Abstract
The neural crest is a unique, transient stem cell population that is critical for craniofacial and ocular development. Understanding the genetics underlying the steps of neural crest development is essential for gaining insight into the pathogenesis of congenital eye diseases. The neural crest cells play an under-appreciated key role in patterning the neural epithelial-derived optic cup. These interactions between neural crest cells within the periocular mesenchyme and the optic cup, while not well-studied, are critical for optic cup morphogenesis and ocular fissure closure. As a result, microphthalmia and coloboma are common phenotypes in human disease and animal models in which neural crest cell specification and early migration are disrupted. In addition, neural crest cells directly contribute to numerous ocular structures including the cornea, iris, sclera, ciliary body, trabecular meshwork, and aqueous outflow tracts. Defects in later neural crest cell migration and differentiation cause a constellation of well-recognized ocular anterior segment anomalies such as Axenfeld–Rieger Syndrome and Peters Anomaly. This review will focus on the genetics of the neural crest cells within the context of how these complex processes specifically affect overall ocular development and can lead to congenital eye diseases.
1. Introduction
The neural crest is a migratory stem cell population which contributes to numerous structures in the anterior segment of the eye, including the cornea, iris, sclera, ciliary body, trabecular meshwork, and aqueous outflow tracts [1,2,3,4,5,6]. The importance of this cell population in anterior segment development is highlighted by examples of potentially blinding diseases such as Axenfeld–Rieger syndrome and Peters anomaly (Figure 1A,B) that are due to genetic defects in neural crest cell migration and differentiation [7,8,9,10,11]. In addition, animal models have demonstrated that neural crest cells have critical cell non-autonomous effects. Disruption of signaling in neural crest cells can lead to alterations in neural epithelial-derived optic cup formation resulting in microphthalmia, anophthalmia, and coloboma (Figure 1C,D) [6,12,13,14]. However, specific interactions between neural crest cells and the neural epithelial-derived optic cup remain undefined.
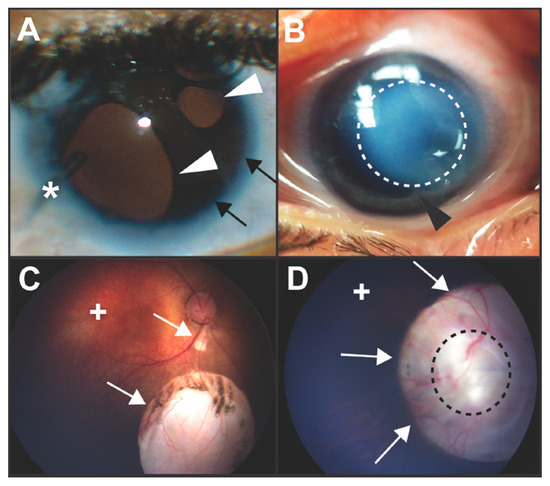
Figure 1.
Clinical images of congenital ocular anomalies. (A) Axenfeld–Rieger syndrome is characterized by Rieger Anomaly (iris hypoplasia resulting in pseudopolycoria and corectopia (white arrowheads)) and Axenfeld Anomaly (anteriorization of Schwalbe’s line of the cornea (posterior embryotoxon, black arrows) with iris adhesions). These defects are due to abnormal migration and differentiation of neural crest cells into the anterior segment of the eye. Over 50% of individuals with Axenfeld–Rieger syndrome develop glaucoma which often requires surgery such as placement of a glaucoma drainage device (asterisk). (B) In Peters anomaly, there is a circumscribed central corneal opacification (outlined by dotted white line) with iris-corneal adhesions (black arrowhead). These anomalies are due to abnormal separation of the lens vesicle from the surface ectoderm resulting in absence of Descemet’s membrane and disruption of neural crest cell migration into the anterior segment. (C,D) Colobomas are due to incomplete closure of the ocular fissure and can affect the iris, zonules, retina, choroid, and optic nerve. Chorioretinal colobomas are inferior to the optic nerve and are characterized by an area that is devoid of retina and choroid (C, white arrows). In these types of coloboma, the macula (+) which accounts for central vision, is typically not affected. Optic nerve colobomas (D, white arrows) can cause severe vision loss especially if the entire optic nerve (outlined by black dotted line) is involved. Although the macula (+) may not be affected, the loss of the ganglion cell axons that comprise the optic nerve limits vision.
Neural crest development is a complex process, which is characterized by several landmark events [15,16,17]. Immediately after gastrulation, the ectodermal germ layer is subdivided and specified through different extracellular signaling systems into two territories: the non-neural ectoderm which gives rise to the epidermis and the neural ectoderm, which forms the central nervous system [17,18,19,20]. These territories are separated by the neural plate border, from which neural crest cells are specified during neural tube closure (Figure 2) [17,20,21]. Unique among stem cell populations, neural crest cells then undergo epithelial-to-mesenchymal transition (EMT), delaminate from the neural tube (Figure 3), and migrate to different regions of the embryo to give rise to a broad range of tissues and cells (Figure 4 and Figure 5) [2,22,23]. Each of these successive processes, neural plate border induction, neural crest specification, neural crest migration, and neural crest differentiation is controlled by overlapping gene regulatory networks [15,17,24].
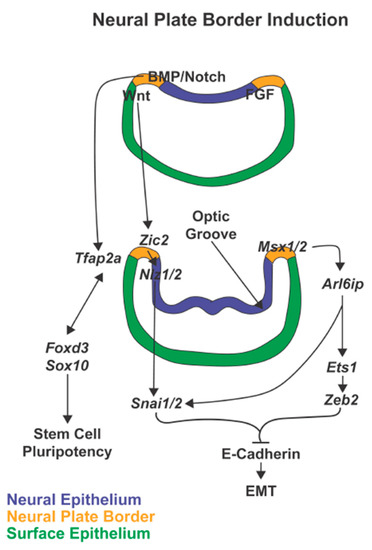
Figure 2.
Neural plate border induction. In neural plate border induction, FGF followed by Notch and BMP signaling are expressed in the neural ectoderm while Wnt is expressed in the non-neural ectoderm. In the next phases of neurulation and optic groove formation, these signals then induce expression of the neural plate border specifiers, Msx1/2, Zic2, and Tfap2. These transcription factors, expressed within the neural plate border, trigger signaling cascades that maintain stem cell pluripotency and prepare the premigratory neural crest cells for epithelial-mesenchymal transition (EMT).
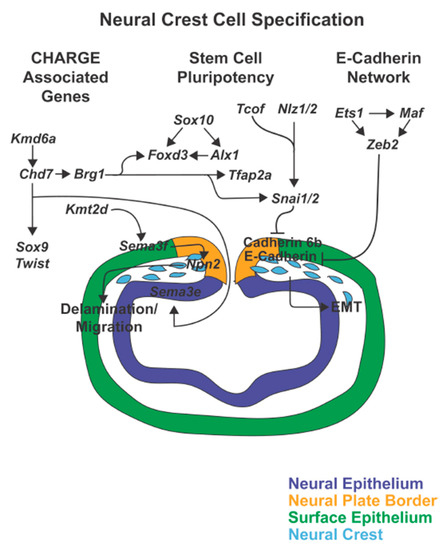
Figure 3.
