Do Transgenerational Epigenetic Inheritance and Immune System Development Share Common Epigenetic Processes?
Abstract
1. Introduction
2. Overview of Epigenetics
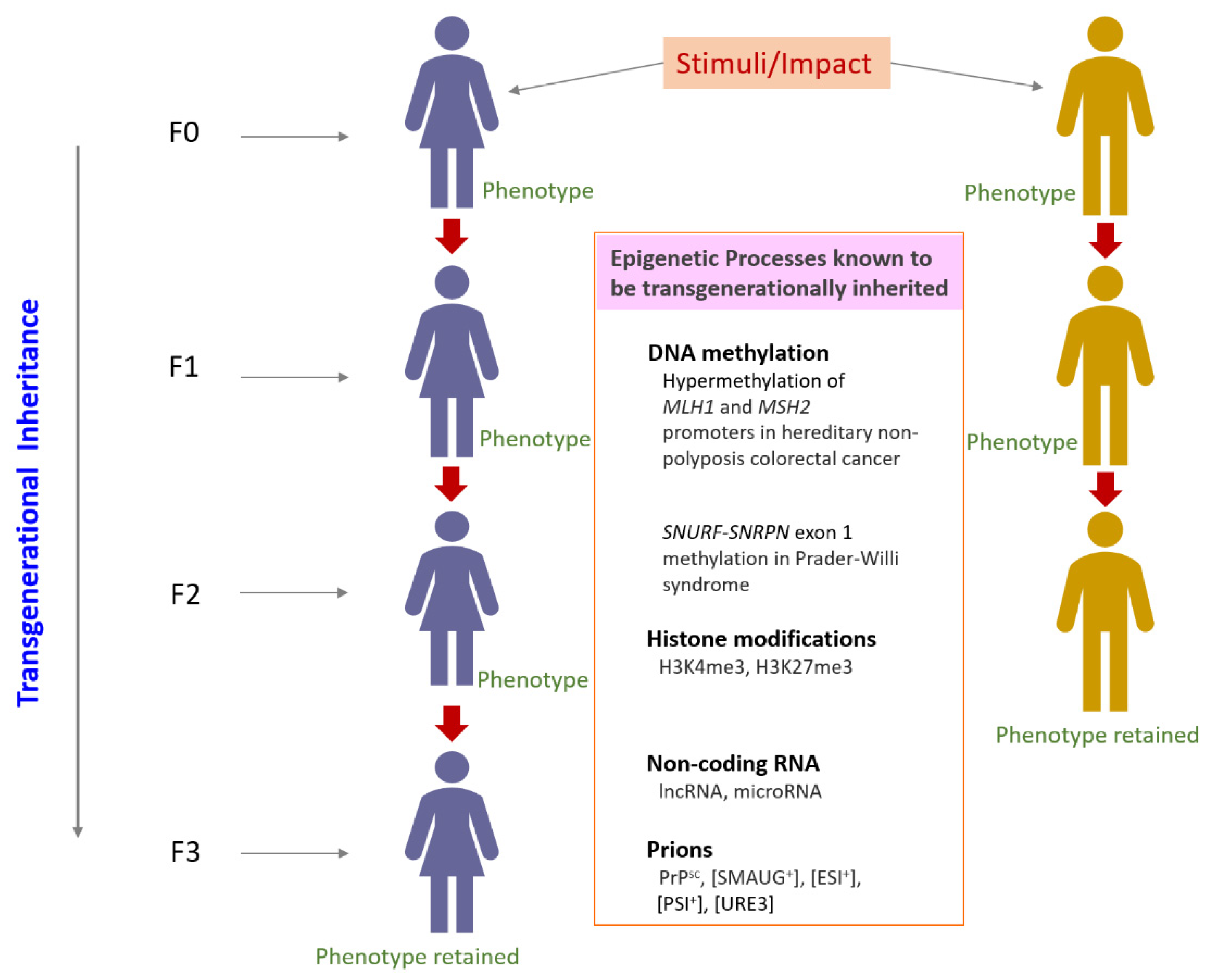
3. Transgenerational Epigenetic Inheritance
3.1. Transgenerational Epigenetic Inheritance in Development
3.2. Transgenerational Epigenetic Inheritance in Disease
3.3. Transgenerational Epigenetic Inheritance and Histone Modifications
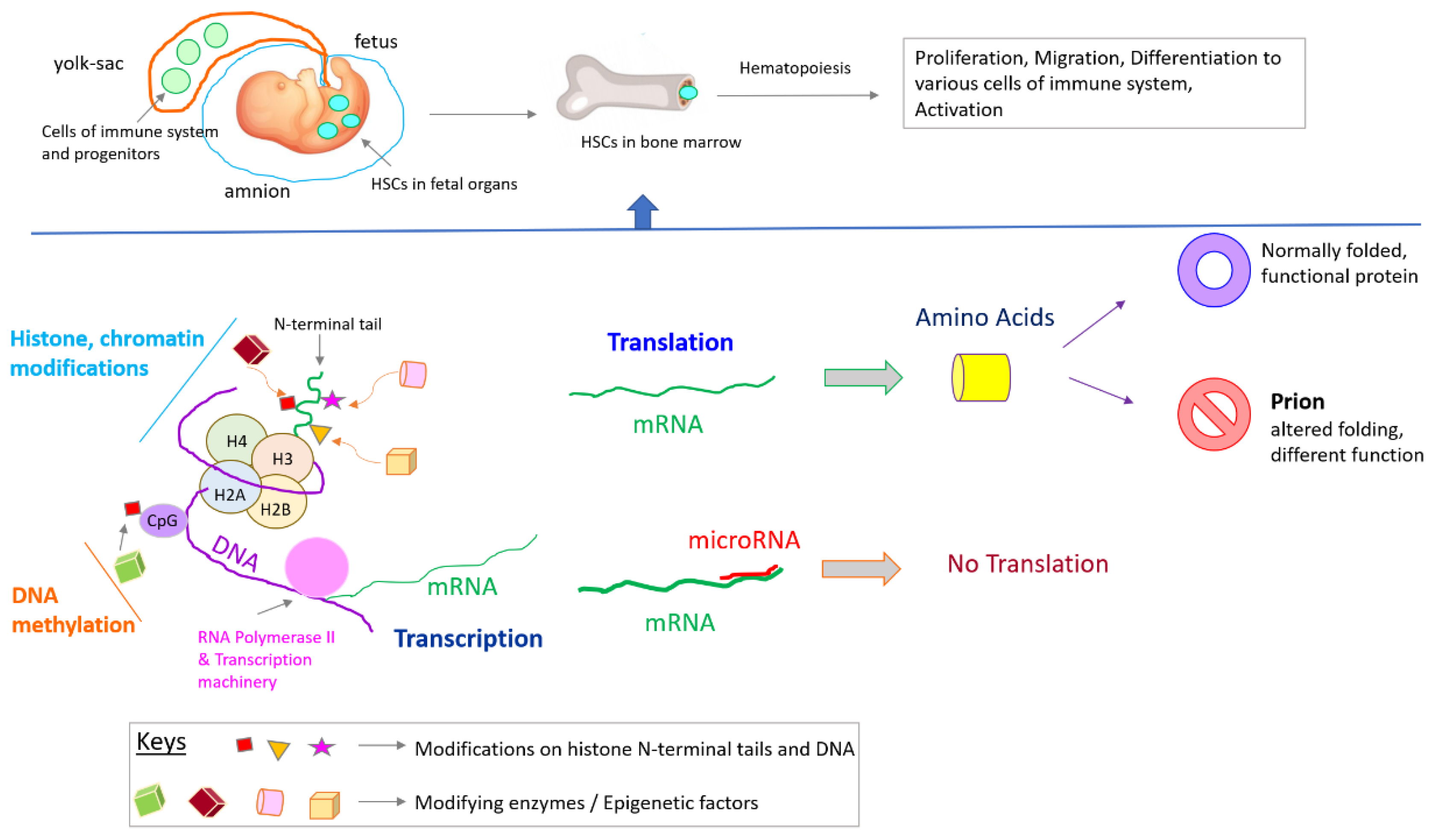
4. Overview of Immune System Development
5. Epigenetics in Immune System Development and Activation
Non-Coding RNA-Mediated Regulation of Epigenetics and the Immune System
6. Epigenetics at the Protein Level: Prions
6.1. Role of Prions in Gene Expression Related to the Immune System
6.2. Prions and Epigenetic Inheritance
6.3. Prions and Transgenerational Epigenetic Inheritance
7. Immune-Related Pathologies Involving Transgenerational Epigenetic Inheritance
8. Epigenetic Inheritance and the Immune System in the Context of Aging
9. Methods to Study Epigenetics and, Hence, Transgenerational Epigenetic Inheritance
10. Clinical Trial on Transgenerational Intervention
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tikhodeyev, O.N. The mechanisms of epigenetic inheritance: How diverse are they? Biol. Rev. 2018, 93, 1987–2005. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.F.; Lehner, B. Intergenerational and transgenerational epigenetic inheritance in animals. Nat. Cell Biol. 2019, 21, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod. Toxicol. 2008, 25, 2–6. [Google Scholar] [CrossRef]
- Heard, E.; Martienssen, R.A. Transgenerational Epigenetic Inheritance: Myths and Mechanisms. Cell 2014, 157, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Bošković, A.; Rando, O.J. Transgenerational Epigenetic Inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef]
- van Otterdijk, S.D.; Michels, K.B. Transgenerational epigenetic inheritance in mammals: How good is the evidence? FASEB J. 2016, 30, 2457–2465. [Google Scholar] [CrossRef]
- E Nilsson, E.; Sadler-Riggleman, I.; Skinner, M.K. Environmentally induced epigenetic transgenerational inheritance of disease. Environ. Epigenet. 2018, 4, dvy016. [Google Scholar] [CrossRef]
- Hackett, J.A.; Surani, M.A. Beyond DNA: Programming and Inheritance of Parental Methylomes. Cell 2013, 153, 737–739. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nat. Cell Biol. 2012, 484, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Constância, M.; Pickard, B.; Kelsey, G.; Reik, W. Imprinting Mechanisms. Genome Res. 1998, 8, 881–900. [Google Scholar] [CrossRef][Green Version]
- Xavier, M.J.; Roman, S.D.; Aitken, R.J.; Nixon, B. Transgenerational inheritance: How impacts to the epigenetic and genetic information of parents affect offspring health. Hum. Reprod. Updat. 2019, 25, 519–541. [Google Scholar] [CrossRef]
- LeGoff, L.; D’Cruz, S.C.; Tevosian, S.; Primig, M.; Smagulova, F. Transgenerational Inheritance of Environmentally Induced Epigenetic Alterations during Mammalian Development. Cells 2019, 8, 1559. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Manikkam, M.; Guerrero-Bosagna, C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 2010, 21, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Bygren, L.O.; Tinghög, P.; Carstensen, J.; Edvinsson, S.; Kaati, G.; E Pembrey, M.; Sjöström, M. Change in paternal grandmothers’ early food supply influenced cardiovascular mortality of the female grandchildren. BMC Genet. 2014, 15, 12. [Google Scholar] [CrossRef] [PubMed]
- E Veenendaal, M.V.; Painter, R.C.; De Rooij, S.R.; Bossuyt, P.M.M.; Post, J.A.M.V.D.; Gluckman, P.D.; A Hanson, M.; Roseboom, T.J. Transgenerational effects of prenatal exposure to the 1944-45 Dutch famine. BJOG Int. J. Obstet. Gynaecol. 2013, 120, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Rando, O.J. Metabolic Inputs into the Epigenome. Cell Metab. 2017, 25, 544–558. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U. Paternal Contributions to Offspring Health: Role of Sperm Small RNAs in Intergenerational Transmission of Epigenetic Information. Front. Cell Dev. Biol. 2019, 7, 215. [Google Scholar] [CrossRef]
- Skinner, M.K.; Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; Nilsson, E.; McBirney, M.; Klukovich, R.; Xie, Y.; Tang, C.; Yan, W. Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease. Epigenet. Chromatin 2018, 11, 1–24. [Google Scholar] [CrossRef]
- Rando, O.J. Daddy Issues: Paternal Effects on Phenotype. Cell 2012, 151, 702–708. [Google Scholar] [CrossRef]
- Bao, J.; Bedford, M.T. Epigenetic regulation of the histone-to-protamine transition during spermiogenesis. Reproduction 2016, 151, R55–R70. [Google Scholar] [CrossRef]
- Rathke, C.; Baarends, W.M.; Awe, S.; Renkawitz-Pohl, R. Chromatin dynamics during spermiogenesis. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1839, 155–168. [Google Scholar] [CrossRef]
