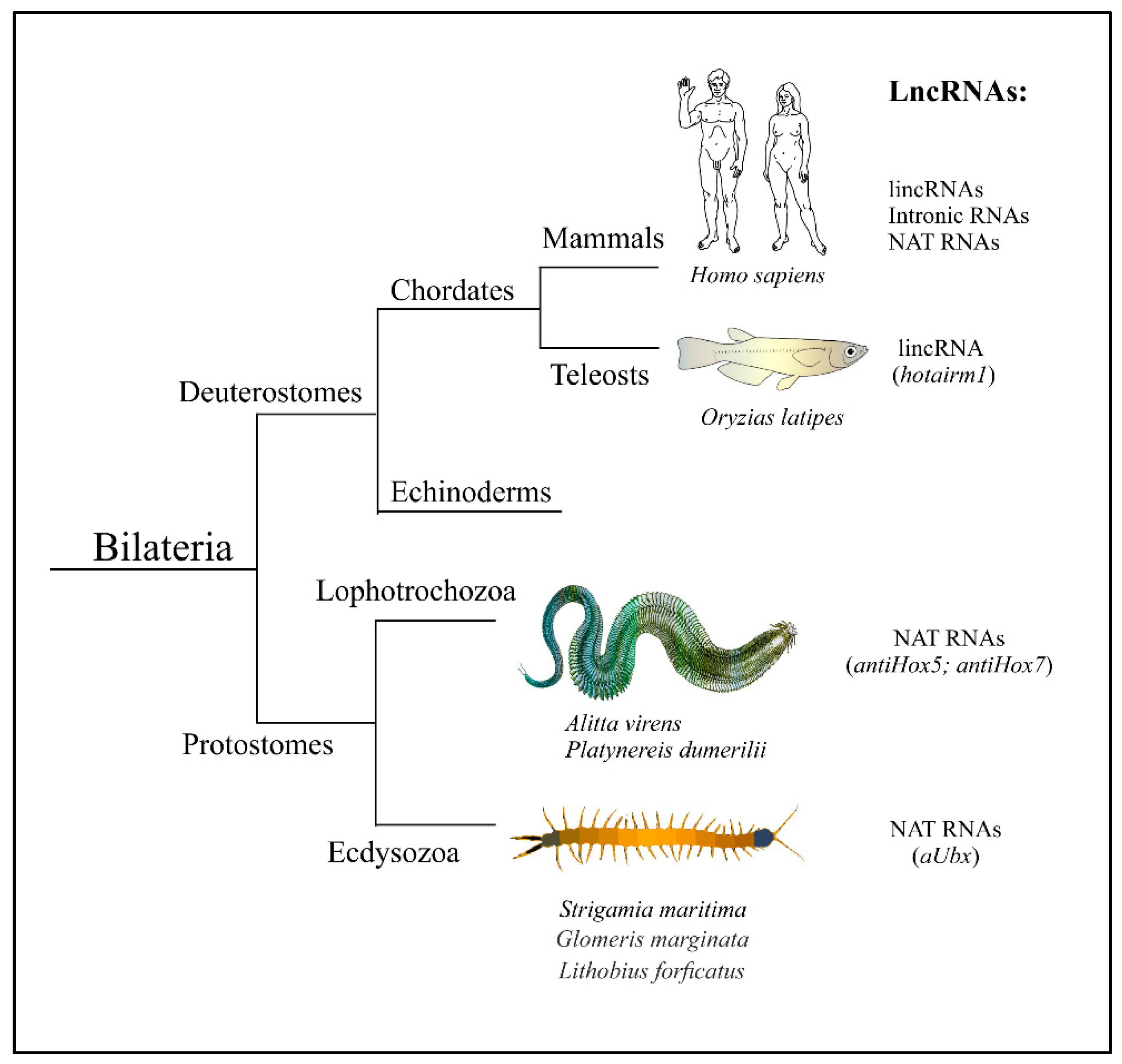
1. Introduction
- Their fundamental role in the ground plan formation (this excludes the loss of Hox genes in most bilaterian animals);
- The simplicity of DNA-consensus for the binding of Hox homeodomain and the ability of Hox genes to form the dimers with cofactors to make this consensus more complicated;
- The complex regulation of their transcription.
2. Antisense LncRNAs in Mammalian Hox Clusters
| As ncRNA | Functions | Mechanism of Work | Localization | Targets | Orthologs | Discovered in | Refs |
|---|---|---|---|---|---|---|---|
| HOTAIRM1 | Control of the cell cycle in the myeloid cell lineage; control of the differentiation of granulocytes; control of neuronal differentiation timing; control of osteogenesis in dental follicle stem cells | Serve as protein scaffolds; Enhancer; Sponges big set miRNAs | Nucleus Cytoplasm | HOXA cluster; NEUROGENIN 2; miR-196b; miR-125b | Chordata | 2009 | [36,39,40,41,42,43,44] |
| HOXA-AS2 | Promotion of proliferation, migration and invasion in many types of tumors; regulation EMT; negative regulates endothelium inflammation | Serve as protein scaffolds; Sponges big set miRNAs | Nucleus Cytoplasm | c-MYC; EGFR; Bax/TRAIL; EZH2/LSD1; PBX3; NF-kB; miR-373 | Primates | 2013 | [45,46,47,48,49] |
| HOXA-AS3 | Control of cell cycle, proliferation, migration and apoptosis in many types of cancer cells; positive regulation of endothelium inflammation; activation the MEK/ERK Signaling Pathway | Stabilization of HOXA6 mRNA, sponges miR-29c and mir-455-5p | Nucleus Cytoplasm | HOXA6; NF-kB; miR-29c | Homo sapiens | 2017 | [50,51,52,53,54] |
| HOXA10-AS | Cell cycle and apoptosis control in glioma, lung adenocarcinoma (LAD), oral cancer and acute myeloid leukemia (AML) cells | ? | Cytoplasm | HOXA10; Wnt pathway; NF-kB | Birds Mammals | 2018 | [55,56,57,58] |
| HOXA11-AS | Control of the menstrual cycle; cell cycle, proliferation, migration and apoptosis control in many types of cancer cells | Serve as protein scaffolds; Sponges big set miRNAs | Nucleus Cytoplasm | HOXA11; TGF-b pathway, LATS1; CyclinD1; CyclinE; CDK4; CDK2 | Rodents Primates Bamboo shark | 2002 | [59] |
| HOTTIP | Control of 5′HOXA genes’ transcription during development. Participation in pathogenesis of almost all types of cancer. | Activates HOXA genes through recruiting of WDR5 и MLL | Nucleus | HOXA7-HOXA13; LSD1; EZH2; IL-6; miR-30b | Rodents Primates Bamboo shark | 2011 | [60,61] |
| HOXB-AS1 | Glioblastoma and endometrial carcinoma and multiple myeloma (MM) promotion | ILF3-mediated activation of HOXB3 and HOXB3 transcription; stabilization of their mRNAs; stabilization of FUT4 mRNA | Nucleus Cytoplasm | HOXB2; HOXB3; Wnt pathway; FUT4; miR-186-5p; miR-149-3p | Homo sapiens | 2019 | [62,63,64] |
| HOXB-AS2 | Potentially participate in the development of atrial fibrillation | ? | ? | ? | Homo sapiens | 2020 | [65] |
| HOXB-AS3 | Control of energetic metabolism in the cell through alternating the isoforms of pyruvate kinase M (PKM); promoting of the cancer processes through repression of p53 transcription; activation of PI3/AKT pathway | Codes the conservative peptide of 53 amino acids long, which is important for PKM splicing; Sponges miRNAs | Nucleus Cytoplasm | PKM; DNMT1; p53; I3K-AKT-mTOR pathway; miR-378a-3p | Homo sapiens Rodents | 2017 | [66,67,68,69,70] |
| HOXB-AS4 | The sequence is differentially methylated in normal and pancreatic cancer cells | ? | ? | ? | Homo sapiens | 2018 | [71] |
| HOXB-AS5 or PRAC2 | Associated with breast cancer (lncRNA) and protstate cancer (protein) | Encodes 140 aa nuclear protein | Nucleus (protein) | I3K-AKT-mTOR pathway (lncRNA) | Artiodactyla Bats Colugo Primates | 2003 (protein) 2017 (lncRNA) | [72,73] |
| HOXC-AS1 | Cholesterol homeostasis participation, inhibition of atherosclerosis; promotion of growth and metastatic formation in several types of malignant tumors | Promotes the transcription and translation of HOXC6; boosts c-MYC mRNA | Nucleus Cytoplasm | HOXC6; miR-590-3p; c-MYC; Wnt pathway; miR-590-3p | Homo sapiens | 2016 | [74,75,76,77,78] |
| HOXC-AS2 | Promotion of growth and metastatic formation in several types of malignant tumors | Promote HOXC13 transcription; can sponge miR-876-5p to affect ZEB1 expression | Nucleus Cytoplasm | HOXC13; ZEB1; miR-876-5p | Homo sapiens | 2019 | [79,80,81,82] |
| HOXC-AS3 | Functions under the direct control of HOXB1. Promotion of growth and metastatic formation in several types of malignant tumors | Promote the transcription of 5’HOXC genes; stabilizes HOXC10 mRNA; can sponge miR-3922-5p, impairs the maturation of miR-96 | Nucleus | HOXC8; HOXC9; HOXC10; HOXC11; HOXC12; HOXC13; YBX1; thymidine kinase 1 (TK1); FOXM1; miR-96; miR-3922-5p | Homo sapiens | 2018 | [37,81,82,83,84,85,86,87,88,89] |
| HOTAIR | Reprogramming of chromatin state to promote cancer metastasis; PRC2 and PRC2-independent induction of transcriptional repression. Promotion of growth and metastatic formation in several types of malignant tumors | Scaffold: A 5′ domain of HOTAIR binds PRC2, whereas a 3′ domain of HOTAIR binds the LSD1/CoREST/REST complex; Sponging big set microRNA | Nucleus Cytoplasm | HOXD cluster (40 Kb in 5′area) HOXA1; HOXA5; HOXC11; p53; p27; E-cadherin; NOTCH1/JAGGED1; SNAIL; GLI2; Protocadherin 10; Wnt pathway; Dozens of miRNAs, critical for proliferation and differentiation control | Rodents Carnivores Primates Marsupials | 2007 | [28,35,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105,106,107,108,109,110,111,112,113,114,115,116,117,118,119] |
| HOXC13-AS | Promotion of proliferation, migration and invasion of cells of several cancer types | Sponging big set microRNA | Cytoplasm | HOXC13; c-MYC; miR-383-3p; miR-497-5p | Homo sapiens | 2019 | [120,121,122,123,124] |