Neural crest cell specification. As the neural tube closes, genes including Sox10, Foxd3, Alx1, Tfap2a, Tcof, Nlz1/2, and Snai1/2 induce neural crest cell identity and maintain stem cell pluripotency. Simultaneously, Tcof, Nlz1/2, and Sna1/2, together with Ets1, Maf, and Zeb2 inhibit E-Cadherin and Cadherin 6b within the premigratory neural crest to trigger epithelial-mesenchymal transition (EMT). Furthermore, the CHARGE associated genes, Chd7, Kmd6a, Brg1, and Kmt2d regulate neural crest cell delamination and initiation of migration.
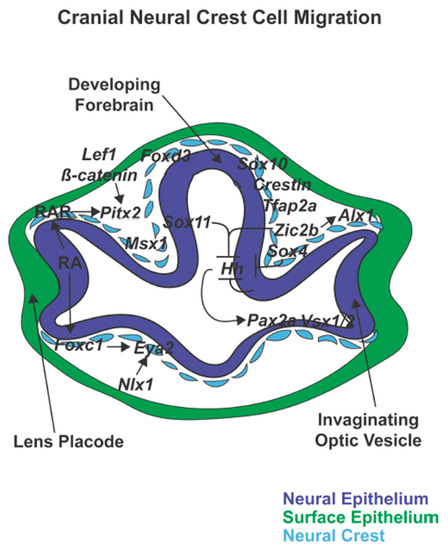
Figure 4.
Cranial neural crest cell migration. Following closure of the neural tube, cranial neural crest cells expressing foxd3, sox10, and crestin migrate between the neural epithelial-derived forebrain and optic vesicle and the surface epithelium. Sox4 within neural crest cells together with Sox11 and Zic2 regulate hh-patterning of the dorsal-ventral retina and subsequent expression of Pax2a and Vsx1/2. In addition, retinoic acid (RA) regulation of Pitx2 and Foxc1 within periocular neural crest cells is important for optic cup and anterior segment development.
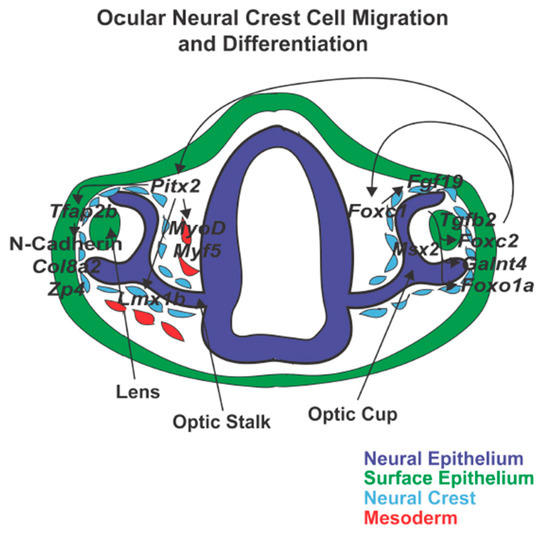
Figure 5.
Ocular neural crest cell migration and differentiation. Neural crest cells surround the developing eye and enter into the ocular anterior segment between the surface epithelium and optic cup and through the ocular fissure. Pitx2 is important for corneal and iridocorneal angle development and function through the regulation of Tfap2b and Lmx1b. In addition, Pitx2 targets mesodermal cells and induces expression of MyoD and Myf5 which triggers extraocular muscle formation. Foxc1 also regulates anterior segment development by targeting Fgf19, Foxc2, GaInt4, and Foxo1a.
Simultaneous with neural tube closure, the optic vesicle extends from the developing forebrain and then invaginates to form a bi-layered cup (Figure 4) [25,26,27]. Neural crest cells, primarily derived from the prosencephalon and mesencephalon, migrate into the periocular mesenchyme that surrounds and supports the optic cup [10,11,28]. Concurrently, the distal edge of the optic cup interacts with the overlying surface ectoderm to induce lens placode formation followed by separation of the lens vesicle [26,29]. Subsequently, neural crest cells gain entry into the eye via two pathways, 1) the ocular fissure on the ventral edge of the cup and 2) between the optic cup and overlying surface ectoderm to give rise to blood vessels and anterior segment structures [6,11,30].
Although neural crest cells sequentially contribute to the corneal endothelium, corneal stroma, iris, ciliary body, and the aqueous outflow system, the pattern by which these cells migrate into the eye differs between species. The neural crest cells migrate in three waves in humans, in two waves in mice, and in a continuous pattern in chicks [1,4,31]. In zebrafish, neural crest cells migrate in at least three waves. Recent studies have shown that there are two distinct sox10-positive cell populations that migrate into the eye 12–18 h after neural crest delamination and a subsequent foxd3-positive cell population that migrates into the anterior segment in the late embryonic and early larval stages [11,32]. Following migration into the anterior segment, signals that regulate terminal neural crest cell differentiation are less well-understood and additional work is needed to identify these gene regulatory networks [33,34].
As a stem cell population, the neural crest requires the ability to detect a variety of signals during specific time frames in order to differentiate into a diverse set of tissues and cells. For example, in zebrafish embryos, neural crest cells in the periocular mesenchyme display both overlapping and individualized expression patterns of foxc1a, foxc1b, eya2, foxd3, pitx2, sox10, lmx1b.1, and lmx1b.2 that correspond with the originating neural crest cell migration streams. This reflects a lack of uniformity within the neural crest cell population in the periocular mesenchyme. However, it is a matter of debate whether the neural crest cell population as a whole is homogeneous or a heterogeneous mixture of cells specified toward particular fates [35,36,37,38]. While neural crest development is a complex process involving numerous steps, here we focus on the genetics of this stem cell population in the context of ocular development.
2. Overview of Neural Crest Signaling Modules
Neural crest development is divided into successive processes which are regulated by overlapping gene regulatory networks that together form signaling modules [15,17,24,39]. While this review will focus on the genes that have been shown to specifically affect eye development, a brief overview of these signaling modules is required.
Early neural plate border induction is driven by Fgf with the activation of pro-neural genes and is then perpetuated by Notch and Bmp signaling in the neural ectoderm and Wnt in the non-neural ectoderm. Together these signals lead to the induction of neural plate border specifiers (Figure 2) [17,19,20,40,41,42,43]. These transcription factors are initially expressed during early gastrulation in the neural plate border and are critical for neural crest specification. Within the pre-migratory neural crest cells, the expression of the neural plate border specifiers Msx1, Zic2, and Tfap2, is maintained along with activation of additional genes required for proper EMT and subsequent migration of neural crest cells [17,44,45,46]. Those genes can be divided into different groups: 1) CHARGE syndrome-associated genes, 2) genes regulating E-Cadherin in neural crest cell EMT, and 3) genes involved in maintaining embryonic stem cell pluripotency and cell lineage-specificity (Figure 3) [46,47,48,49,50]. The CHARGE syndrome-associated genes regulate cell delamination from the edge of the neural tube via epigenetic modifications. The genes involved in the regulation of E-Cadherin signal the switch from E-Cadherin to N-Cadherin expression, an essential step for preparing cells for migration [47,51,52]. During these processes, however, the cells must maintain their stem cell pluripotency as well as the ability to respond properly to external signals to differentiate into specific cell lineages [46,53]. Once migratory, additional transcription factors direct neural crest cell migration from the edge of the neural tube to destinations throughout the body (Figure 4) [28,54]. Differentiation signals at the end locations, such as the ocular anterior segment, then regulate the formation of the diverse set of structures and tissues (Figure 5) [46].