- Ihara, M.; Meyer-Ficca, M.L.; Leu, N.A.; Rao, S.; Li, F.; Gregory, B.D.; Zalenskaya, I.A.; Schultz, R.M.; Meyer, R.G. Paternal Poly (ADP-ribose) Metabolism Modulates Retention of Inheritable Sperm Histones and Early Embryonic Gene Expression. PLoS Genet. 2014, 10, e1004317. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Zhang, H.; Purwar, J.; Carrell, D.T.; Cairns, B.R. Distinctive chromatin in human sperm packages genes for embryo development. Nat. Cell Biol. 2009, 460, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Ben Maamar, M.; Sadler-Riggleman, I.; Beck, D.; Skinner, M.K. Epigenetic Transgenerational Inheritance of Altered Sperm Histone Retention Sites. Sci. Rep. 2018, 8, 5308. [Google Scholar] [CrossRef] [PubMed]
- Hammoud, S.S.; Nix, D.A.; Hammoud, A.O.; Gibson, M.; Cairns, B.R.; Carrell, D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011, 26, 2558–2569. [Google Scholar] [CrossRef] [PubMed]
- Skinner, M.K.; Manikkam, M.; Tracey, R.; Guerrero-Bosagna, C.; Haque, M.; E Nilsson, E. Ancestral dichlorodiphenyltrichloroethane (DDT) exposure promotes epigenetic transgenerational inheritance of obesity. BMC Med. 2013, 11, 228. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, M.; Haque, M.M.; Guerrero-Bosagna, C.; Nilsson, E.E.; Skinner, M.K. Pesticide Methoxychlor Promotes the Epigenetic Transgenerational Inheritance of Adult-Onset Disease through the Female Germline. PLoS ONE 2014, 9, e102091. [Google Scholar] [CrossRef]
- Guerrero-Bosagna, C.; Savenkova, M.; Haque, M.; Nilsson, E.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Altered Sertoli Cell Transcriptome and Epigenome: Molecular Etiology of Male Infertility. PLoS ONE 2013, 8, e59922. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, E.; Larsen, G.; Manikkam, M.; Guerrero-Bosagna, C.; Savenkova, M.I.; Skinner, M.K. Environmentally Induced Epigenetic Transgenerational Inheritance of Ovarian Disease. PLoS ONE 2012, 7, e36129. [Google Scholar] [CrossRef]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef]
- Scoville, S.D.; Freud, A.G.; Caligiuri, M.A. Modeling Human Natural Killer Cell Development in the Era of Innate Lymphoid Cells. Front. Immunol. 2017, 8, 360. [Google Scholar] [CrossRef] [PubMed]
- Rosales, C. Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front. Physiol. 2018, 9, 113. [Google Scholar] [CrossRef] [PubMed]
- Görgens, A.; Radtke, S.; Möllmann, M.; Cross, M.; Dürig, J.; Horn, P.A.; Giebel, B. Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. Cell Rep. 2013, 3, 1539–1552. [Google Scholar] [CrossRef]
- Park, J.-E.; Jardine, L.; Gottgens, B.; Teichmann, S.A.; Haniffa, M. Prenatal development of human immunity. Science 2020, 368, 600–603. [Google Scholar] [CrossRef] [PubMed]
- Wolf, A.A.; Yáñez, A.; Barman, P.K.; Goodridge, H.S. The Ontogeny of Monocyte Subsets. Front. Immunol. 2019, 10, 1642. [Google Scholar] [CrossRef] [PubMed]
- Geissmann, F.; Manz, M.G.; Jung, S.; Sieweke, M.H.; Merad, M.; Ley, K. Development of Monocytes, Macrophages, and Dendritic Cells. Science 2010, 327, 656–661. [Google Scholar] [CrossRef]
- LeBien, T.W.; Tedder, T.F. B lymphocytes: How they develop and function. Blood 2008, 112, 1570–1580. [Google Scholar] [CrossRef] [PubMed]
- Krystel-Whittemore, M.; Dileepan, K.N.; Wood, J.G. Mast Cell: A Multi-Functional Master Cell. Front. Immunol. 2016, 6, 620. [Google Scholar] [CrossRef]
- Kumar, B.V.; Connors, T.J.; Farber, D.L. Human T Cell Development, Localization, and Function throughout Life. Immunity 2018, 48, 202–213. [Google Scholar] [CrossRef]
- Germain, R.N. T-cell development and the CD4–CD8 lineage decision. Nat. Rev. Immunol. 2002, 2, 309–322. [Google Scholar] [CrossRef]
- Koch, U.; Radtke, F. Mechanisms of T Cell Development and Transformation. Annu. Rev. Cell Dev. Biol. 2011, 27, 539–562. [Google Scholar] [CrossRef] [PubMed]
- Caramalho, Í.; Nunes-Cabaço, H.; Foxall, R.B.; Sousa, A.E. Regulatory T-Cell Development in the Human Thymus. Front. Immunol. 2015, 6, 395. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.-M.; Botting, R.A.; Stephenson, E.; Green, K.; Webb, S.; Jardine, L.; Calderbank, E.F.; Polanski, K.; Goh, I.; Efremova, M.; et al. Decoding human fetal liver haematopoiesis. Nat. Cell Biol. 2019, 574, 365–371. [Google Scholar] [CrossRef]
- Ginhoux, F.; Guilliams, M. Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 2016, 44, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Slayton, W.B.; Li, Y.; A Calhoun, D.; E Juul, S.; Iturraspe, J.; Braylan, R.C.; Christensen, R.D. The first-appearance of neutrophils in the human fetal bone marrow cavity. Early Hum. Dev. 1998, 53, 129–144. [Google Scholar] [CrossRef]
- Haynes, B.F.; Heinly, C.S. Early human T cell development: Analysis of the human thymus at the time of initial entry of hematopoietic stem cells into the fetal thymic microenvironment. J. Exp. Med. 1995, 181, 1445–1458. [Google Scholar] [CrossRef]
- Busslinger, M.; Tarakhovsky, A. Epigenetic control of immunity. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef]
- Nina, S.; Bruno, S.-S.; Anita, Q.G. Chapter 4—Epigenetic mechanisms in the regulation of lymphocyte differentiation. Transl. Epigenet. 2020, 16, 77–116. [Google Scholar] [CrossRef]