| HAGLR or HOXD-AS1 | Impairment of HAGLR regulation (up or down) to promote growth and metastatic formation of several types of malignant tumors | Binds to WDR5 and EZH2 for activation and repression of target genes | Nucleus Cytoplasm | HOXD3; JAK2/STAT3 pathway; Wnt pathway; Ras/ERK pathway; TGF-β pathway; p57; miR-133a-3p; miR-133b; miR-130a; miR-185-5p | Rodents Primates | 2014 | [125,126,127,128,129,130,131,132,133,134,135,136,137,138] |
| HOXD-AS2 | Downregulation of HOXD-AS2 significantly promotes the progression of gastric cancer | ? | Cytoplasm | HOXD8; PI3K/Akt pathway | Homo sapiens | 2018 | [139,140,141,142,143,144,145,146,147,148,149] |
3. Antisense LncRNAs in Hox Clusters of Protostomian Animals
4. The Implications for the Uprise of Antisense LncRNAs in Hox Clusters and the Reasons for Their Evolutionary Maintenance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| bx | bithotax |
| ceRNA | competing endogenous RNA |
| eve | even-skipped |
| GBM | glioblastoma |
| HOTAIRM1 | HOXA Transcript Antisense RNA, Myeloid-Specific 1 |
| iab | infraabdominal |
| lincRNA | intergenic lncRNA |
| lncRNA | long non-coding RNA |
| LSD1 | Lysine-Specific Demethylase 1 |
| NAT | Natural Antisense Transcript |
| PKM | pyruvate kinase M |
| PMC | primary mesenchymal cell |
| PRC2 | Polycomb Repressive Complex 2 |
| RA | retinoic acid |
| smORF | small open reading frames |
| SMRT | single-molecule real-time |
| TUs | transcriptional units |
References
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 7, 565–570. [Google Scholar] [CrossRef]
- Maslakov, G.P.; Kulishkin, N.S.; Surkova, A.A.; Kulakova, M.A. Maternal Transcripts of Hox Genes are Found in Oocytes of Platynereis dumerilii (Annelida, Nereididae). J. Dev. Biol. 2021. Unpublished work. [Google Scholar]
- Awgulewitsch, A. Hox in hair growth and development. Naturwissenschaften 2003, 90, 193–211. [Google Scholar] [CrossRef] [PubMed]
- Brun, A.C.; Björnsson, J.M.; Magnusson, M.; Larsson, N.; Leveén, P.; Ehinger, M.; Nilsson, E.; Karlsson, S. Hoxb4-deficient mice undergo normal hematopoietic development but exhibit a mild proliferation defect in hematopoietic stem cells. Blood 2004, 103, 4126–4133. [Google Scholar] [CrossRef]
- Argiropoulos, B.; Humphries, R.K. Hox genes in hematopoiesis and leukemogenesis. Oncogene 2007, 26, 6766–6776. [Google Scholar] [CrossRef] [PubMed]
- Morgan, R.; Whiting, K. Differential expression of HOX genes upon activation of leukocyte sub-populations. Int. J. Hematol. 2008, 87, 246–249. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stansbury, M.S.; Moczek, A.P. The function of Hox and appendage-patterning genes in the development of an evolutionary novelty, the Photuris firefly lantern. Proc. Biol. Sci. 2014, 281, 20133333. [Google Scholar] [CrossRef]
- Horabin, J.I. Long noncoding RNAs as metazoan developmental regulators. Chromosome Res. 2013, 21, 673–684. [Google Scholar] [CrossRef]
- Brosnan, C.A.; Voinnet, O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009, 21, 416–425. [Google Scholar] [CrossRef]
- Ponting, C.P.; Belgard, T.G. Transcribed dark matter: Meaning or myth? Hum. Mol. Genet. 2010, 19, R162–R168. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Valverde, S.L.; Calcino, A.D.; Degnan, B.M. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genom. 2015, 16, 387. [Google Scholar] [CrossRef]
- Gaiti, F.; Fernandez-Valverde, S.L.; Nakanishi, N.; Calcino, A.D.; Yanai, I.; Tanurdzic, M.; Degnan, B.M. Dynamic and Widespread lncRNA Expression in a Sponge and the Origin of Animal Complexity. Mol. Biol. Evol. 2015, 32, 2367–2382. [Google Scholar] [CrossRef] [PubMed]
- Vickaryous, M.K.; Hall, B.K. Human cell type diversity, evolution, development, and classification with special reference to cells derived from the neural crest. Biol. Rev. Camb. Philos. Soc. 2006, 81, 425–455. [Google Scholar] [CrossRef]
- Pertea, M.; Shumate, A.; Pertea, G.; Varabyou, A.; Breitwieser, F.P.; Chang, Y.-C.; Madugundu, A.K.; Pandey, A.; Salzberg, S.L. CHESS: A new human gene catalog curated from thousands of large-scale RNA sequencing experiments reveals extensive transcriptional noise. Genome Biol. 2018, 19, 208. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Trapnell, C.; Goff, L.; Koziol, M.; Tazon-Vega, B.; Regev, A.; Rinn, J.L. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011, 25, 1915–1927. [Google Scholar] [CrossRef]
- Sarropoulos, I.; Marin, R.; Cardoso-Moreira, M.; Kaessmann, H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature 2019, 571, 510–514. [Google Scholar] [CrossRef]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; RIKEN Genome Exploration Research Group; Genome Science Group (Genome Network Project Core Group); FANTOM Consortium; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar] [PubMed]
- Lipshitz, H.D.; Peattie, D.A.; Hogness, D.S. Novel transcripts from the Ultrabithorax domain of the bithorax complex. Genes Dev. 1987, 1, 307–322. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113, 2526–2534. [Google Scholar] [CrossRef]
- Li, L.; Liu, B.; Wapinski, O.L.; Tsai, M.C.; Qu, K.; Zhang, J.; Carlson, J.C.; Lin, M.; Fang, F.; Gupta, R.A.; et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013, 5, 3–12. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, X.; Frazão, J.B.; Condino-Neto, A.; Newburger, P.E. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J. Cell Biochem. 2013, 114, 2375–2383. [Google Scholar] [CrossRef]
- Pradeepa, M.M.; McKenna, F.; Taylor, G.C.A.; Bengani, H.; Grimes, G.R.; Wood, A.J.; Bhatia, B.; Bickmore, W.A. Psip1/p52 regulates posterior Hoxa genes through activation of lncRNA Hottip. PLoS Genet. 2017, 13, e1006677. [Google Scholar] [CrossRef]
- Duboule, D. The rise and fall of Hox gene clusters. Development 2007, 134, 2549–2560. [Google Scholar] [CrossRef]
- Engström, P.G.; Suzuki, H.; Ninomiya, N.; Akalin, A.; Sessa, L.; Lavorgna, G.; Brozzi, A.; Luzi, L.; Tan, S.L.; Yang, L.; et al. Complex Loci in human and mouse genomes. PLoS Genet. 2006, 2, e47. [Google Scholar] [CrossRef] [PubMed]
- Bedford, M.; Arman, E.; Orr-Urtreger, A.; Lonai, P. Analysis of the Hoxd-3 gene: Structure and localization of its sense and natural antisense transcripts. DNA Cell Biol. 1995, 14, 295–304. [Google Scholar] [CrossRef]