Common amongst these signaling modules is the repetition of genes in different steps in neural crest development. Furthermore, many of these genes are also expressed in other neural ectodermal-derived tissues. The redundancy of pathways shows the fluidity of these processes but also complicates the determination of critical functions of these genes especially within the context of end-organ development. In the following sections, genes important for both neural crest cell and eye development will be discussed.
3. Neural Plate Border Genes
3.1. Msx Gene Family
The Msx genes encode transcription factors that share homology with the muscle segment homeobox (msh) genes in drosophila. As a gene family, they are expressed in diverse tissues throughout embryogenesis, although are best characterized for their role in cranial neural crest cells and thereby craniofacial development [55,56]. The three identified members in the family, Msx1, Msx2, and Msx3, show a functional redundancy where individual gene disruption leads to modest defects while multiple Msx gene disruption causes severe abnormalities in neural plate border and neural crest specification [57,58,59,60,61]. Msx1 and Msx2 have a continued requirement during specification of the pre-migratory neural crest cells by activating Arl6ip and further downstream targets Snai1/2, Foxd3, Ets1, and Zeb2 (Figure 2) [58]. This pathway leads to the inhibition of E-Cadherin expression, which is essential in triggering EMT [62]. In addition to these early roles, Msx1 is then expressed in the periocular mesenchyme of mice and chick, while Msx2 is found later in development in surface-ectoderm-derived corneal epithelium and lens and neuro-epithelial-derived retina [63,64]. However, knockdown of Msx1 and Msx2 results in misshapen and enlarged optic cups in mice, but microphthalmia in zebrafish [65]. This phenotypic variation could be due to species differences in the role of Msx genes in the forebrain and neural crest cell development. Further studies are required to investigate the detailed function of Msx1 and Msx2 during optic cup development and the identification of specific targets of Msx genes within the optic vesicle. Abnormal optic cup morphology and colobomas are commonly observed in mice and zebrafish in which expression of genes important in neural crest specification has been genetically disrupted [6,12,13,14]. The mechanisms underlying this interaction and downstream targets within the optic vesicle are further discussed in this review in the context of genes with this shared phenotype.
3.2. Zic Gene Family
Members of the Zic (zinc finger of the cerebellum) gene family are important regulators of neuroectodermal and neural crest development. While there are five homologs in chick, mice, and humans and seven in zebrafish, Zic1, Zic2 and Zic3 are the most well-studied [66,67,68]. Zic1 and Zic3 work together in neural ectoderm induction which is required for both neural and neural crest cell fates [69,70,71]. However, their primary role, as evidenced by human pathology and animal knockdown and knockout models, is within forebrain development rather than in neural crest-derived tissues [72,73,74,75]. In contrast to Zic1 and Zic3, Zic2 is a direct target of Wnt-signaling within the anterior neural plate by mid-gastrulation and initiates neural plate border specification [76,77]. In mice, this Zic2-regulated step is required for the generation of the appropriate number of neural crest cells [78,79]. Subsequently, in mice and zebrafish, Zic2 inhibits E-Cadherin via Nlz1/2 and Snai1/2 in order to trigger EMT and neural tube exit (Figure 2).
In zebrafish, the expression of zic2b persists within neural crest cells while zic2a is expressed within a restricted domain in the distal optic stalk [76,77,79,80]. Although the combined zic2a and zic2b knockout resulted in coloboma with periocular hemorrhage and edema, the main driver of this phenotype was the loss of zic2b [79]. Loss of Zic2b results in decreased alx1 expression and fewer neural crest cells within the periocular mesenchyme, including the crestin-negative neural crest cells in the area adjacent to the ocular fissure [79]. In addition, zic2b restricts hedgehog (hh) signaling in the neural epithelial-derived pre-optic diencephalon, optic stalk, and ventral retina (Figure 4) [79]. Similarly in zebrafish, sox4 and sox11 act by limiting hh expression within the ventral forebrain adjacent to the optic vesicle. Furthermore, knockdown of Sox4 or Sox11 results in colobomas in zebrafish [81,82]. While both of these SoxC transcription factors are expressed in the forebrain, sox4 is also expressed in neural crest cells in the periocular mesenchyme and together with zic2b may have a cell non-autonomous effects on optic cup development [82]. Down-regulation of zic2b, sox4, or sox11 causes mis-localization of pax2a, vsx1, and vsx2, which are necessary for dorsal and ventral patterning of the retina and subsequent ocular fissure closure [79,81,82]. Although later in development during retinal ganglion cell differentiation, zic2 antagonizes the expression of sox4 and sox11 in patterning axonal projections, the interplay between zic2 and sox4 in the neural crest has not been determined [83].
3.3. TFAP2 Genes
The Tfap2 genes encode for the transcription factor AP-2 family members of proteins, which are expressed in both non-neural ectoderm and neural crest cells during embryogenesis [84,85]. The three genes, Tfap2a, Tfap2b, and Tfap2c, show overlapping, but not identical expression within Xenopus and mouse neural crest cells, while only tfap2a and tfap2c are expressed in zebrafish neural crest cells demonstrating species differences in gene function [84,85,86,87,88]. Of the three genes, Tfap2a is most critical for neural crest cell and craniofacial development. This is highlighted by the human Branchio-Oculo-Facial Syndrome, which is associated with TFAP2A mutations and is characterized by craniofacial (cleft lip, cleft palate, ear malformations) and ocular anomalies (anophthalmia, microphthalmia, cataract, and coloboma) [89,90]. Within the neural plate border, Bmp signaling induces the expression of Tfap2a and Tfap2c [91]. While Tfap2a is a key regulator of Foxd3 in neural crest specification, Tfap2c expression is short-lived and redundant with Tfap2a [17,85]. In the pre-migratory neural crest cells, Tfap2a expression is independent of Foxd3. However, in early migratory cells, Tfap2a expression becomes synergistic with and dependent on Foxd3 and Sox10 [85,92,93] (Figure 2). Later during migration, Tfap2a and Foxd3 expression segregates into distinct populations accounting for differences in expression of further downstream neural crest markers and mutant phenotypes [92,93]. In post-migratory human, mouse, and zebrafish neural crest cells, Tfap2a is expressed in the nasal process, palate, and tooth buds [94]. Similar to Zic2b, Tfap2a is also expressed in the periocular mesenchyme that surrounds the early optic vesicle in mouse and zebrafish (Figure 4) [94,95]. While Tfap2a is expressed in the zebrafish and mouse corneal and lens epithelium and retina, this is after optic cup morphogenesis including ocular fissure closure. Like humans, Tfap2a zebrafish and mouse mutants display microphthalmia and coloboma, indicating that neural crest-specific expression has cell non-autonomous effects on the optic cup [94,95]. However, it is unclear whether Tfap2a functions through the same hedgehog mediated pathway as Zic2.