- Stephen, J.T.; Jasmine, L.; Brendan, E.R. Chapter 5—Epigenetics mechanisms driving immune memory cell differentiation and function. Transl. Epigenet. 2020, 16, 117–137. [Google Scholar]
- Lara-Astiaso, D.; Weiner, A.; Lorenzo-Vivas, E.; Zaretsky, I.; Jaitin, D.A.; David, E.; Keren-Shaul, H.; Mildner, A.; Winter, D.; Jung, S.; et al. Chromatin state dynamics during blood formation. Science 2014, 345, 943–949. [Google Scholar] [CrossRef]
- Suarez-Alvarez, B.; Baragano Raneros, A.; Ortega, F.; Lopez-Larrea, C. Epigenetic modulation of the immune function: A potential target for tolerance. Epigenetics 2013, 8, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Weng, N.-P.; Araki, Y.; Subedi, K. The molecular basis of the memory T cell response: Differential gene expression and its epigenetic regulation. Nat. Rev. Immunol. 2012, 12, 306–315. [Google Scholar] [CrossRef]
- Araki, Y.; Wang, Z.; Zang, C.; Wood, W.H.; Schones, D.; Cui, K.; Roh, T.-Y.; Lhotsky, B.; Wersto, R.P.; Peng, W.; et al. Genome-wide Analysis of Histone Methylation Reveals Chromatin State-Based Regulation of Gene Transcription and Function of Memory CD8+ T Cells. Immunity 2009, 30, 912–925. [Google Scholar] [CrossRef] [PubMed]
- Gjoneska, E.; Pfenning, A.R.; Mathys, H.; Quon, G.; Kundaje, A.; Tsai, L.-H.; Kellis, M. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nat. Cell Biol. 2015, 518, 365–369. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Mut, J.V.; Graff, J. Epigenetic Alterations in Alzheimer’s Disease. Front. Behav. Neurosci. 2015, 9, 347. [Google Scholar] [CrossRef]
- Gutierrez, M.J.; Nino, G.; Hong, X.; Wang, X. Epigenetic Dynamics of the Infant Immune System Reveals a Tumor Necrosis Factor Superfamily Signature in Early Human Life. Epigenomes 2020, 4, 12. [Google Scholar] [CrossRef]
- van der Heijden, C.; Noz, M.P.; Joosten, L.A.B.; Netea, M.G.; Riksen, N.P.; Keating, S.T. Epigenetics and Trained Immunity. Antioxid. Redox Signal. 2018, 29, 1023–1040. [Google Scholar] [CrossRef]
- Álvarez-Errico, D.; Vento-Tormo, R.; Sieweke, M.H.; Ballestar, E. Epigenetic control of myeloid cell differentiation, identity and function. Nat. Rev. Immunol. 2015, 15, 7–17. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Álvarez-Errico, D.; Rodríguez-Ubreva, J.; Ballestar, E. Gains of DNA methylation in myeloid terminal differentiation are dispensable for gene silencing but influence the differentiated phenotype. FEBS J. 2014, 282, 1815–1825. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Ramírez, J.; Lukin, K.; Hagman, J. From hematopoietic progenitors to B cells: Mechanisms of lineage restriction and commitment. Curr. Opin. Immunol. 2010, 22, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Santos, P.; Arumemi, F.; Park, K.S.; Borghesi, L.; Milcarek, C. Transcriptional and epigenetic regulation of B cell development. Immunol. Res. 2011, 50, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Parra, M. Epigenetic events during B lymphocyte development. Epigenetics 2009, 4, 462–468. [Google Scholar] [CrossRef]
- Lee, S.-T.; Xiao, Y.; Muench, M.O.; Xiao, J.; Fomin, M.E.; Wiencke, J.K.; Zheng, S.; Dou, X.; De Smith, A.; Chokkalingam, A.; et al. A global DNA methylation and gene expression analysis of early human B-cell development reveals a demethylation signature and transcription factor network. Nucleic Acids Res. 2012, 40, 11339–11351. [Google Scholar] [CrossRef] [PubMed]
- Teperek-Tkacz, M.; Pasque, V.; Gentsch, G.; Ferguson-Smith, A.C. Epigenetic reprogramming: Is deamination key to active DNA demethylation? Reproduction 2011, 142, 621–632. [Google Scholar] [CrossRef]
- Kuraoka, M.; Holl, T.M.; Liao, D.; Womble, M.; Cain, D.W.; Reynolds, A.E.; Kelsoe, G. Activation-induced cytidine deaminase mediates central tolerance in B cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11560–11565. [Google Scholar] [CrossRef] [PubMed]
- Su, S.-T.; Ying, H.-Y.; Chiu, Y.-K.; Lin, F.-R.; Chen, M.-Y.; Lin, K.-I. Involvement of Histone Demethylase LSD1 in Blimp-1-Mediated Gene Repression during Plasma Cell Differentiation. Mol. Cell. Biol. 2009, 29, 1421–1431. [Google Scholar] [CrossRef]
- Shin, H.M.; Kapoor, V.N.; Guan, T.; Kaech, S.M.; Welsh, R.M.; Berg, L.J. Epigenetic Modifications Induced by Blimp-1 Regulate CD8+ T Cell Memory Progression during Acute Virus Infection. Immunity 2013, 39, 661–675. [Google Scholar] [CrossRef]
- Fu, S.-H.; Yeh, L.-T.; Chu, C.-C.; Yen, B.L.-J.; Sytwu, H.-K. New insights into Blimp-1 in T lymphocytes: A divergent regulator of cell destiny and effector function. J. Biomed. Sci. 2017, 24, 1–17. [Google Scholar] [CrossRef]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef]
- Reiner, S.L. Epigenetic control in the immune response. Hum. Mol. Genet. 2005, 14, R41–R46. [Google Scholar] [CrossRef] [PubMed]
- Placek, K.; Schultze, J.L.; Aschenbrenner, A.C. Epigenetic reprogramming of immune cells in injury, repair, and resolution. J. Clin. Investig. 2019, 129, 2994–3005. [Google Scholar] [CrossRef] [PubMed]