- Hsieh-Li, H.M.; Witte, D.P.; Weinstein, M.; Branford, W.; Li, H.; Small, K.; Potter, S.S. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development 1995, 121, 1373–1385. [Google Scholar] [CrossRef]
- Sessa, L.; Breiling, A.; Lavorgna, G.; Silvestri, L.; Casari, G.; Orlando, V. Noncoding RNA synthesis and loss of Polycomb group repression accompanies the colinear activation of the human HOXA cluster. RNA 2007, 13, 223–239. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Mainguy, G.; Koster, J.; Woltering, J.; Jansen, H.; Durston, A. Extensive polycistronism and antisense transcription in the mammalian Hox clusters. PLoS ONE 2007, 2, e356. [Google Scholar] [CrossRef]
- Simeone, A.; Pannese, M.; Acampora, D.; D’Esposito, M.; Boncinelli, E. At least three human homeoboxes on chromosome 12 belong to the same transcription unit. Nucleic Acids Res. 1988, 16, 5379–5390. [Google Scholar] [CrossRef]
- Shiga, Y.; Sagawa, K.; Takai, R.; Sakaguchi, H.; Yamagata, H.; Hayashi, S. Transcriptional readthrough of Hox genes Ubx and Antp and their divergent post-transcriptional control during crustacean evolution. Evol. Dev. 2006, 8, 407–414. [Google Scholar] [CrossRef] [PubMed]
- Brena, C.; Chipman, A.D.; Minelli, A.; Akam, M. Expression of trunk Hox genes in the centipede Strigamia maritima: Sense and anti-sense transcripts. Evol. Dev. 2006, 8, 252–265. [Google Scholar] [CrossRef] [PubMed]
- Janssen, R.; Budd, G.E. Gene expression suggests conserved aspects of Hox gene regulation in arthropods and provides additional support for monophyletic Myriapoda. Evodevo 2010, 1, 4. [Google Scholar] [CrossRef]
- Degani, N.; Ainbinder, E.; Ulitsky, I. Highly conserved and cis-acting lncRNAs produced from paralogous regions in the center of HOXA and HOXB clusters in the endoderm lineage. bioRxiv 2020. [Google Scholar] [CrossRef]
- Diederichs, S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014, 30, 121–123. [Google Scholar] [CrossRef]
- Herrera-Úbeda, C.; Marín-Barba, M.; Navas-Pérez, E.; Gravemeyer, J.; Albuixech-Crespo, B.; Wheeler, G.N.; Garcia-Fernàndez, J. Microsyntenic Clusters Reveal Conservation of lncRNAs in Chordates Despite Absence of Sequence Conservation. Biology 2019, 8, 61. [Google Scholar] [CrossRef]
- Wang, X.; Sun, Y.; Xu, T.; Qian, K.; Huang, B.; Zhang, K.; Song, Z.; Qian, T.; Shi, J.; Li, L. HOXB13 promotes proliferation, migration, and invasion of glioblastoma through transcriptional upregulation of lncRNA HOXC-AS3. J. Cell Biochem. 2019, 120, 15527–15537. [Google Scholar] [CrossRef]
- Latgé, G.; Poulet, C.; Bours, V.; Josse, C.; Jerusalem, G. Natural Antisense Transcripts: Molecular Mechanisms and Implications in Breast Cancers. Int. J. Mol. Sci. 2018, 19, 123. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, J.; Hong, H.; Chen, D.; Deng, L.; Zhang, X.; Ling, J.; Wu, L. lncRNA HOTAIRM1 promotes osteogenesis of hDFSCs by epigenetically regulating HOXA2 via DNMT1 in vitro. J. Cell Physiol. 2020, 235, 8507–8519. [Google Scholar] [CrossRef] [PubMed]
- Rea, J.; Menci, V.; Tollis, P.; Santini, T.; Armaos, A.; Garone, M.G.; Iberite, F.; Cipriano, A.; Tartaglia, G.G.; Rosa, A.; et al. HOTAIRM1 regulates neuronal differentiation by modulating NEUROGENIN 2 and the downstream neurogenic cascade. Cell Death Dis. 2020, 11, 527. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Weissman, S.M.; Newburger, P.E. Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol. 2014, 11, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Beyá, M.; Brunet, S.; Nomdedéu, J.; Pratcorona, M.; Cordeiro, A.; Gallardo, D.; Escoda, L.; Tormo, M.; Heras, I.; Ribera, J.M.; et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015, 6, 31613–31627. [Google Scholar] [PubMed]
- Shi, T.; Guo, D.; Xu, H.; Su, G.; Chen, J.; Zhao, Z.; Shi, J.; Wedemeyer, M.; Attenello, F.; Zhang, L.; et al. HOTAIRM1, an enhancer lncRNA, promotes glioma proliferation by regulating long-range chromatin interactions within HOXA cluster genes. Mol. Biol. Rep. 2020, 47, 2723–2733. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Q.; Sun, N.; Zhang, C.S.; Li, N.; Wu, B.; Zhang, J.L. Inactivation of lncRNA HOTAIRM1 caused by histone methyltransferase RIZ1 accelerated the proliferation and invasion of liver cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8767–8777. [Google Scholar]
- Fang, Y.; Wang, J.; Wu, F.; Song, Y.; Zhao, S.; Zhang, Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget 2017, 8, 46090–46103. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Zhang, S.; An, J.; Zhang, J.; Huang, J.; Jin, Y. HOXA-AS2 Promotes Proliferation and Induces Epithelial-Mesenchymal Transition via the miR-520c-3p/GPC3 Axis in Hepatocellular Carcinoma. Cell Physiol. Biochem. 2018, 50, 2124–2138. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, Y.; Yu, J.; Du, J.; Guo, R.; Feng, Y.; Zhong, G.; Jiang, Y.; Lin, J. LncRNA HOXA-AS2 represses endothelium inflammation by regulating the activity of NF-κB signaling. Atherosclerosis 2019, 281, 38–46. [Google Scholar] [CrossRef]
- Xiao, S.; Song, B. LncRNA HOXA-AS2 promotes the progression of prostate cancer via targeting miR-509-3p/PBX3 axis. Biosci. Rep. 2020, 40, BSR20193287. [Google Scholar] [CrossRef]
- Wang, J.; Su, Z.; Lu, S.; Fu, W.; Liu, Z.; Jiang, X.; Tai, S. LncRNA HOXA-AS2 and its molecular mechanisms in human cancer. Clin. Chim. Acta 2018, 485, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, C.; Cai, J.; Yang, F.; Liang, T.; Yan, X.; Wang, H.; Wang, W.; Chen, J.; Jiang, T. Upregulation of long noncoding RNA HOXA-AS3 promotes tumor progression and predicts poor prognosis in glioma. Oncotarget 2017, 8, 53110–53123. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, Y.; Yan, L.; Zhang, M.; Yu, X.; Du, W.; Wang, S.; Li, Q.; Chen, H.; Zhang, Y.; et al. Increased levels of the long noncoding RNA, HOXA-AS3, promote proliferation of A549 cells. Cell Death Dis. 2018, 9, 707. [Google Scholar] [CrossRef]