4. CHARGE Syndrome Associated Genes
The CHARGE syndrome-associated genes regulate epigenetic mechanisms to govern the correct delamination of pre-migratory neural crest cells from the edge of the neural tube [96]. Chd7, which is central to this signaling cascade, is a helicase DNA-binding protein. In humans, autosomal dominant mutations of CHD7 are associated with CHARGE syndrome, a constellation of congenital abnormalities including Coloboma, Heart defects, choanal Atresia, Retarded growth and development, Genital hypoplasia, and Ear anomalies [97,98,99]. In mice and Xenopus, Chd7 is expressed in pre-migratory and migratory neural crest cells and is sensitive to gene dosage. Mice heterozygous for Chd7 mutation or Xenopus in which the protein was knocked down with morpholino oligonucleotides showed disorganization and reduction of neural crest cells [48,100]. In neural crest cells, Chd7 activates Sox9, Twist, and Snai1/2 to further direct migration (Figure 3). Additionally, Chd7 works with the BAF (SWI/SNF) complex, which is responsible for chromatin remodeling. Within the BAF complex, Brg1 (Smarca4a) encodes a core ATPase, and knockout of this gene down-regulates Tfap2a, Foxd3, and Snai1/2 expression [100]. In addition, in zebrafish, chd7 regulates sema3e, which is expressed in the hindbrain, and over expression of this sema3e rescues the chd7 knockout phenotype [101]. Consistent with this, mutations in SEMA3E have been identified in patients with CHARGE syndrome and knockout of sema3e in zebrafish phenocopies chd7 knockout phenotype.
Colobomas are a prominent feature of CHARGE syndrome, however, it is unclear whether this eye defect is due to neural ectoderm-derived optic cup malformations and/or neural crest defects [102,103]. In addition to its role in the neural crest cells, Chd7 expression in the neural ectoderm is required for optic cup and stalk development [103]. Hence, additional studies utilizing tissue-specific conditional knockouts are required to investigate the role of Chd7 in the neural ectoderm and neural crest cells in eye development.
Furthermore, modifications of methylation status by the H3 lysine 4 (H3K4) methylase (encoded by the Kmt2d gene) and the X-linked histone H3 lysine 27 (H3K27) demethylase (encoded by the Kmd6a/Utx gene) are also important in regulating neural crest cell delamination and subsequent migration [104,105,106]. Mutations in KMT2D or KMD6A are associated with Kabuki Syndrome, which shares characteristics to CHARGE syndrome including microphthalmia and coloboma [107,108,109]. In Xenopus, Kmt2d loss of function mutations recapitulate Kabuki syndrome and demonstrate that this methylase is required for dispersion of the migrating neural crest cells [105]. Furthermore, knockdown of Kmt2d in neural crest cells leads to decreased sema3F expression in the branchial arches such that over expression of sema3F rescues the knockdown. However, this effect is likely indirect as sema3F is expressed in adjacent non-neural crest cells and acts as the secreted ligand for the Neuropilin2 (Npn2) receptor, which is expressed in the neural crest (Figure 3). [105,110,111]. Similarly, neural crest-specific conditional knockout of Kmd6a in mice phenocopies Kabuki syndrome. However, only a small set of targets within those cells are related to histone demethylation [104]. Most prominently within neural crest cells, Kmd6a regulates Chd7 expression and is a target of Notch signaling, the Wnt/β-Catenin pathway, and p53-based targets [112]. While both Notch and Wnt/β-Catenin have defined roles in early neural plate border and subsequent neural crest specification, there may be cross regulation between p53 and Kmd6a as variant p53 alleles modify the phenotypic severity of Kmd6a mutations [112]. Unlike Chd7 which has roles in both the neural ectoderm and neural crest cells, Kmt2d and Kmd6a both appear to have neural crest cell-specific functions. Thus, the eye phenotype seen in Kabuki syndrome is likely due to cell non-autonomous effects of the neural crest cells on the optic cup, however, the specific mechanism downstream of these genes that underlie this phenotype has yet to be determined.
4.1. E-Cadherin in Neural Crest Cell EMT
Initiation of EMT is a key step in the specification of pre-migratory neural crest cells [23]. The separation of cell-cell interactions and specifically the deconstruction of the tight and adherens junctions between epithelial cells triggers these morphological changes. With dissolution of the adherens junction, E-Cadherin protein is cleaved from the plasma membrane and degraded while E-Cadherin mRNA transcription is down-regulated. This state releases β-Catenin from the plasma membrane allowing it to accumulate in the nucleus and activate transcription in response to Wnt signaling [47,113].
The transcription factor, Zeb2 (Zinc Finger E-Box Binding Homeobox 2) is an important inhibitor of E-Cadherin in the initiation of EMT in neural crest cells. Expression of Zeb2 may be regulated by Msx1 via Arl6ip1 and Ets1 [114,115]. Arl6ip1 encodes ADP ribosylation factor-like 6 interacting protein, which interacts with Arl6 to regulate intracellular trafficking [116]. Interestingly, human mutations in ARL6 are associated with Bardet–Biedel syndrome, a disease characterized by retinal degeneration, but not neural crest defects [117,118]. However, in zebrafish, knockdown of Arl6ip1 reduces expression of foxd3, snai1b, sox10, and crestin and inhibits specification of neural crest sublineages [119]. Ets1 encodes an E26 transformation-specific transcription factor that targets Zeb2 in the pre-migratory neural crest cells (Figure 3) [120]. Ets1 in non-neural crest cells has also been shown to interact with Maf, a basic leucine zipper transcription factor, in which human mutations cause Aymé-Gripp Syndrome [121,122]. This syndrome is characterized by craniofacial abnormalities, sensorineural hearing loss, short stature, developmental delay, and eye anomalies including congenital cataracts and colobomas [122,123]. In mice, Mafb is expressed in the neural crest cells and can bind to the Sox9 promotor [124]. Thus, Maf is also likely active within neural crest cells and may act together with Ets1 to regulate Zeb2 during neural crest specification.
However, Zeb2 is expressed in both the neural tube and pre-migratory neural crest cells. In chick, knockdown of Zeb2 causes prolonged maintenance of E-Cadherin in migratory neural crest cells. This disrupts delamination resulting in aggregation of adherent neural crest cells along the neural tube edge [17,125]. Similarly in mice and zebrafish, knockout of Zeb2 causes neural crest cell migration defects and a phenotype similar to the corresponding ZEB2 haplosufficient human Mowat–Wilson syndrome [126]. This congenital disease is characterized by neural crest abnormalities (craniofacial and heart anomalies and Hirschsprung disease) as well as neural defects (epilepsy, genesis of the corpus callosum, and developmental delay) [127,128]. While eye abnormalities including coloboma are also present, it is not clear whether these are due to defects in neural and/or neural crest cell development.
Snai1/2 encode zinc finger transcription factors that inhibit cadherin expression and trigger EMT. Within the pre-migratory neural crest cells, Snai1 expression is induced by Wnt signaling and Zic1/2, along with Foxd3, Sox10, and Ets1 is an early specifier of neural crest cells [45,119,129,130,131]. Wnt signaling regulates Snai1 expression via the Tcof1 (Treacle ribosome biogenesis factor 1) gene which is expressed within the pre-migratory and migratory neural crest cells and encodes the Treacle protein (Figure 3) [131]. The Treacle protein resides within the nucleolus and is involved in ribosomal DNA gene transcription. Autosomal dominant mutations in TCOF1 result in Treacher Collins Syndrome, which is associated with severe craniofacial abnormalities (micrognathia, microtia, midface hypoplasia, zygomatic hypoplasia) and ocular anomalies including coloboma [132,133]. In mice, Tcof1 is expressed in the frontonasal process and the maxillary and mandibular mesenchyme, and haploinsufficency causes fewer migrating neural crest cells and recapitulation of the human disease [134]. In addition to Wnt signaling, Snai1/2 is also regulated by Zic1/2 through the Nlz1/2 genes, which encode for the NET family of zinc finger transcription repressors [130]. Nlz1 is expressed on the neural ectoderm side of the neural plate border and appears to demarcate the neural crest cell delamination line [135,136]. Through this action, Nlz1 indirectly signals via Snai1/2 the pre-migratory neural crest cells which should undergo EMT [135].