- Joh, R.I.; Palmieri, C.M.; Hill, I.T.; Motamedi, M. Regulation of histone methylation by noncoding RNAs. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1839, 1385–1394. [Google Scholar] [CrossRef]
- Ansel, K.M. RNA regulation of the immune system. Immunol. Rev. 2013, 253, 5–11. [Google Scholar] [CrossRef]
- Chen, C.-Z.; Li, L.; Lodish, H.F.; Bartel, D.P. MicroRNAs Modulate Hematopoietic Lineage Differentiation. Science 2004, 303, 83–86. [Google Scholar] [CrossRef]
- Schwarz, D.S.; Zamore, P.D. Why do miRNAs live in the miRNP? Genes Dev. 2002, 16, 1025–1031. [Google Scholar] [CrossRef]
- Rana, T.M. Illuminating the silence: Understanding the structure and function of small RNAs. Nat. Rev. Mol. Cell Biol. 2007, 8, 23–36. [Google Scholar] [CrossRef]
- Pratt, A.J.; MacRae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, B.; Wu, T.; Skogerbo, G.; Zhu, X.; Guo, X.; He, S.; Chen, R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009, 10, 12. [Google Scholar] [CrossRef]
- Hawkins, P.G.; Morris, K.V. RNA and transcriptional modulation of gene expression. Cell Cycle 2008, 7, 602–607. [Google Scholar] [CrossRef] [PubMed]
- Humphries, B.; Wang, Z.; Yang, C. MicroRNA Regulation of Epigenetic Modifiers in Breast Cancer. Cancers 2019, 11, 897. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Tuddenham, L.; Wheeler, G.; Ntounia-Fousara, S.; Waters, J.; Hajihosseini, M.K.; Clark, I.; Dalmay, T. The cartilage specific microRNA-140 targets histone deacetylase 4 in mouse cells. FEBS Lett. 2006, 580, 4214–4217. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; E Callis, T.; Hammond, S.M.; Conlon, F.L.; Wang, D.-Z. The Role of MicroRNA-1 and MicroRNA-133 in Skeletal Muscle Proliferation and Differentiation. Nat. Genet. 2005, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Sang, M.; Meng, L.; Gu, L.; Liu, S.; Li, J.; Geng, C. miR-92b promotes autophagy and suppresses viability and invasion in breast cancer by targeting EZH2. Int. J. Oncol. 2018, 53, 1505–1515. [Google Scholar] [CrossRef]
- Monticelli, S.; Ansel, K.M.; Xiao, C.; Socci, N.D.; Krichevsky, A.M.; Thai, T.-H.; Rajewsky, N.; Marks, D.S.; Sander, C.; Rajewsky, K.; et al. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 2005, 6, R71. [Google Scholar] [CrossRef]
- Montagner, S.; Orlandi, E.M.; Merante, S.; Monticelli, S. The role of miRNAs in mast cells and other innate immune cells. Immunol. Rev. 2013, 253, 12–24. [Google Scholar] [CrossRef]
- De Yébenes, V.G.; Bartolomé-Izquierdo, N.; Ramiro, A.R. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013, 253, 25–39. [Google Scholar] [CrossRef]
- Beaulieu, A.M.; Bezman, N.A.; Lee, J.E.; Matloubian, M.; Sun, J.C.; Lanier, L.L. MicroRNA function in NK-cell biology. Immunol. Rev. 2013, 253, 40–52. [Google Scholar] [CrossRef] [PubMed]
- Dooley, J.; Linterman, M.A.; Liston, A. MicroRNA regulation of T-cell development. Immunol. Rev. 2013, 253, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Jeker, L.T.; Bluestone, J.A. MicroRNA regulation of T-cell differentiation and function. Immunol. Rev. 2013, 253, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Pagani, M.; Rossetti, G.; Panzeri, I.; De Candia, P.; Bonnal, R.J.P.; Rossi, R.L.; Geginat, J.; Abrignani, S. Role of microRNAs and long-non-coding RNAs in CD4+T-cell differentiation. Immunol. Rev. 2013, 253, 82–96. [Google Scholar] [CrossRef]
- Chen, J.-Q.; Papp, G.; Szodoray, P.; Zeher, M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun. Rev. 2016, 15, 1171–1180. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, M.; Riba, M.; Garcia Manteiga, J.M.; Tian, L.; Vigano, V.; Rossetti, G.; Pagani, M.; Xiao, C.; Liston, A.; Stupka, E.; et al. miR-17 approximately 92 family clusters control iNKT cell ontogenesis via modulation of TGF-beta signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E8286–E8295. [Google Scholar] [CrossRef]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenet. 2019, 11, 1–15. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Bayne, E.H.; Allshire, R.C. RNA-directed transcriptional gene silencing in mammals. Trends Genet. 2005, 21, 370–373. [Google Scholar] [CrossRef]
- Gapp, K.; Jawaid, A.; Sarkies, P.; Bohacek, J.; Pelczar, P.; Prados, J.; Farinelli, L.; Miska, E.A.; Mansuy, I.M. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat. Neurosci. 2014, 17, 667–669. [Google Scholar] [CrossRef]
- Rodgers, A.B.; Morgan, C.P.; Leu, N.A.; Bale, T.L. Transgenerational epigenetic programming via sperm microRNA recapitulates effects of paternal stress. Proc. Natl. Acad. Sci. USA 2015, 112, 13699–13704. [Google Scholar] [CrossRef]
- Schuster, A.; Skinner, M.K.; Yan, W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ. Epigenet. 2016, 2. [Google Scholar] [CrossRef]
- Duempelmann, L.; Skribbe, M.; Bühler, M. Small RNAs in the Transgenerational Inheritance of Epigenetic Information. Trends Genet. 2020, 36, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Lin, H. An epigenetic activation role of Piwi and a Piwi-associated piRNA in Drosophila melanogaster. Nat. Cell Biol. 2007, 450, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L. lncRNAs: Linking RNA to Chromatin. Cold Spring Harb. Perspect. Biol. 2014, 6, a018614. [Google Scholar] [CrossRef] [PubMed]