- Tong, Y.; Wang, M.; Dai, Y.; Bao, D.; Zhang, J.; Pan, H. LncRNA HOXA-AS3 Sponges miR-29c to Facilitate Cell Proliferation, Metastasis, and EMT Process and Activate the MEK/ERK Signaling Pathway in Hepatocellular Carcinoma. Hum. Gene Ther. Clin. Dev. 2019, 30, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, D.; Liu, Y.; Yu, J.; Qiao, L.; Lin, S.; Chen, D.; Zhong, G.; Lu, X.; Wang, Y.; et al. Long Noncoding RNA HOXA-AS3 Integrates NF-κB Signaling to Regulate Endothelium Inflammation. Mol. Cell Biol. 2019, 39, e00139-19. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, Q.; Zhang, G.; Wang, H.; Zhu, Z.; Chen, L. LncRNA HOXA-AS3 promotes the malignancy of glioblastoma through regulating miR-455-5p/USP3 axis. J. Cell Mol. Med. 2020, 24, 11755–11767. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.Y.; Cui, J.; Li, D.H.; Li, Q.; Hong, X.Y. HOXA10-AS: A novel oncogenic long non-coding RNA in glioma. Oncol. Rep. 2018, 40, 2573–2583. [Google Scholar] [CrossRef] [PubMed]
- Sheng, K.; Lu, J.; Zhao, H. ELK1-induced upregulation of lncRNA HOXA10-AS promotes lung adenocarcinoma progression by increasing Wnt/β-catenin signaling. Biochem. Biophys. Res. Commun. 2018, 501, 612–618. [Google Scholar] [CrossRef]
- Al-Kershi, S.; Bhayadia, R.; Ng, M.; Verboon, L.; Emmrich, S.; Gack, L.; Schwarzer, A.; Strowig, T.; Heckl, D.; Klusmann, J.H. The stem cell-specific long noncoding RNA HOXA10-AS in the pathogenesis of KMT2A-rearranged leukemia. Blood Adv. 2019, 3, 4252–4263. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Cong, B.; Chen, Q.; Liu, L.; Luan, X.; Du, J.; Cao, M. Silencing lncRNA HOXA10-AS decreases cell proliferation of oral cancer and HOXA10-antisense RNA can serve as a novel prognostic predictor. J. Int. Med. Res. 2020, 48. [Google Scholar] [CrossRef]
- Wei, C.; Zhao, L.; Liang, H.; Zhen, Y.; Han, L. Recent advances in unraveling the molecular mechanisms and functions of HOXA11-AS in human cancers and other diseases (Review). Oncol. Rep. 2020, 43, 1737–1754. [Google Scholar] [CrossRef]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Dashti, S.; Taheri, M. The HOTTIP (HOXA transcript at the distal tip) lncRNA: Review of oncogenic roles in human. Biomed. Pharmacother. 2020, 127, 110158. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Zhang, X.; Wang, C. LncRNA HOXB-AS1 promotes cell growth in multiple myeloma via FUT4 mRNA stability by ELAVL1. J. Cell Biochem. 2020, 121, 4043–4051. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Mao, Y.; Su, Z.; Du, J.; Ye, L.; Xu, F. HOXB-AS1 accelerates the tumorigenesis of glioblastoma via modulation of HOBX2 and HOBX3 at transcriptional and posttranscriptional levels. J. Cell Physiol. 2021, 236, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Qiu, M.; Jiang, L.; Liu, K. Long Noncoding RNA HOXB-AS1 Is Upregulated in Endometrial Carcinoma and Sponged miR-149-3p to Upregulate Wnt10b. Technol. Cancer Res. Treat. 2020, 19. [Google Scholar] [CrossRef]
- Shi, X.; Shao, X.; Liu, B.; Lv, M.; Pandey, P.; Guo, C.; Zhang, R.; Zhang, Y. Genome-wide screening of functional long noncoding RNAs in the epicardial adipose tissues of atrial fibrillation. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165757. [Google Scholar] [CrossRef]
- Huang, J.Z.; Chen, M.; Chen, D.; Gao, X.C.; Zhu, S.; Huang, H.; Hu, M.; Zhu, H.; Yan, G.R. A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol. Cell. 2017, 68, 171–184. [Google Scholar] [CrossRef]
- Zhang, X.M.; Chen, H.; Zhou, B.; Zhang, Q.Y.; Liao, Y.; Wang, J.S.; Wang, Z.H. lncRNA HOXB-AS3 promotes hepatoma by inhibiting p53 expression. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6784–6792. [Google Scholar]
- Jiang, W.; Kai, J.; Li, D.; Wei, Z.; Wang, Y.; Wang, W. lncRNA HOXB-AS3 exacerbates proliferation, migration, and invasion of lung cancer via activating the PI3K-AKT pathway. J. Cell Physiol. 2020, 235, 7194–7203. [Google Scholar] [CrossRef]
- Huang, H.H.; Chen, F.Y.; Chou, W.C.; Hou, H.A.; Ko, B.S.; Lin, C.T.; Tang, J.L.; Li, C.C.; Yao, M.; Tsay, W.; et al. Long non-coding RNA HOXB-AS3 promotes myeloid cell proliferation and its higher expression is an adverse prognostic marker in patients with acute myeloid leukemia and myelodysplastic syndrome. BMC Cancer 2019, 19, 617. [Google Scholar] [CrossRef]
- Xu, S.; Jia, G.; Zhang, H.; Wang, L.; Cong, Y.; Lv, M.; Xu, J.; Ruan, H.; Jia, X.; Xu, P.; et al. LncRNA HOXB-AS3 promotes growth, invasion and migration of epithelial ovarian cancer by altering glycolysis. Life Sci. 2021, 264, 118636. [Google Scholar] [CrossRef]
- Ishihara, H.; Yamashita, S.; Amano, R.; Kimura, K.; Hirakawa, K.; Ueda, T.; Murakami, Y.; Tamori, A.; Tanabe, K.; Kawada, N.; et al. Pancreatic Cancer Cell Fraction Estimation in a DNA Sample. Oncology 2018, 95, 370–379. [Google Scholar] [CrossRef]
- Olsson, P.; Motegi, A.; Bera, T.K.; Lee, B.; Pastan, I. PRAC2: A new gene expressed in human prostate and prostate cancer. Prostate 2003, 56, 123–130. [Google Scholar] [CrossRef]
- Rui, J.; Chunming, Z.; Binbin, G.; Na, S.; Shengxi, W.; Wei, S. IL-22 promotes the progression of breast cancer through regulating HOXB-AS5. Oncotarget 2017, 8, 103601–103612. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Hu, Y.W.; Zhao, J.J.; Ma, X.; Zhang, Y.; Guo, F.X.; Kang, C.M.; Lu, J.B.; Xiu, J.C.; Sha, Y.H.; et al. Long Noncoding RNA HOXC-AS1 Suppresses Ox-LDL-Induced Cholesterol Accumulation Through Promoting HOXC6 Expression in THP-1 Macrophages. DNA Cell Biol. 2016, 35, 722–729. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, X.; Lin, Z.; Zou, W.; Liu, Y.; Qian, H.; Jia, J. HOXC-AS1-MYC regulatory loop contributes to the growth and metastasis in gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 502. [Google Scholar] [CrossRef] [PubMed]
- Takayama, K.I.; Fujimura, T.; Suzuki, Y.; Inoue, S. Identification of long non-coding RNAs in advanced prostate cancer associated with androgen receptor splicing factors. Commun. Biol. 2020, 3, 393. [Google Scholar] [CrossRef]