Snai1 represses E-cadherin within the pre-migratory cells and continues to be expressed in the migratory cells, including in the periocular mesenchyme [137]. Snai2 is induced by Wnt-signaling following neural plate border specification and represses Cadherin 6b in the pre-migratory neural crest cells along the dorsal neural tube (Figure 3) [138]. Cadherin 6b, like E-Cadherin, needs to be down regulated in order to initiate temporal and spatial emigration of the neural crest cells from the edge of the neural tube [139]. Snai1 mutants display colobomas, and the specific effects of this gene in regulating EMT suggests that this action in the neural crest cells indirectly affects ocular fissure closure [140]. In contrast, Snai2 mutants do not appear to have eye abnormalities [141]. This may be related to either the lack of Snai2 expression within the periocular mesenchyme or compensation of function by Snai1. Additional studies are required to further understand the role of the Snai genes specifically on ocular development.
4.2. Maintaining Embryonic Stem Cell Pluripotency and Cell Lineage-Specificity
Despite acquiring mesenchymal properties and ultimately migratory capacity, the neural crest cells need to remain in a pluripotent state, as well as to be governed towards a specific cell fate. While neural plate border specifiers show continued expression, additional factors, including Sox10, Foxd3, Snai1/2, and Ets1 are activated at this stage to maintain the stem cell qualities of the pre-migratory neural crest cells [53,142]. However, during migration and post-migration, these transcription factors guide the segregation of sublineages, and eventually the loss of expression triggers terminal differentiation.
Sox10 encodes for a SRY-related HMG box transcription factor that shuttles proteins between the cytoplasm and nucleus in order to regulate cell fate [143,144]. During migration, Sox10 is one of the earliest specific markers for undifferentiated neural crest cells, however, in post-migratory cells, Sox10 directs cells toward melanocyte and glial differentiation [143,144,145,146]. In the zebrafish and mouse craniofacial region, Sox10 is expressed within the pharyngeal arches and in the periocular mesenchyme [145,147,148,149,150]. In zebrafish, sox10 continues to be expressed within the mandibular and maxillary cartilage during the early larval stage but is absent in juveniles [11]. While sox10 positive cells enter into the developing eye, its expression is short-lived and it is unclear to which ocular tissues these neural crest cells contribute [11,32]. In humans, autosomal dominant nonsense and frame shift mutations in SOX10, are associated with Waardenburg syndrome, which is primarily characterized by pigmentation abnormalities and mild facial dysmorphism [151,152]. Similarly, the colorless zebrafish mutants, which were generated through an ENU mutagenesis screen, have various point mutations in the sox10 allele resulting in pigmentation abnormalities, However, the mutant fish show normal craniofacial and ocular development and survive to adulthood [147]. In contrast, morpholino oligonucleotide knockdown of Sox10 in zebrafish disrupts early neural crest specification resulting in significantly fewer migrating cells. Severely affected animals have severe craniofacial abnormalities, the eyes are microphthalmic with colobomas and do not survive past hatching (unpublished data, personal observation). Furthermore, homozygous Sox10 mutations in mice are lethal in embryonic or early perinatal stages [153,154]. These phenotypic variations may be due to the effects of gene dosage and the amount of residual activity in the heterozygous versus homozygous states. Additional studies are required to further investigate these discrepancies and the role of Sox10 in neural crest cells and optic vesicle development.
Foxd3 encodes a forkhead winged-helix transcription factor, which is involved in changing stem cells from a naive state to a primed pluripotent state. Foxd3 works by modifying chromatin structure to either decrease recruitment or repress activation of enhancers [38]. Like Sox10, Foxd3 is an early marker for neural crest cells but directs post-migratory differentiation towards neuronal, bone, and cartilage fates [155,156]. Foxd3 is also expressed within the pharyngeal arches and periocular mesenchyme [30,92]. In zebrafish, foxd3-positive cells enter into the developing eye later than the smaller population of sox10-positive cells. During the embryonic into early larval stages, foxd3-positive cells populate the iris and cornea, however, expression is down-regulated by mid-larval stage [11]. A variant allele of human FOXD3 has been associated with pigmentation abnormalities (vitiligo). However, this mutation, which is located within the promoter region, has been shown to increase transcriptional activity in vitro and may also be involved in autoimmune processes such as Hashimoto’s thyroiditis [157]. To date, no haploinsufficient mutations of FOXD3 have been identified in human diseases. This correlates with the animal Foxd3 knockout and knockdown models, which show severe neural crest defects especially involving the peripheral nervous system and cartilages that are incompatible with survival. In zebrafish, morpholino oligonucleotide Foxd3 knockdown causes microphthalmia with impaired neural crest cell migration into the eye [158,159,160,161]. Interestingly, one clinical study showed that FOXD3 variants of unknown pathogenic significance were identified in a handful of patients with aniridia and Peters Anomaly [162]. Nonetheless, further investigation is required to determine the role of Foxd3 in eye development and specifically in the regulation of neural crest cell interactions with the optic cup.
The Alx family of genes encodes for paired-class homeobox transcription factors, and all three members, Alx1, Alx3, and Alx4 play various roles in neural crest development [163]. Alx1 coregulates Sox10 and Foxd3 during early neural crest cell specification and continues to be expressed throughout migration. In addition, Alx1 is expressed in the periocular mesenchyme [164]. In humans, mutations of ALX1 result in frontonasal dysplasia and eye abnormalities including microphthalmia and anophthalmia [165,166]. Correspondingly, knockout of alx1 in zebrafish also causes microphthalmia and coloboma. In contrast, alx3 and alx4 play later roles in post-migratory neural crest cells and are only expressed in limited areas of the periocular mesenchyme and eye [164]. Subsequently, human mutations in either ALX3 or ALX4 cause mild frononasal dysplasia, but no ocular abnormalities [167,168]. Thus, early disruption of neural crest cell specification and maintenance of pluripotency by Alx1 is required for normal optic cup development and ocular fissure closure. This effect of Alx1, is likely due to cell non-autonomous effects on the optic cup mediated through neural crest cells. However, downstream targets and molecular events need to be further defined.
5. Cranial Neural Crest Cell Migration Gene Regulation
After delamination from the edge of the neural tube, neural crest cells follow distinct pathways throughout the body. The cranial neural crest cells originate between the diencephalon and the rhombencephalon to eventually contribute to much of the craniofacial skeleton (frontal, nasal, mandible, maxillary, zygomatic), and numerous eye structures (corneal stroma, corneal endothelium, trabecular meshwork, sclera, iris stroma, uveal melanocytes, ciliary body muscle) [6,34,169,170,171]. Cranial neural crest cells which originate from the diencephalon and anterior mesencephalon migrate in streams dorsal and ventral to the eye to populate the periocular mesenchyme and frontonasal process [11]. Cells from the posterior mesencephalon and rhombencephalon successively migrate into the pharyngeal arches [11,172]. Within these streams of cranial neural crest cells, molecular markers are already distinguishing into subpopulations. Crestin, a well-defined marker for migratory neural crest cells is expressed in the dorsal, but not ventral periocular mesenchyme [173]. In zebrafish, while there is initially coexpression of sox10 and foxd3 during early migration into the pharyngeal arches and periocular mesenchyme, the timing of the down regulation of these transcription factors varies depending on the triggering of terminal differentiation [11,174]. During this process, the expression of additional transcription factors including pitx2, foxc1, eya2, and lmx1b is upregulated in cranial neural crest cells in order to regulate migration to final destinations and differentiation (Figure 4) [174].