- Wutz, A.; Gribnau, J. X inactivation Xplained. Curr. Opin. Genet. Dev. 2007, 17, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Morey, C.; Navarro, P.; Debrand, E.; Avner, P.; Rougeulle, C.; Clerc, P. The region 3′ to Xist mediates X chromosome counting and H3 Lys-4 dimethylation within the Xist gene. EMBO J. 2004, 23, 594–604. [Google Scholar] [CrossRef]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef]
- Wurm, A.A.; Pina, C. Long Non-coding RNAs as Functional and Structural Chromatin Modulators in Acute Myeloid Leukemia. Front. Oncol. 2019, 9, 899. [Google Scholar] [CrossRef]
- Hanly, D.J.; Esteller, M.; Berdasco, M. Interplay between long non-coding RNAs and epigenetic machinery: Emerging targets in cancer? Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170074. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2007, 10, 228–236. [Google Scholar] [CrossRef] [PubMed]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, A.K.; Jarosz, D.F. More than Just a Phase: Prions at the Crossroads of Epigenetic Inheritance and Evolutionary Change. J. Mol. Biol. 2018, 430, 4607–4618. [Google Scholar] [CrossRef]
- Prusiner, S.B. Molecular Structure, Biology, and Genetics of Prions. Adv. Virus Res. 1988, 35, 83–136. [Google Scholar] [CrossRef]
- Westaway, D.; Carlson, G.A.; Prusiner, S.B. Unraveling prion diseases through molecular genetics. Trends Neurosci. 1989, 12, 221–227. [Google Scholar] [CrossRef]
- Prusiner, S.B.; McKinley, M.P.; Bowman, K.A.; Bolton, D.C.; Bendheim, P.E.; Groth, D.F.; Glenner, G.G. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983, 35, 349–358. [Google Scholar] [CrossRef]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nat. Cell Biol. 2004, 428, 323–328. [Google Scholar] [CrossRef]
- Safar, J.; Wille, H.; Itri, V.; Groth, D.; Serban, H.; Torchia, M.; Cohen, F.E.; Prusiner, S.B. Eight prion strains have PrP(Sc) molecules with different conformations. Nat. Med. 1998, 4, 1157–1165. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; O Chernoff, Y.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and Variability of [PSI] Prion Factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef]
- Manjrekar, J.; Shah, H. Protein-based inheritance. Semin. Cell Dev. Biol. 2020, 97, 138–155. [Google Scholar] [CrossRef] [PubMed]
- Zajkowski, T.; Lee, M.D.; Mondal, S.S.; Carbajal, A.; Dec, R.; Brennock, P.D.; Piast, R.W.; E Snyder, J.; Bense, N.B.; Dzwolak, W.; et al. The Hunt for Ancient Prions: Archaeal Prion-Like Domains Form Amyloid-Based Epigenetic Elements. Mol. Biol. Evol. 2021, 38, 2088–2103. [Google Scholar] [CrossRef] [PubMed]
- Shlyakhtina, Y.; Moran, K.L.; Portal, M.M. Asymmetric Inheritance of Cell Fate Determinants: Focus on RNA. Non-Coding RNA 2019, 5, 38. [Google Scholar] [CrossRef]
- True, H.L.; Lindquist, S.L. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nat. Cell Biol. 2000, 407, 477–483. [Google Scholar] [CrossRef]
- Oamen, H.P.; Lau, Y.; Caudron, F. Prion-like proteins as epigenetic devices of stress adaptation. Exp. Cell Res. 2020, 396, 112262. [Google Scholar] [CrossRef]
- Liu, B.; Larsson, L.; Franssens, V.; Hao, X.; Hill, S.M.; Andersson, V.; Höglund, D.; Song, J.; Yang, X.; Öling, D.; et al. Segregation of Protein Aggregates Involves Actin and the Polarity Machinery. Cell 2011, 147, 959–961. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.X.; Chen, Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Chen, J.; Xu, H.; Liu, S.; Jiang, Q.-X.; Halfmann, R.; Chen, Z.J. Prion-like Polymerization Underlies Signal Transduction in Antiviral Immune Defense and Inflammasome Activation. Cell 2014, 156, 1207–1222. [Google Scholar] [CrossRef]
- Li, X.; Rayman, J.B.; Kandel, E.R.; Derkatch, I.L. Functional Role of Tia1/Pub1 and Sup35 Prion Domains: Directing Protein Synthesis Machinery to the Tubulin Cytoskeleton. Mol. Cell 2014, 55, 305–318. [Google Scholar] [CrossRef]
- Gilks, N.; Kedersha, N.; Ayodele, M.; Shen, L.; Stoecklin, G.; Dember, L.M.; Anderson, P. Stress Granule Assembly Is Mediated by Prion-like Aggregation of TIA-1. Mol. Biol. Cell 2004, 15, 5383–5398. [Google Scholar] [CrossRef]
- Goldrath, A.W.; Luckey, C.J.; Park, R.; Benoist, C.; Mathis, D. The molecular program induced in T cells undergoing homeostatic proliferation. Proc. Natl. Acad. Sci. USA 2004, 101, 16885–16890. [Google Scholar] [CrossRef]
- Isaacs, J.D.; Jackson, G.S.; Altmann, D.M. The role of the cellular prion protein in the immune system. Clin. Exp. Immunol. 2006, 146, 1–8. [Google Scholar] [CrossRef]
- Li, R.; Liu, D.; Zanusso, G.; Liu, T.; Fayen, J.D.; Huang, J.-H.; Petersen, R.B.; Gambetti, P.; Sy, M.-S. The Expression and Potential Function of Cellular Prion Protein in Human Lymphocytes. Cell. Immunol. 2001, 207, 49–58. [Google Scholar] [CrossRef]
- Zhang, C.C.; Steele, A.D.; Lindquist, S.; Lodish, H.F. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc. Natl. Acad. Sci. USA 2006, 103, 2184–2189. [Google Scholar] [CrossRef]