- Zhou, C.; An, N.; Cao, C.; Wang, G. lncRNA HOXC-AS1 promotes gastric cancer via binding eIF4AIII by activating Wnt/β-catenin signaling. J. Gene Med. 2020, 22, e3202. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Gao, Y.; Fan, Y.; Zhang, G.; Zhang, Y. Molecular Mechanism of 73HOXC-AS1-Activated Wntβ-Catenin Signaling and eIF4AIII in Promoting Progression of Gastric Cancer. Biomed Res. Int. 2021, 2021, 8814843. [Google Scholar]
- Dong, N.; Guo, J.; Han, S.; Bao, L.; Diao, Y.; Lin, Z. Positive feedback loop of lncRNA HOXC-AS2/miR-876-5p/ZEB1 to regulate EMT in glioma. Onco Targets Ther. 2019, 12, 7601–7609. [Google Scholar] [CrossRef]
- Turner, D.C.; Gorski, P.P.; Maasar, M.F.; Seaborne, R.A.; Baumert, P.; Brown, A.D.; Kitchen, M.O.; Erskine, R.M.; Dos-Remedios, I.; Voisin, S.; et al. DNA methylation across the genome in aged human skeletal muscle tissue and muscle-derived cells: The role of HOX genes and physical activity. Sci. Rep. 2020, 10, 15360. [Google Scholar] [CrossRef]
- Fu, T.; Ji, X.; Bu, Z.; Zhang, J.; Wu, X.; Zong, X.; Fan, B.; Jia, Z.; Ji, J. Identification of key long non-coding RNAs in gastric adenocarcinoma. Cancer Biomark. 2020, 27, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Li, J.; Li, J.M.; Liu, G.Y.; Wang, Y.S. HOXC-AS2 mediates the proliferation, apoptosis, and migration of non-small cell lung cancer by combining with HOXC13 gene. Cell Cycle 2021, 20, 236–246. [Google Scholar] [CrossRef]
- Yang, Z.; Hu, T. Long noncoding RNA HOXC-AS3 facilitates the progression of invasive mucinous adenocarcinomas of the lung via modulating FUS/FOXM1. Vitr. Cell Dev. Biol. Anim. 2020, 56, 15–23. [Google Scholar] [CrossRef]
- Zhang, E.; He, X.; Zhang, C.; Su, J.; Lu, X.; Si, X.; Chen, J.; Yin, D.; Han, L.; De, W. A novel long noncoding RNA HOXC-AS3 mediates tumorigenesis of gastric cancer by binding to YBX1. Genome Biol. 2018, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Han, H.; Song, S.; Fan, G.; Xu, H.; Zhou, W.; Qiu, Y.; Qian, C.; Wang, Y.; Yuan, Z.; et al. HOXC10 Regulates Osteogenesis of Mesenchymal Stromal Cells Through Interaction with Its Natural Antisense Transcript lncHOXC-AS3. Stem Cells 2019, 37, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.H.; Jiang, J.; Zhang, W.; Sun, L.; Li, X.J.; Li, C.; Ge, Q.D.; Zhuang, Z.G. A Novel lncRNA HOXC-AS3 Acts as a miR-3922-5p Sponge to Promote Breast Cancer Metastasis. Cancer Investig. 2020, 38, 1–12. [Google Scholar] [CrossRef]
- Enteghami, M.; Ghorbani, M.; Zamani, M.; Galehdari, H. HOXC10 is significantly overexpressed in colorectal cancer. Biomed. Rep. 2020, 13, 18. [Google Scholar] [CrossRef]
- Su, J.; Yu, B.; Zhang, C.; Yi, P.; Li, H.; Xu, C.; Cao, L.; Chen, P.; Li, M.; Shen, K.; et al. Long noncoding RNA HOXC-AS3 indicates a poor prognosis and regulates tumorigenesis by binding to YBX1 in breast cancer. Am. J. Transl. Res. 2020, 12, 6335–6350. [Google Scholar]
- Yang, B.; Sun, L.; Liang, L. LncRNA HOXC-AS3 Suppresses the Formation of Mature miR-96 in Ovarian Cancer Cells to Promote Cell Proliferation. Reprod. Sci. 2021. [Google Scholar] [CrossRef] [PubMed]
- Spitale, R.C.; Tsai, M.C.; Chang, H.Y. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics 2011, 6, 539–543. [Google Scholar] [CrossRef]
- Battistelli, C.; Cicchini, C.; Santangelo, L.; Tramontano, A.; Grassi, L.; Gonzalez, F.J.; de Nonno, V.; Grassi, G.; Amicone, L.; Tripodi, M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 2017, 36, 942–955. [Google Scholar] [CrossRef]
- He, S.; Liu, S.; Zhu, H. The sequence, structure and evolutionary features of HOTAIR in mammals. BMC Evol. Biol. 2011, 11, 102. [Google Scholar] [CrossRef]
- Schorderet, P.; Duboule, D. Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet. 2011, 7, e1002071. [Google Scholar] [CrossRef]
- Amândio, A.R.; Necsulea, A.; Joye, E.; Mascrez, B.; Duboule, D. Hotair Is Dispensible for Mouse Development. PLoS Genet. 2016, 12, e1006232. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef]
- Hung, T.; Chang, H.Y. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol. 2010, 7, 582–585. [Google Scholar] [CrossRef]
- Schiavo, G.; D’Antò, V.; Cantile, M.; Procino, A.; Di Giovanni, S.; Valletta, R.; Terracciano, L.; Baumhoer, D.; Jundt, G.; Cillo, C. Deregulated HOX genes in ameloblastomas are located in physical contiguity to keratin genes. J. Cell Biochem. 2011, 112, 3206–3215. [Google Scholar] [CrossRef]
- Wutz, A. RNA-mediated silencing mechanisms in mammalian cells. Prog. Mol. Biol. Transl. Sci. 2011, 101, 351–376. [Google Scholar]
- Guil, S.; Soler, M.; Portela, A.; Carrère, J.; Fonalleras, E.; Gómez, A.; Villanueva, A.; Esteller, M. Intronic RNAs mediate EZH2 regulation of epigenetic targets. Nat. Struct. Mol. Biol. 2012, 19, 664–670. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, L.; Wang, Y.; Li, H.; Ren, X.; Wei, F.; Yu, W.; Wang, X.; Zhang, L.; Yu, J.; et al. Long noncoding RNA HOTAIR involvement in cancer. Tumour Biol. 2014, 35, 9531–9538. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Sun, H.; He, B.; Pan, Y.; Gao, T.; Chen, J.; Ying, H.; Liu, X.; Wang, F.; Xu, Y.; et al. Prognostic value of long non-coding RNA HOTAIR in various cancers. PLoS ONE 2014, 9, e110059. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Chen, J.; Tang, W. The molecular mechanism of HOTAIR in tumorigenesis, metastasis, and drug resistance. Acta Biochim. Biophys. Sin. 2014, 46, 1011–1015. [Google Scholar] [CrossRef]
- Dong, C.; Liu, S.; Lv, Y.; Zhang, C.; Gao, H.; Tan, L.; Wang, H. Long non-coding RNA HOTAIR regulates proliferation and invasion via activating Notch signalling pathway in retinoblastoma. J. Biosci. 2016, 41, 677–687. [Google Scholar] [CrossRef]
- Kalwa, M.; Hänzelmann, S.; Otto, S.; Kuo, C.C.; Franzen, J.; Joussen, S.; Fernandez-Rebollo, E.; Rath, B.; Koch, C.; Hofmann, A.; et al. The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res. 2016, 44, 10631–10643. [Google Scholar] [CrossRef] [PubMed]