Pitx2 encodes for a paired-like homeodomain transcription factor first identified within the pituitary as a regulator of prolactin. However, extensive animal studies have shown Pitx2 to be critical for establishing left-right axis and subsequent asymmetrical development of the heart, lungs, and gastrointestinal system [175,176,177]. Thus, complete knockout of Pitx2 in mice is embryonic lethal due to severe cardiac abnormalities. Nevertheless, these mutants displayed severe eye abnormalities including microphthalmia and coloboma [175,176]. Tissue specific-knockouts demonstrated that these ocular defects are due to cell non-autonomous effects of Pitx2 within neural crest cells on optic cup development [178]. In humans, autosomal dominant mutations in PITX2 are most commonly associated with Axenfeld–Rieger Syndrome, which consists of a distinct set of systemic, craniofacial, and ocular anomalies that stem from disruption of neural crest cell migration and differentiation. Systemic abnormalities include Hirschsprungs disease (absence of enteric innervation of segments of intestine) and cardiac outflow tract anomalies while mid-face hypoplasia with oligo/microdontia are the most common craniofacial findings. In addition, extraocular muscle and eyelid abnormalities have been associated with Axenfeld–Rieger Syndrome [179,180]. Within the eye, there are bridging strands that connect the iris to an anteriorized Schwalbe’s line, which demarcates the outer edge of the corneal endothelium. In addition, the iris is hypoplastic resulting in corectopia (irregular pupil) and pseudopolycoria (multiple pupils) [181,182,183,184]. Interestingly, in a smaller subset of patients, optic nerve abnormalities including coloboma and morning glory appearance are present (unpublished data, personal observation). The greatest source of morbidity is vision loss due to glaucoma that affects approximately 50–75% of patients and is often refractory to medical treatment [185]. Less commonly, PITX2 mutations are associated with Peters Anomaly, which is characterized by a central corneal opacity that is due to incomplete separation of the lens vesicle from the corneal epithelium and failure of neural crest cells to migrate into the anterior segment [186].
Pitx2 is first expressed in neural crest cells after migration has been initiated and together with the morphogen, retinoic acid (RA), directs the migrating neural crest cells along different streams into the pharyngeal arches and periocular mesenchyme [176,187]. Knockdown of Pitx2 in zebrafish did not alter the number of delaminating neural crest cells but disrupted migratory pathways into the craniofacial region resulting in apoptosis of these cells [10]. This resulted in mandibular and maxillary malformations as well as ocular anterior segment dysgenesis, coloboma, and microphthalmia [10,176]. Pitx2 knockout mice and mice in which RA fails to promote Pitx2 expression (i.e., knockout of multiple RA receptors), show a more severe eye phenotype. This includes anophthalmia, microphthalmia, and defects of the optic nerve such that the eyes attach directly to the ventral hypothalamus [175,178,188]. Conditional knockout of Pitx2 within the neural crest shows a similar phenotype, indicating that these ocular defects are due to cell non-autonomous function of Pitx2 in neural crest cells [178]. Pitx2 expression in the periocular mesenchyme activates signals that cause the morphogenetic extension of the optic stalk such that failure of this step results in a foreshortened optic stalk and microphthalmia/anophthalmia. Subsequently, Pitx2 expression is predominantly maintained within the periocular mesenchyme where it is required for corneal differentiation [178]. In mice, knockout of Pitx2 may cause incompetence of the ocular neural crest cells to form corneal endothelium and stroma resulting in malformed and thickened corneas. [176,178,189]. Importantly, Pitx2 integrates input from RA and Wnt signaling to trigger ocular neural crest cell migration and differentiation (Figure 4) [176,187]. RA, which is produced by the dorsal and ventral retina, targets the periocular mesenchyme via RA receptors (RAR) α and γ [34,176,190,191,192]. Within these periocular neural crest cells, RA directly upregulates the expression of Pitx2 through binding to the RA response elements. Furthermore, while the Wnt pathway does not initiate activation, the signaling components, β-catenin, and Lef1, bind to the Pitx2 promotor and are subsequently required for maintaining Pitx2 expression. However, Pitx2 targets Dkk2, an extracellular antagonist that moderates Wnt signaling activity, to form a regulatory feedback loop within the periocular neural crest cells [187,193,194].
Additional downstream targets of Pitx2 includes Lmx1b, a LIM-homeodomain transcription factor that is expressed in periocular mesenchyme and is later enriched in presumptive anterior segment structures (iris, cornea, trabecular meshwork), the ocular fissure, and the hyaloid vasculature (Figure 5) [195]. Lmx1b regulates migration and survival of neural crest cells in the anterior segment and Lmx1b mutant mice have microphthalmic eyes with iris and ciliary body hypoplasia [195]. In humans, autosomal dominant mutations in LMX1B cause Nail–Patella Syndrome which is strongly associated with open-angle glaucoma [196]. However, unlike Axenfeld–Rieger syndrome, affected individuals do not exhibit congenital ocular abnormalities, which suggests an ongoing requirement for the LMX1B gene in post-natal maintenance of the structure and function of the trabecular meshwork and sclera. Interestingly, as with Pitx2, morpholino oligonucleotide knockdown of Lmx1b in zebrafish causes colobomas indicating a cell non-autonomous role for these two factors on optic cup development [197]. Another Pitx2 target within neural crest cells is Tfap2b (Figure 5). In contrast to Tfap2a, which is important in neural border induction, Tfap2b is expressed in the periocular mesenchyme in mice and plays a Lmx1b-independent role in anterior chamber development [198]. Neural crest-specific Tfap2b knockout mice show mal-developed iris, cornea, trabecular meshwork, and ciliary body resulting in a closed angle-configuration. Loss of Tfap2b expression causes absence of N-Cadherin in the corneal endothelium and disorganization of junctional complexes and corneal edema. Furthermore, these Tfap2b mutants also have decreased expression of the cornea-specific collagen, Col8a2, and the corneal endothelial marker, Zp4 leading to decreased cell proliferation [199]. While Lmx1b and Tfap2b have both been identified as Pitx2 targets, additional studies are required to identify additional downstream factors within the anterior segment.
Although strabismus and blepharophimosis are rarely reported in Axenfeld–Rieger syndrome, in animal models periocular neural crest cells are essential for extraocular muscle formation and organization [179,180,200]. Both RA and Pitx2 regulate expression of muscle-specific transcription factors such as Myf5, Myog, and Myod1 that are required for extraocular muscle myogenesis and survival [201,202]. Furthermore, Foxl2 in periocular neural crest cells activates expression of smooth muscle alpha actin (encoded by the Acta2 gene) in extraocular muscles in mice, and human mutations in FOXL2 are associated with blepharophimosis syndrome. While Myf5, Myog, and Myod1 induce myoblast differentiation and eventual Acta2 expression, a direct connection at the molecular level between Pitx2 and Foxl2 in the periocular mesenchyme has not been established. In contrast, Pitx2 knockdown in zebrafish, while affecting the jaw musculature, did not alter expression of myod nor organization of the extraocular muscles. However, the ocular phenotype in pitx2 knockout mice causes anophthalmia or severe microphthalmia, which is worse than in zebrafish or humans. The eye itself is known to be important in regulating extraocular muscle development such that genetic, toxic, or surgical loss of the optic vesicle causes disorganization and in some cases agenesis of the extraocular muscles [200,203]. Thus, the extraocular muscle phenotype seen in pitx2 knockout mice may be directly or indirectly related to neural crest cell signaling.