- Zhang, C.C.; Lodish, H.F. Murine hematopoietic stem cells change their surface phenotype during ex vivo expansion. Blood 2005, 105, 4314–4320. [Google Scholar] [CrossRef]
- Ballerini, C.; Gourdain, P.; Bachy, V.; Blanchard, N.; Levavasseur, E.; Grégoire, S.; Fontes, P.; Aucouturier, P.; Hivroz, C.; Carnaud, C. Functional Implication of Cellular Prion Protein in Antigen-Driven Interactions between T Cells and Dendritic Cells. J. Immunol. 2006, 176, 7254–7262. [Google Scholar] [CrossRef] [PubMed]
- Bakkebø, M.K.; Mouillet-Richard, S.; Espenes, A.; Goldmann, W.; Tatzelt, J.; Tranulis, M.A. The Cellular Prion Protein: A Player in Immunological Quiescence. Front. Immunol. 2015, 6, 450. [Google Scholar] [CrossRef] [PubMed]
- Siberchicot, C.; Gault, N.; Déchamps, N.; Barroca, V.; Aguzzi, A.; Roméo, P.-H.; Radicella, J.P.; Bravard, A.; Bernardino-Sgherri, J. Prion protein deficiency impairs hematopoietic stem cell determination and sensitizes myeloid progenitors to irradiation. Haematology 2019, 105, 1216–1222. [Google Scholar] [CrossRef]
- Reiten, M.R.; Bakkebø, M.K.; Brun-Hansen, H.; Lewandowska-Sabat, A.M.; Olsaker, I.; Tranulis, M.A.; Espenes, A.; Boysen, P. Hematological shift in goat kids naturally devoid of prion protein. Front. Cell Dev. Biol. 2015, 3, 44. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hitchins, M.P.; Wong, J.J.; Suthers, G.; Suter, C.M.; Martin, D.I.; Hawkins, N.J.; Ward, R.L. Inheritance of a Cancer-AssociatedMLH1Germ-Line Epimutation. N. Engl. J. Med. 2007, 356, 697–705. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.L.; Yuen, S.T.; Kong, C.K.; Chan, Y.W.; Chan, A.S.Y.; Ng, W.F.; Tsui, W.Y.; Lo, M.W.S.; Tam, W.Y.; Li, V.S.W.; et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nat. Genet. 2006, 38, 1178–1183. [Google Scholar] [CrossRef]
- Buiting, K.; Groß, S.; Lich, C.; Gillessen-Kaesbach, G.; El-Maarri, O.; Horsthemke, B. Epimutations in Prader-Willi and Angelman Syndromes: A Molecular Study of 136 Patients with an Imprinting Defect. Am. J. Hum. Genet. 2003, 72, 571–577. [Google Scholar] [CrossRef]
- Hansen, K.H.; Bracken, A.P.; Pasini, D.; Dietrich, N.; Gehani, S.S.; Monrad, A.; Rappsilber, J.; Lerdrup, M.; Helin, K. A model for transmission of the H3K27me3 epigenetic mark. Nat. Cell Biol. 2008, 10, 1291–1300. [Google Scholar] [CrossRef]
- Brykczynska, U.; Hisano, M.; Erkek, S.; Ramos, L.; Oakeley, E.J.; Roloff, T.C.; Beisel, C.; Schübeler, D.; Stadler, M.B.; Peters, A.H.F.M. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat. Struct. Mol. Biol. 2010, 17, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, P.; Saadat, A. Neurodegeneration and epigenetics: A review. Neurología 2021. [Google Scholar] [CrossRef]
- Halfmann, R.; Lindquist, S. Epigenetics in the Extreme: Prions and the Inheritance of Environmentally Acquired Traits. Science 2010, 330, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zolotarev, N.; Yang, C.-Y.; Rambold, A.; Mittler, G.; Grosschedl, R. A Prion-like Domain in Transcription Factor EBF1 Promotes Phase Separation and Enables B Cell Programming of Progenitor Chromatin. Immunity 2020, 53, 1151–1167.e6. [Google Scholar] [CrossRef]
- Hirsch, T.Z.; Martin-Lannerée, S.; Reine, F.; Hernandez-Rapp, J.; Herzog, L.; Dron, M.; Privat, N.; Passet, B.; Halliez, S.; Villa-Diaz, A.; et al. Epigenetic Control of the Notch and Eph Signaling Pathways by the Prion Protein: Implications for Prion Diseases. Mol. Neurobiol. 2019, 56, 2159–2173. [Google Scholar] [CrossRef] [PubMed]
- Harvey, Z.H.; Chen, Y.; Jarosz, D.F. Protein-Based Inheritance: Epigenetics beyond the Chromosome. Mol. Cell 2018, 69, 195–202. [Google Scholar] [CrossRef]
- Young, C.S.H.; Cox, B.S. Extrachromosomal elements in a super-suppression system of yeast I. A nuclear gene controlling the inheritance of the extrachromosomal elements. Heredity 1971, 26, 413–422. [Google Scholar] [CrossRef][Green Version]
- Tuite, M.F.; Staniforth, G.L.; Cox, B.S. [PSI(+)] turns 50. Prion 2015, 9, 318–332. [Google Scholar] [CrossRef][Green Version]
- Cox, B.S. Ψ, A cytoplasmic suppressor of super-suppressor in yeast. Heredity 1965, 20, 505–521. [Google Scholar] [CrossRef]
- Tyedmers, J.; Madariaga, M.L.; Lindquist, S. Prion Switching in Response to Environmental Stress. PLoS Biol. 2008, 6, e294. [Google Scholar] [CrossRef] [PubMed]
- Griswold, C.K.; Masel, J. Complex Adaptations Can Drive the Evolution of the Capacitor [PSI+], Even with Realistic Rates of Yeast Sex. PLoS Genet. 2009, 5, e1000517. [Google Scholar] [CrossRef] [PubMed]
- Schwimmer, C.; Masison, D.C. Antagonistic Interactions between Yeast [PSI+] and [URE3] Prions and Curing of [URE3] by Hsp70 Protein Chaperone Ssa1p but Not by Ssa2p. Mol. Cell. Biol. 2002, 22, 3590–3598. [Google Scholar] [CrossRef]
- Chakravarty, A.K.; Smejkal, T.; Itakura, A.; Garcia, D.M.; Jarosz, D.F. A Non-Amyloid Prion Particle that Activates a Heritable Gene Expression Program. SSRN Electron. J. 2019, 77, 251–265.e9. [Google Scholar] [CrossRef]
- Harvey, Z.H.; Futia, R.A.; Jarosz, D.F. A Prion Epigenetic Switch Establishes an Active Chromatin State. SSRN Electron. J. 2019, 180, 928–940.e14. [Google Scholar] [CrossRef]