- Pawłowska, E.; Szczepanska, J.; Blasiak, J. The Long Noncoding RNA HOTAIR in Breast Cancer: Does Autophagy Play a Role? Int. J. Mol. Sci. 2017, 18, 2317. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, H.; Liu, Y.; Quan, F.; Zhang, X.; Yu, L. LncRNA HOTAIR mediates TGF-β2-induced cell growth and epithelial-mesenchymal transition in human lens epithelial cells. Acta Biochim. Biophys. Sin. 2018, 50, 1028–1037. [Google Scholar] [CrossRef]
- Botti, G.; Scognamiglio, G.; Aquino, G.; Liguori, G.; Cantile, M. LncRNA HOTAIR in Tumor Microenvironment: What Role? Int. J. Mol. Sci. 2019, 20, 2279. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.J.; Zhang, C.H.; Chen, Z.W.; Wang, Z.X.; Yang, D.C.; Zhang, F.L.; Feng, K.H. LncRNA HOTAIR inhibited osteogenic differentiation of BMSCs by regulating Wnt/β-catenin pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7232–7246. [Google Scholar] [PubMed]
- Bai, J.Y.; Jin, B.; Ma, J.B.; Liu, T.J.; Yang, C.; Chong, Y.; Wang, X.; He, D.; Guo, P. HOTAIR and androgen receptor synergistically increase GLI2 transcription to promote tumor angiogenesis and cancer stemness in renal cell carcinoma. Cancer Lett. 2021, 498, 70–79. [Google Scholar] [CrossRef]
- Zhang, J.; Qiu, W.Q.; Zhu, H.; Liu, H.; Sun, J.H.; Chen, Y.; Shen, H.; Qian, C.L.; Shen, Z.Y. HOTAIR contributes to the carcinogenesis of gastric cancer via modulating cellular and exosomal miRNAs level. Cell Death Dis. 2020, 11, 780. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Dashti, S.; Farsi, M.; Taheri, M. HOX transcript antisense RNA: An oncogenic lncRNA in diverse malignancies. Exp. Mol. Pathol. 2021, 118, 104578. [Google Scholar] [CrossRef]
- Zhou, Y.H.; Cui, Y.H.; Wang, T.; Luo, Y. Long non-coding RNA HOTAIR in cervical cancer: Molecular marker, mechanistic insight, and therapeutic target. Adv. Clin. Chem. 2020, 97, 117–140. [Google Scholar]
- Ren, M.M.; Xu, S.; Wei, Y.B.; Yang, J.J.; Yang, Y.N.; Sun, S.S.; Li, Y.J.; Wang, P.Y.; Xie, S.Y. Roles of HOTAIR in lung cancer susceptibility and prognosis. Mol. Genet. Genomic Med. 2020, 8, e1299. [Google Scholar] [CrossRef]
- Mozdarani, H.; Ezzatizadeh, V.; Rahbar Parvaneh, R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 2020, 18, 152. [Google Scholar] [CrossRef]
- Battistelli, C.; Garbo, S.; Riccioni, V.; Montaldo, C.; Santangelo, L.; Vandelli, A.; Strippoli, R.; Tartaglia, G.G.; Tripodi, M.; Cicchini, C. Design and Functional Validation of a Mutant Variant of the LncRNA HOTAIR to Counteract Snail Function in Epithelial-to-Mesenchymal Transition. Cancer Res. 2021, 81, 103–113. [Google Scholar]
- Cantile, M.; Di Bonito, M.; De Bellis, M.T.; Botti, G. Functional Interaction among lncRNA HOTAIR and MicroRNAs in Cancer and Other Human Diseases. Cancers 2021, 13, 570. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Asila, A.; Deng, Y.; Liao, J.; Liu, Z.; Fang, R. Osteopontin-induced lncRNA HOTAIR expression is involved in osteoarthritis by regulating cell proliferation. BMC Geriatr. 2021, 21, 57. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Lu, W.; Lou, W.; Wang, L.; Xu, Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J. Cell Physiol. 2019, 234, 12809–12820. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Q.; Rui, Y.; Zhang, C.; Wang, W.; Gu, J.; Tang, J.; Ding, Y. HOXC13-AS promotes breast cancer cell growth through regulating miR-497-5p/PTEN axis. J. Cell Physiol. 2019, 234, 22343–22351. [Google Scholar] [CrossRef]
- Liu, N.; Wang, Z.; Liu, D.; Xie, P. HOXC13-AS-miR-122-5p-SATB1-C-Myc feedback loop promotes migration, invasion and EMT process in glioma. Onco Targets Ther. 2019, 12, 7165–7173. [Google Scholar] [CrossRef]
- Zhou, J.F.; Shi, Y.T.; Wang, H.G.; Yang, X.Z.; Wu, S.N. Overexpression of long noncoding RNA HOXC13-AS and its prognostic significance in hepatocellular carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7369–7374. [Google Scholar] [PubMed]
- Li, W.; Zhu, Q.; Zhang, S.; Liu, L.; Zhang, H.; Zhu, D. HOXC13-AS accelerates cell proliferation and migration in oral squamous cell carcinoma via miR-378g/HOXC13 axis. Oral Oncol. 2020, 111, 104946. [Google Scholar] [CrossRef] [PubMed]
- Yarmishyn, A.A.; Batagov, A.O.; Tan, J.Z.; Sundaram, G.M.; Sampath, P.; Kuznetsov, V.A.; Kurochkin, I.V. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genom. 2014, 15, S7. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhuang, C.; Liu, Y.; Chen, M.; Chen, Y.; Chen, Z.; He, A.; Lin, J.; Zhan, Y.; Liu, L.; et al. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J. Exp. Clin. Cancer Res. 2016, 35, 99. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Ma, J.; Cai, D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipogenesis. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, J.; Zhou, Z.; He, Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. 2017, 39. [Google Scholar] [CrossRef]
- Zhang, Y.; Dun, Y.; Zhou, S.; Huang, X.H. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/β-catenin signaling pathway. Biomed. Pharmacother. 2017, 96, 1216–1221. [Google Scholar] [CrossRef]
- Gu, P.; Chen, X.; Xie, R.; Han, J.; Xie, W.; Wang, B.; Dong, W.; Chen, C.; Yang, M.; Jiang, J.; et al. lncRNA HOXD-AS1 Regulates Proliferation and Chemo-Resistance of Castration-Resistant Prostate Cancer via Recruiting WDR5. Mol. Ther. 2017, 25, 1959–1973. [Google Scholar] [CrossRef]
- Hu, Y.C.; Wang, A.M.; Lu, J.K.; Cen, R.; Liu, L.L. Long noncoding RNA HOXD-AS1 regulates proliferation of cervical cancer cells by activating Ras/ERK signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5049–5055. [Google Scholar]
- Chen, Y.; Zhao, F.; Cui, D.; Jiang, R.; Chen, J.; Huang, Q.; Shi, J. HOXD-AS1/miR-130a sponge regulates glioma development by targeting E2F8. Int. J. Cancer 2018, 142, 2313–2322. [Google Scholar] [CrossRef]