FoxC1 encodes for a forkhead transcription factor, that functions in craniofacial and ocular neural crest cell migration and differentiation [204]. Like Pitx2, Foxc1 is not required for delamination, but critical for survival and migration of neural crest cells [205]. Foxc1 is then expressed within the neural crest cells of the periocular mesenchyme and later within the presumptive anterior segment structures [204]. Furthermore, autosomal dominant mutations in FOXC1 also primarily cause Axenfeld–Rieger syndrome [9,206]. In Foxc1 knockout mice, the cornea is thickened and disorganized, and there is failure of the lens to separate from the cornea, which is more reminiscent of Peters Anomaly [207]. Knockdown of the zebrafish homolog Foxc1a also results in maldevelopment of the cornea and iris and causes optic nerve hypoplasia and colobomas. In addition, the zebrafish show cerebral hemorrhages suggesting a separate role of Foxc1a in blood vessel integrity. Correspondingly, patients with FOXC1 mutations have also been found to have early-onset cerebrovascular disease [205].
Foxc1, like Pitx2, is regulated by RA within the periocular mesenchyme. Furthermore, Foxc1 and Pitx2 are coexpressed within neural crest cells and have been found to regulate each other’s expression. In addition, the transcriptional regulator protein, Pawr (PRKC apoptosis Wilm’s tumor 1 regulator) modulates the abilities of Pitx2 and Foxc1 to activate target genes [193]. Tgfβ2 signaling from the lens which targets its receptor (Tgfbr2) in the periocular mesenchyme also regulates Foxc1 and Pitx2 expression and interestingly, Tgfβ2 knockout mice share a Peters Anomaly-like phenotype similar to Foxc1 knockout mice [208,209,210]. However, the main effect of Tgfβ2 signaling is likely post-migration in directing Foxc1 and Pitx2 positive cells toward corneal endothelial and stromal differentiation.
Within neural crest cells, Foxc1 has been shown to target genes including Eya2, Ffg19, Foxo1a, Foxc2, and Galnt4 (Figure 5). Eya2 is a transcription factor that is expressed in the periocular mesenchyme and ocular fissure in mice and zebrafish. Eya2 expression, which is increased by foxc1a, RA, and nlx1, promotes remodeling of the periocular mesenchyme by inducing apoptosis of neural crests cells [211,212,213]. In presumptive ocular tissues, Foxc1 binds to the Fgf19 promoter, activating gene transcription which directs both the development and maintenance of anterior segment structures via ERK1/2 signaling [214]. Foxc1 also has been shown to regulate the forkhead transcription factor Foxo1a. Within zebrafish and cultured human trabecular meshwork cells, decreased Foxc1 reduces Foxo1a expression, which impaired response to oxidative stress and ultimately survivability [215]. In mouse periocular mesenchyme, Foxc1 together with Pitx2 and Wnt-signaling interact with Foxc2 to cooperatively regulate early corneal development. Foxc2 is required within neural crest cells to promote corneal cell identity and demarcate corneal and scleral tissues [216]. However, it has yet to be shown whether Foxc2 functions in the same role in human eye development. Galnt4 encodes an enzyme which initiates mucin-type O-linked glycosylation and ultimately changes the gel-like properties of mucin secreted from epithelial cells [212]. Although the specific function of Galnt4 within the neural crest cells is unknown, the involvement of glycosylases in altering extracellular matrices and mediating adhesion, suggests a role in promoting migration. Taken together, Foxc1 has a variety of targets within the neural crest cells and eye. However, additional studies are required to identify additional effectors and determine how these genes work together to regulate neural crest cell migration and differentiation.
6. Summary/Conclusions
Neural crest development is a complex process that involves multiple steps and numerous gene regulatory networks [15,17,24,39]. Genetic disruption can trigger a cascade of downstream effects that can lead to widespread systemic, craniofacial, and ocular abnormalities. Eye development in particular is exquisitely sensitive to proper neural crest cell development.
While microphthalmia, anophthalmia, and coloboma are typically associated with disruption of neural epithelial gene function in the forebrain and optic cup development, human disease and animal models have demonstrated that perturbation of neural crest specification or migration often results in a similar phenotype [6,12,13,14]. As discussed in this review, this common phenotype of microphthalmia and coloboma is exhibited when there is genetic disruption of neural plate border genes (Msx1/2, Zic2, and Tfap2) or neural crest specification genes [59,65,160]. While these eye malformations are within the neural epithelial-derived optic cup, it is clear from expression analysis and conditional knockout models that the neural crest cells within the periocular mesenchyme have cell non-autonomous effects on the optic cup. The neural crest cells restrict hedgehog signaling within the optic stalk and cup, which is critical for proper patterning of the retina [81,82,217]. Failure to establish the dorsal-ventral axis of the retina impairs normal optic cup growth and prevents ocular fissure closure. However, additional studies specifically assessing these interactions between periocular neural crest cells and the optic cup are required to fully understand the underlying mechanisms.
In addition, genetic disruption of later neural crest cell migration and differentiation can have significant effects on eye development. The Pitx2 and Foxc1 genes are the most well-studied as human autosomal dominant mutations in either is more often associated with Axenfeld–Rieger Syndrome and less commonly Peters Anomaly [9,184,206]. Although on rare occasions these diseases can be accompanied by optic nerve abnormalities such as Morning Glory Disc and colobomas, the majority of cases are limited to the anterior segment. However, gene knockout and knockdown animal models exhibit optic cup abnormalities. This discrepancy may be due to greater sensitivity of gene dosage effects on optic cup patterning compared to ocular neural crest cell migration and differentiation. Nevertheless, Pitx2 and Foxc1 are key regulators of eye development, and further studies are required to identify downstream targets both within the optic cup and neural crest cells.
While congenital eye anomalies are overall rarely occurring in approximately 1:5000 to 1:10,000 live births, understanding the genetics regulating neural crest cell development and ultimately the pathogenesis of these diseases is important [218]. Insight gained from improving our knowledge on neural crest genetics will lead to breakthroughs in stem cell and gene therapy treatments for these potentially blinding diseases.