- Van Cauwenbergh, O.; Di Serafino, A.; Tytgat, J.; Soubry, A. Transgenerational epigenetic effects from male exposure to endocrine-disrupting compounds: A systematic review on research in mammals. Clin. Epigenet. 2020, 12, 1–23. [Google Scholar] [CrossRef]
- Skinner, M.K.; Guerrero-Bosagna, C. Environmental signals and transgenerational epigenetics. Epigenomics 2009, 1, 111–117. [Google Scholar] [CrossRef]
- Gangisetty, O.; Palagani, A.; Sarkar, D.K. Transgenerational inheritance of fetal alcohol exposure adverse effects on immune gene interferon-Upsilon. Clin. Epigenet. 2020, 12, 70. [Google Scholar] [CrossRef]
- Doehner, W.; Praße, L.; Wolpers, J.; Brückner, M.K.; Ueberham, U.; Arendt, T. Transgenerational transmission of an anticholinergic endophenotype with memory dysfunction. Neurobiol. Aging 2017, 51, 19–30. [Google Scholar] [CrossRef]
- Velazquez, R.; Ferreira, E.; Winslow, W.; Dave, N.; Piras, I.S.; Naymik, M.; Huentelman, M.J.; Tran, A.; Caccamo, A.; Oddo, S. Maternal choline supplementation ameliorates Alzheimer’s disease pathology by reducing brain homocysteine levels across multiple generations. Mol. Psychiatry 2020, 25, 2620–2629. [Google Scholar] [CrossRef]
- Takamatsu, Y.; Ho, G.; Waragai, M.; Wada, R.; Sugama, S.; Takenouchi, T.; Masliah, E.; Hashimoto, M. Transgenerational Interaction of Alzheimer’s Disease with Schizophrenia through Amyloid Evolvability. J. Alzheimer’s Dis. 2019, 68, 473–481. [Google Scholar] [CrossRef] [PubMed]
- Gate, D.; Saligrama, N.; Leventhal, O.; Yang, A.C.; Unger, M.S.; Middeldorp, J.; Chen, K.; Lehallier, B.; Channappa, D.; Santos, M.B.D.L.; et al. Clonally expanded CD8 T cells patrol the cerebrospinal fluid in Alzheimer’s disease. Nat. Cell Biol. 2020, 577, 399–404. [Google Scholar] [CrossRef] [PubMed]
- Knudsen, T.M.; Rezwan, F.I.; Jiang, Y.; Karmaus, W.; Svanes, C.; Holloway, J.W. Transgenerational and intergenerational epigenetic inheritance in allergic diseases. J. Allergy Clin. Immunol. 2018, 142, 765–772. [Google Scholar] [CrossRef] [PubMed]
- Weber-Stadlbauer, U. Epigenetic and transgenerational mechanisms in infection-mediated neurodevelopmental disorders. Transl. Psychiatry 2017, 7, e1113. [Google Scholar] [CrossRef] [PubMed]
- Kiss, D.; Ambeskovic, M.; Montina, T.; Metz, G.A.S. Stress transgenerationally programs metabolic pathways linked to altered mental health. Cell. Mol. Life Sci. 2016, 73, 4547–4557. [Google Scholar] [CrossRef]
- Jasiulionis, M.G. Abnormal Epigenetic Regulation of Immune System during Aging. Front. Immunol. 2018, 9, 197. [Google Scholar] [CrossRef]
- Heyn, H.; Li, N.; Ferreira, H.J.; Moran, S.; Pisano, D.G.; Gomez, A.; Diez, J.; Sanchez-Mut, J.V.; Setien, F.; Carmona, F.J.; et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. USA 2012, 109, 10522–10527. [Google Scholar] [CrossRef]
- Dozmorov, M.G.; Coit, P.; Maksimowicz-McKinnon, K.; Sawalha, A.H. Age-associated DNA methylation changes in naive CD4+T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics 2017, 9, 429–445. [Google Scholar] [CrossRef]
- McEwen, L.M.; Morin, A.M.; Edgar, R.D.; MacIsaac, J.L.; Jones, M.J.; Dow, W.H.; Rosero-Bixby, L.; Kobor, M.S.; Rehkopf, D.H. Differential DNA methylation and lymphocyte proportions in a Costa Rican high longevity region. Epigenet. Chromatin 2017, 10, 1–14. [Google Scholar] [CrossRef]
- Chambers, S.M.; A Shaw, C.; Gatza, C.; Fisk, C.J.; A Donehower, L.; A Goodell, M. Aging Hematopoietic Stem Cells Decline in Function and Exhibit Epigenetic Dysregulation. PLoS Biol. 2007, 5, e201. [Google Scholar] [CrossRef]
- Papalexi, E.; Satija, R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018, 18, 35–45. [Google Scholar] [CrossRef]
- Satpathy, A.T.; Saligrama, N.; Buenrostro, J.D.; Wei, Y.; Wu, B.; Rubin, A.J.; Granja, J.M.; Lareau, C.A.; Li, R.; Qi, Y.; et al. Transcript-indexed ATAC-seq for precision immune profiling. Nat. Med. 2018, 24, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Northrup, D.L.; Zhao, K. Application of ChIP-Seq and related techniques to the study of immune function. Immunity 2011, 34, 830–842. [Google Scholar] [CrossRef] [PubMed]
- Maslova, A.; Ramirez, R.N.; Ma, K.; Schmutz, H.; Wang, C.; Fox, C.; Ng, B.; Benoist, C.; Mostafavi, S.; Project, I.G. Deep learning of immune cell differentiation. Proc. Natl. Acad. Sci. USA 2020, 117, 25655–25666. [Google Scholar] [CrossRef] [PubMed]
- Bartosovic, M.; Kabbe, M.; Castelo-Branco, G. Single-cell CUT&Tag profiles histone modifications and transcription factors in complex tissues. Nat. Biotechnol. 2021, 1–11. [Google Scholar] [CrossRef]
- González-Serna, D.; Villanueva-Martin, G.; Acosta-Herrera, M.; Márquez, A.; Martín, J. Approaching Shared Pathophysiology in Immune-Mediated Diseases through Functional Genomics. Genes 2020, 11, 1482. [Google Scholar] [CrossRef]