- Gu, W.; Zhang, E.; Song, L.; Tu, L.; Wang, Z.; Tian, F.; Aikenmu, K.; Chu, G.; Zhao, J. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed. Pharmacother. 2018, 106, 890–895. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Zhang, X.; Huang, Q.; Diao, Y.; Yin, H.; Liu, H. Long non-coding RNA HOXD-AS1 in cancer. Clin. Chim. Acta 2018, 487, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Jing, H.; Li, Y.; Lv, X. Long noncoding RNA HOXD-AS1 promotes non-small cell lung cancer migration and invasion through regulating miR-133b/MMP9 axis. Biomed. Pharmacother. 2018, 106, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Zhao, L.; Wang, L.; Ou-Yang, W.; Hu, S.S.; Li, W.L.; Ai, M.L.; Wang, Y.Q.; Han, Y.; Li, T.T.; et al. Nuclear lncRNA HOXD-AS1 suppresses colorectal carcinoma growth and metastasis via inhibiting HOXD3-induced integrin β3 transcriptional activating and MAPK/AKT signalling. Mol. Cancer 2019, 18, 31. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Nie, W.; Wang, Z.; Zhang, H.; Li, Y.; Fang, X. Lnc HAGLR Promotes Colon Cancer Progression Through Sponging miR-185-5p and Activating CDK4 and CDK6 in vitro and in vivo. Onco Targets Ther. 2020, 13, 5913–5925. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.X.; Ding, W. LncRNA HAGLR accelerates femoral neck fracture healing through negatively regulating miRNA-19a-3p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4080–4087. [Google Scholar] [PubMed]
- Bae, E.; Calhoun, V.C.; Levine, M.; Lewis, E.B.; Drewell, R.A. Characterization of the intergenic RNA profile at abdominal-A and Abdominal-B in the Drosophila bithorax complex. Proc. Natl. Acad. Sci. USA 2002, 99, 16847–16852. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, Z.; Wu, F.; Yin, B.; Jiang, T.; Qiang, B.; Yuan, J.; Han, W.; Peng, X. Long noncoding RNA HOXD-AS2 regulates cell cycle to promote glioma progression. J. Cell Biochem. 2018. [Google Scholar] [CrossRef]
- Yao, L.; Ye, P.C.; Tan, W.; Luo, Y.J.; Xiang, W.P.; Liu, Z.L.; Fu, Z.M.; Lu, F.; Tang, L.H.; Xiao, J.W. Decreased expression of the long non-coding RNA HOXD-AS2 promotes gastric cancer progression by targeting HOXD8 and activating PI3K/Akt signaling pathway. World J. Gastrointest. Oncol. 2020, 12, 1237–1254. [Google Scholar] [CrossRef] [PubMed]
- Galis, F.; Van Dooren, T.J.; Feuth, J.D.; Metz, J.A.; Witkam, A.; Ruinard, S.; Steigenga, M.J.; Wijnaendts, L.C. Extreme selection in humans against homeotic transformations of cervical vertebrae. Evolution 2006, 60, 2643–2654. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, Y.; Lu, Y.; Li, B.; Chen, K.; Li, C. Genome-wide Identification and Characterization of Long Non-coding RNAs in Tribolium castaneum. Insect Sci. 2020. [Google Scholar] [CrossRef]
- Wu, Y.; Cheng, T.; Liu, C.; Liu, D.; Zhang, Q.; Long, R.; Zhao, P.; Xia, Q. Systematic Identification and Characterization of Long Non-Coding RNAs in the Silkworm, Bombyx mori. PLoS ONE 2016, 11, e0147147. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, J.; Sun, Y.; Li, S.; Gao, Y.; Yu, Y.; Liu, C.; Wang, Q.; Lv, X.; Zhang, X.; et al. Penaeid shrimp genome provides insights into benthic adaptation and frequent molting. Nat. Commun. 2019, 10, 356. [Google Scholar] [CrossRef]
- Gutekunst, J.; Andriantsoa, R.; Falckenhayn, C.; Hanna, K.; Stein, W.; Rasamy, J.; Lyko, F. Clonal genome evolution and rapid invasive spread of the marbled crayfish. Nat. Ecol. Evol. 2018, 2, 567–573. [Google Scholar] [CrossRef]
- Tang, B.; Zhang, D.; Li, H.; Jiang, S.; Zhang, H.; Xuan, F.; Ge, B.; Wang, Z.; Liu, Y.; Sha, Z.; et al. Chromosome-level genome assembly reveals the unique genome evolution of the swimming crab (Portunus trituberculatus). Gigascience 2020, 9, giz161. [Google Scholar] [CrossRef] [PubMed]
- Chebbi, M.A.; Becking, T.; Moumen, B.; Giraud, I.; Gilbert, C.; Peccoud, J.; Cordaux, R. The Genome of Armadillidium vulgare (Crustacea, Isopoda) Provides Insights into Sex Chromosome Evolution in the Context of Cytoplasmic Sex Determination. Mol. Biol. Evol. 2019, 36, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Thomas, G.W.C.; Dohmen, E.; Hughes, D.S.T.; Murali, S.C.; Poelchau, M.; Glastad, K.; Anstead, C.A.; Ayoub, N.A.; Batterham, P.; Bellair, M.; et al. Gene content evolution in the arthropods. Genome Biol. 2020, 21, 15. [Google Scholar] [CrossRef]
- Pang, Y.; Mao, C.; Liu, S. Encoding activities of non-coding RNAs. Theranostics 2018, 8, 2496–2507. [Google Scholar] [CrossRef]
- Mori, K.; Weng, S.M.; Arzberger, T.; May, S.; Rentzsch, K.; Kremmer, E.; Schmid, B.; Kretzschmar, H.A.; Cruts, M.; Van Broeckhoven, C.; et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 2013, 339, 1335–1338. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Dostie, J. Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res. 2017, 45, 1091–1104. [Google Scholar] [PubMed]
- Jenkins, A.M.; Waterhouse, R.M.; Muskavitch, M.A. Long non-coding RNA discovery across the genus anopheles reveals conserved secondary structures within and beyond the Gambiae complex. BMC Genom. 2015, 16, 337. [Google Scholar] [CrossRef] [PubMed]
- Legeai, F.; Derrien, T. Identification of long non-coding RNAs in insects genomes. Curr. Opin. Insect Sci. 2015, 7, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Chang, Z.X.; Ajayi, O.E.; Guo, D.Y.; Wu, F. Genome-wide characterization and developmental expression profiling of long non-coding RNAs in Sogatella furcifera. Insect Sci. 2020, 27, 987–997. [Google Scholar] [CrossRef]
- Guan, R.; Li, H.; Zhang, H.; An, S. Comparative analysis of dsRNA-induced lncRNAs in three kinds of insect species. Arch. Insect Biochem. Physiol. 2020, 103, e21640. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, C.; Sharma, S.; Meghwanshi, K.K.; Patel, S.; Mehta, P.; Shukla, N.; Do, D.N.; Rajpurohit, S.; Suravajhala, P.; Shukla, J.N. Long Non-Coding RNAs in Insects. Animals 2021, 11, 1118. [Google Scholar] [CrossRef]
- Ronshaugen, M.; Biemar, F.; Piel, J.; Levine, M.; Lai, E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005, 19, 2947–2952. [Google Scholar] [CrossRef]
- Garaulet, D.L.; Lai, E.C. Hox miRNA regulation within the Drosophila Bithorax complex: Patterning behavior. Mech. Dev. 2015, 138, 151–159. [Google Scholar] [CrossRef]