- Rehman, S.; Aatif, M.; Rafi, Z.; Khan, M.Y.; Shahab, U.; Ahmad, S.; Farhan, M. Effect of non-enzymatic glycosylation in the epigenetics of cancer. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Kizuka, Y. Epigenetic Regulation of and by Glycosylation. In Glycoscience: Biology and Medicine; Endo, T., Seeberger, P.H., Hart, G.W., Wong, C.-H., Taniguchi, N., Eds.; Springer: Tokyo, Japan, 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Sawalha, A.H.; Zhao, M.; Coit, P.; Lu, Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020, 215, 108410. [Google Scholar] [CrossRef]
- Sen, R.; Garbati, M.R.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385. [Google Scholar] [CrossRef]
- Cardenas, A.; Rifas-Shiman, S.L.; Sordillo, J.E.; DeMeo, D.L.; Baccarelli, A.A.; Hivert, M.-F.; Gold, D.R.; Oken, E. DNA methylation architecture of the ACE2 gene in nasal cells of children. Sci. Rep. 2021, 11, 1–9. [Google Scholar] [CrossRef]
- Jit, B.P.; Qazi, S.; Arya, R.; Srivastava, A.; Gupta, N.; Sharma, A. An immune epigenetic insight to COVID-19 infection. Epigenomics 2021, 13, 465–480. [Google Scholar] [CrossRef]
- Li, S.; Wu, B.; Ling, Y.; Guo, M.; Qin, B.; Ren, X.; Wang, C.; Yang, H.; Chen, L.; Liao, Y.; et al. Epigenetic Landscapes of Single-Cell Chromatin Accessibility and Transcriptomic Immune Profiles of T Cells in COVID-19 Patients. Front. Immunol. 2021, 12, 625881. [Google Scholar] [CrossRef] [PubMed]
- Kotlyar, A.M.; Grechukhina, O.; Chen, A.; Popkhadze, S.; Grimshaw, A.; Tal, O.; Taylor, H.S.; Tal, R. Vertical transmission of coronavirus disease 2019: A systematic review and meta-analysis. Am. J. Obstet. Gynecol. 2021, 224, 35–53.e3. [Google Scholar] [CrossRef] [PubMed]
- Roth, O.; Beemelmanns, A.; Barribeau, S.M.; Sadd, B.M. Recent advances in vertebrate and invertebrate transgenerational immunity in the light of ecology and evolution. Heredity 2018, 121, 225–238. [Google Scholar] [CrossRef]
- Feeney, A.; Nilsson, E.; Skinner, M.K. Epigenetics and transgenerational inheritance in domesticated farm animals. J. Anim. Sci. Biotechnol. 2014, 5, 48. [Google Scholar] [CrossRef] [PubMed]
- Berghof, T.V.L.; Parmentier, H.K.; Lammers, A. Transgenerational epigenetic effects on innate immunity in broilers: An underestimated field to be explored? Poult. Sci. 2013, 92, 2904–2913. [Google Scholar] [CrossRef] [PubMed]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 1–11. [Google Scholar] [CrossRef]
- Sánchez, A.L.; Pascual-Pardo, D.; Furci, L.; Roberts, M.R.; Ton, J. Costs and Benefits of Transgenerational Induced Resistance in Arabidopsis. Front. Plant Sci. 2021, 12, 644999. [Google Scholar] [CrossRef] [PubMed]
- Heitmueller, M.; Billion, A.; Dobrindt, U.; Vilcinskas, A.; Mukherjee, K. Epigenetic Mechanisms Regulate Innate Immunity against Uropathogenic and Commensal-Like Escherichia coli in the Surrogate Insect Model Galleria mellonella. Infect. Immun. 2017, 85, e00336-17. [Google Scholar] [CrossRef]
- Norouzitallab, P.; Baruah, K.; Biswas, P.; Vanrompay, D.; Bossier, P. Probing the phenomenon of trained immunity in invertebrates during a transgenerational study, using brine shrimp Artemia as a model system. Sci. Rep. 2016, 6, 21166. [Google Scholar] [CrossRef] [PubMed]
- Norouzitallab, P.; Biswas, P.; Baruah, K.; Bossier, P. Multigenerational immune priming in an invertebrate parthenogenetic Artemia to a pathogenic Vibrio campbellii. Fish Shellfish. Immunol. 2015, 42, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Gegner, J.; Baudach, A.; Mukherjee, K.; Halitschke, R.; Vogel, H.; Vilcinskas, A. Epigenetic Mechanisms Are Involved in Sex-Specific Trans-Generational Immune Priming in the Lepidopteran Model Host Manduca sexta. Front. Physiol. 2019, 10, 137. [Google Scholar] [CrossRef]
- Grossniklaus, U.; Kelly, W.G.; Kelly, B.; Ferguson-Smith, A.C.; Pembrey, M.; Lindquist, S. Transgenerational epigenetic inheritance: How important is it? Nat. Rev. Genet. 2013, 14, 228–235. [Google Scholar] [CrossRef] [PubMed]
- Jablonka, E.; Raz, G. Transgenerational Epigenetic Inheritance: Prevalence, Mechanisms, and Implications for the Study of Heredity and Evolution. Q. Rev. Biol. 2009, 84, 131–176. [Google Scholar] [CrossRef]
- David, C.; Andrea, C.G. Chapter 26—Transgenerational Epigenetics: Current Controversies and Debates. In Transgenerational Epigenetics; Academic Press: Cambridge, MA, USA, 2014; pp. 371–390. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sen, R.; Barnes, C. Do Transgenerational Epigenetic Inheritance and Immune System Development Share Common Epigenetic Processes? J. Dev. Biol. 2021, 9, 20. https://doi.org/10.3390/jdb9020020
Sen R, Barnes C. Do Transgenerational Epigenetic Inheritance and Immune System Development Share Common Epigenetic Processes? Journal of Developmental Biology. 2021; 9(2):20. https://doi.org/10.3390/jdb9020020
Chicago/Turabian StyleSen, Rwik, and Christopher Barnes. 2021. "Do Transgenerational Epigenetic Inheritance and Immune System Development Share Common Epigenetic Processes?" Journal of Developmental Biology 9, no. 2: 20. https://doi.org/10.3390/jdb9020020
APA StyleSen, R., & Barnes, C. (2021). Do Transgenerational Epigenetic Inheritance and Immune System Development Share Common Epigenetic Processes? Journal of Developmental Biology, 9(2), 20. https://doi.org/10.3390/jdb9020020