- Hui, J.H.; Marco, A.; Hunt, S.; Melling, J.; Griffiths-Jones, S.; Ronshaugen, M. Structure, evolution and function of the bi-directionally transcribed iab-4/iab-8 microRNA locus in arthropods. Nucleic Acids Res. 2013, 41, 3352–3361. [Google Scholar] [CrossRef]
- Negre, B.; Ruiz, A. HOM-C evolution in Drosophila: Is there a need for Hox gene clustering? Trends Genet. 2007, 23, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Shippy, T.D.; Ronshaugen, M.; Cande, J.; He, J.; Beeman, R.W.; Levine, M.; Brown, S.J.; Denell, R.E. Analysis of the Tribolium homeotic complex: Insights into mechanisms constraining insect Hox clusters. Dev. Genes Evol. 2008, 218, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Chen, X.; Peng, J.; Yang, C.; Peng, M.; Zhu, W.; Xie, D.; He, P.; Wei, P.; Lin, Y.; et al. Single-molecule long-read sequencing facilitates shrimp transcriptome research. Sci. Rep. 2018, 8, 16920. [Google Scholar] [CrossRef] [PubMed]
- Wan, H.; Jia, X.; Zou, P.; Zhang, Z.; Wang, Y. The Single-molecule long-read sequencing of Scylla paramamosain. Sci. Rep. 2019, 9, 12401. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, B.M.; Lee, B.Y.; Souissi, S.; Park, H.G.; Lee, J.S. Identification of Hox genes and rearrangements within the single homeobox (Hox) cluster (192.8 kb) of the cyclopoid copepod (Paracyclopina nana). J. Exp. Zool. B Mol. Dev. Evol. 2016, 326, 105–109. [Google Scholar] [CrossRef]
- Kim, D.H.; Lee, B.Y.; Kim, H.S.; Jeong, C.B.; Hwang, D.S.; Kim, I.C.; Lee, J.S. Identification and characterization of homeobox (Hox) genes and conservation of the single Hox cluster (324.6 kb) in the water flea Daphnia magna. J. Exp. Zool. B Mol. Dev. Evol. 2018, 330, 76–82. [Google Scholar] [CrossRef]
- Chipman, A.; Ferrier, D.; Brena, C.; Qu, J.; Hughes, D.S.T.; Schröder, R.; Torres-Oliva, M.; Znassi, N.; Jiang, H.; Almeida, F.; et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014, 12, e1002005. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Nong, W.; So, W.L.; Barton-Owen, T.; Li, Y.; Leung, T.C.N.; Li, C.; Baril, T.; Wong, A.Y.P.; Swale, T.; et al. Millipede genomes reveal unique adaptations during myriapod evolution. PLoS Biol. 2020, 18, e3000636. [Google Scholar] [CrossRef]
- Bakalenko, N.I.; Novikova, E.L.; Nesterenko, A.Y.; Kulakova, M.A. Hox gene expression during postlarval development of the polychaete Alitta virens. Evodevo 2013, 4, 13. [Google Scholar] [CrossRef] [PubMed]
- Novikova, E.L.; Bakalenko, N.I.; Nesterenko, A.Y.; Kulakova, M.A. Expression of Hox genes during regeneration of nereid polychaete Alitta (Nereis) virens (Annelida, Lophotrochozoa). Evodevo 2013, 4, 14. [Google Scholar] [CrossRef]
- Novikova, E.L.; Bakalenko, N.I.; Kulakova, M.A. Annelids win again: The first evidence of Hox antisense transcription in Spiralia. bioRxiv 2021. [Google Scholar] [CrossRef]
- Andreeva, T.F.; Cook, C.; Korchagina, N.M.; Akam, M.; Dondya, A.K. Cloning and analysis of structural organization of Hox genes in the polychaete Nereis virens. Ontogenez 2001, 32, 225–233. (In Russian) [Google Scholar] [PubMed]
- Deschamps, J.; Duboule, D. Embryonic timing, axial stem cells, chromatin dynamics, and the Hox clock. Genes Dev. 2017, 31, 1406–1416. [Google Scholar] [CrossRef]
- Iimura, T.; Pourquié, O. Hox genes in time and space during vertebrate body formation. Dev. Growth Differ. 2007, 49, 265–275. [Google Scholar] [CrossRef]
- Hara, Y.; Yamaguchi, K.; Onimaru, K.; Kadota, M.; Koyanagi, M.; Keeley, S.D.; Tatsumi, K.; Tanaka, K.; Motone, F.; Kageyama, Y.; et al. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2018, 2, 1761–1771. [Google Scholar] [CrossRef]
- Murray, S.C.; Mellor, J. Using both strands: The fundamental nature of antisense transcription. Bioarchitecture 2016, 6, 12–21. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Xu, Z.; Wei, W.; Gagneur, J.; Clauder-Münster, S.; Smolik, M.; Huber, W.; Steinmetz, L.M. Antisense expression increases gene expression variability and locus interdependency. Mol. Syst. Biol. 2011, 7, 468. [Google Scholar] [CrossRef] [PubMed]
- Fusco, G. Trunk segment numbers and sequential segmentation in myriapods. Evol. Dev. 2005, 7, 608–617. [Google Scholar] [CrossRef]

| LncRNA | Length (nt) | Type of LncRNA | Position in the Hox Cluster |
|---|---|---|---|
| HOXA Cluster | |||
| HOTAIRM1 | 4000; 1052; 783 | Linc | HOXA1→HOXA2 |
| HOXA-AS2 | 1048 | Linc or NAT | HOXA3→HOXA4 |
| HOXA-AS3 | 3918; 3992 | Linc or NAT | HOXA4→A5,→A6→HOXA7 |
| HOXA10-AS | 1161 | Linc or NAT | HOXA9→HOXA10 |
| HOXA11-AS | 1628 | Linc or NAT | HOXA11→HOXA13 |
| HOTTIP | 4665 | Linc or NAT | HOXA13→EVX1 |
| HOXB Cluster | |||
| HOXB-AS1 | 797 | Linc or NAT | HOXB2→HOXB3 |
| HOXB-AS2 | 3594 | NAT RNA | HOXB3 |
| HOXB-AS3 | 785; 611; 549; 545; 514; 452; 446; 336 | Linc or NAT * | HOXB4→B5→HOXB6 |
| HOXB-AS4 | 543; 513 | Linc | HOXB9→HOXA13 |
| PRAC2 | 1193; 560; 518; 503; 448 | Linc * | HOXB9→HOXA13 |
| HOXC Cluster | |||
| HOXC-AS1 | 548 | Intronic | HOXC9 |
| HOXC-AS2 | 504 | Linc or NAT | HOXC9→HOXC10 |
| HOXC-AS3 | 368 | Linc or NAT | HOXC10→HOXC11 |
| HOTAIR | 2370; 2364; 2337 | Linc | HOXC11→HOXC12 |
| HOXC13-AS | 1408 | Linc or NAT | HOXC9→ |
| HOXD Cluster | |||
| HAGLR | 4086; 4037; 4007; 3942; 3923; 3905; 3893; 3891; 3821; 3812; 3794; 3782 | Linc or NAT | HOXD1→HOXD3 |
| HOXD-AS2 | 692 | Linc or NAT | HOXD4→D8→D9→HOXD10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Novikova, E.L.; Kulakova, M.A. There and Back Again: Hox Clusters Use Both DNA Strands. J. Dev. Biol. 2021, 9, 28. https://doi.org/10.3390/jdb9030028
Novikova EL, Kulakova MA. There and Back Again: Hox Clusters Use Both DNA Strands. Journal of Developmental Biology. 2021; 9(3):28. https://doi.org/10.3390/jdb9030028
Chicago/Turabian StyleNovikova, Elena L., and Milana A. Kulakova. 2021. "There and Back Again: Hox Clusters Use Both DNA Strands" Journal of Developmental Biology 9, no. 3: 28. https://doi.org/10.3390/jdb9030028
APA StyleNovikova, E. L., & Kulakova, M. A. (2021). There and Back Again: Hox Clusters Use Both DNA Strands. Journal of Developmental Biology, 9(3), 28. https://doi.org/10.3390/jdb9030028