Author Contributions
Manuscript preparation and editing: J.W. and B.L.B. All authors have read and agree to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hay, E. Development of the vertebrate cornea. Int. Rev. Cytol. 1979, 63, 263–322. [Google Scholar]
- Beebe, D.C.; Coats, J.M. The lens organizes the anterior segment: Specification of neural crest cell differentiation in the avian eye. Dev. Biol. 2000, 220, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Cvekl, A.; Tamm, E.R. Anterior eye development and ocular mesenchyme: New insights from mouse models and human disease. BioEssays 2004, 26, 374–386. [Google Scholar] [CrossRef] [PubMed]
- Gage, P.J.; Rhoades, W.; Prucka, S.K.; Hjalt, T. Fate maps of neural crest and mesoderm in the mammalian eye. Invest. Ophthalmol. Vis. Sci. 2005, 46, 4200–4208. [Google Scholar] [CrossRef] [PubMed]
- Kish, P.E.; Bohnsack, B.L.; Gallina, D.; Kasprick, D.S.; Kahana, A. The eye as an organizer of craniofacial development. Genesis 2011, 49, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Bohnsack, B.L. Neural crest derivatives in ocular development: Discerning the eye of the storm. Birth Defects Res. C Embryo Today 2015, 105, 87–95. [Google Scholar] [CrossRef]
- Cook, C.S.; Sulik, K.K. Keratolenticular dysgenesis (Peters’ anomaly) as a result of acute embryonic insult during gastrulation. J. Pediatr. Ophthalmol. Strabismus 1988, 25, 60–66. [Google Scholar]
- Ozeki, H.; Shirai, S.; Ikeda, K.; Ogura, Y. Anomalies associated with Axenfeld-Rieger syndrome. Graefes Arch. Clin. Exp. Ophthalmol. 1999, 237, 730–734. [Google Scholar] [CrossRef]
- Strungaru, M.H.; Dinu, I.; Walter, M.A. Genotype-phenotype correlations in Axenfeld-Rieger malformation and glaucoma patients with FOXC1 and PITX2 mutations. Invest. Ophthalmol. Vis. Sci. 2007, 48, 228–237. [Google Scholar] [CrossRef]
- Chawla, B.; Schley, E.; Williams, A.L.; Bohnsack, B.L. Retinoic acid and pitx2 regulate early neural crest survival and migration in craniofacial and ocular development. Birth Defects Res. B Dev. Reprod. Toxicol. 2016, 107, 126–135. [Google Scholar] [CrossRef]
- Eason, J.; Williams, A.L.; Chawla, B.; Apsey, C.; Bohnsack, B.L. Differences in neural crest sensitivity to ethanol account for the infrequency of anterior segment defects in the eye compared with craniofacial anomalies in a zebrafish model of fetal alcohol syndrome. Birth Defects Res. 2017, 109, 1212–1227. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Taylor, A.E.; Sowden, J.C.; Ragge, N.K.; Russell-Eggitt, I.; Rahi, J.S.; Gilbert, C.E.; Surveillance of Eye Anomalies (SEA-UK) Special Interest Group. Anophthalmos, microphthalmos, and typical coloboma in the United Kingdom: A prospective study of incidence and risk. Invest. Ophthalmol. Vis. Sci. 2011, 52, 558–564. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.P.; Taylor, A.E.; Sowden, J.C.; Ragge, N.; Russell-Eggitt, I.; Rahi, J.S.; Gilbert, C.E.; Surveillance of Eye Anomalies Special Interest Group. Anophthalmos, microphthalmos, and coloboma in the United Kingdom: Clinical features, results of investigations, and early management. Ophthalmology 2012, 119, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Bryan, C.D.; Casey, M.A.; Pfeiffer, R.L.; Jones, B.W.; Kwan, K.M. Optic cup morphogenesis requires neural crest-mediated basement membrane assembly. Development 2020, 147, dev181420. [Google Scholar] [CrossRef] [PubMed]
- Betancur, P.; Bronner-Fraser, M.; Sauka-Spengler, T. Assembling neural crest regulatory circuits into a gene regulatory network. Annu. Rev. Cell Dev. Biol. 2010, 26, 581–603. [Google Scholar] [CrossRef]
- Mayor, R.; Theveneau, E. The neural crest. Development 2013, 140, 2247–2251. [Google Scholar] [CrossRef]
- Simões-Costa, M.; Bronner, M.E. Establishing neural crest identity: A gene regulatory recipe. Development 2015, 142, 2420257. [Google Scholar] [CrossRef]
- Ozair, M.Z.; Kintner, C.; Brivanlou, A.H. Neural induction and early patterning in vertebrates. Wiley Interdiscip. Rev. Dev. Biol. 2013, 2, 479–498. [Google Scholar] [CrossRef]
- Groves, A.K.; LaBonne, C. Setting appropriate boundaries: Fate, patterning and competence at the neural plate border. Dev. Biol. 2014, 389, 2–12. [Google Scholar] [CrossRef]
- Schille, C.; Schambony, A. Signaling pathways and tissue interactions in neural plate border formation. Neurogenesis (Austin) 2017, 4, e1292783. [Google Scholar] [CrossRef]
- Sauka-Spengler, T.; Bronner-Fraser, M. Development and evolution of the migratory neural crest: A gene regulatory perspective. Curr. Opin. Genet. Dev. 2006, 16, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Creuzet, S.; Vincent, C.; Couly, G. Neural crest derivatives in ocular and periocular structures. Int. J. Dev. Biol. 2005, 49, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Theveneau, E.; Mayor, R. Neural crest delamination and migration: From epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 2012, 366, 34–54. [Google Scholar] [CrossRef] [PubMed]
- Gammill, L.S.; Bronner-Fraser, M. Neural crest specification: Migrating into genomics. Nat. Rev. Neurosci. 2003, 4, 795–805. [Google Scholar] [CrossRef]
- Adler, R.; Canto-Soler, M.V. Molecular mechanisms of optic vesicle development: Complexities, ambiguities, and controversies. Dev. Biol. 2007, 305, 1–13. [Google Scholar] [CrossRef]
- Fuhrmann, S. Eye morphogenesis and patterning of the optic vesicle. Curr. Top. Dev. Biol. 2010, 93, 61–84. [Google Scholar]
- Graw, J. Eye development. Curr. Top. Dev. Biol. 2010, 90, 343–386. [Google Scholar]
- Kulesa, P.M.; Bailey, C.M.; Kasemeier-Kulesa, J.C.; McLennan, R. Cranial neural crest migration: New rules for an old road. Dev. Biol. 2010, 344, 543–554. [Google Scholar] [CrossRef]
- Canto-Soler, M.V.; Adler, R. Optic cup and lens development requires Pax6 expression in the early optic vesicle during a narrow time window. Dev. Biol. 2006, 294, 119–132. [Google Scholar] [CrossRef]
- Williams, A.L.; Eason, J.; Chawla, B.; Bohnsack, B.L. Cyp1b1 regulates ocular fissure closure through a retinoic acid-independent pathway. Invest. Ophthalmol. Vis. Sci. 2017, 58, 1084–1097. [Google Scholar] [CrossRef]
- Johnston, M.C.; Noden, D.M.; Hazelton, R.D.; Coulombre, J.L.; Coulombre, A. Origins of avian ocular and periocular tissues. Exp. Eye Res. 1979, 29, 27–43. [Google Scholar] [CrossRef]
- Takamiya, M.; Stegmaier, J.; Kobitski, A.Y.; Schott, B.; Weger, B.D.; Margariti, D.; Cereceda Delgado, A.R.; Gourain, V.; Scherr, T.; Yang, L.; et al. Pax6 organizes the anterior eye segment by guiding two distinct neural crest waves. PLoS Genet. 2020, 16, e1008774. [Google Scholar] [CrossRef] [PubMed]
- Akula, M.; Park, J.W.; West-Mays, J.A. Relationship between neural crest cell specification and rare ocular diseases. J. Neurosci. Res. 2019, 97, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.L.; Bohnsack, B.L. What’s retinoic acid got to do with it? Retinoic acid regulation of the neural crest in craniofacial and ocular development. Genesis 2019, 57, e23308. [Google Scholar] [CrossRef] [PubMed]
- Harris, M.L.; Erickson, C.A. Lineage specification in neural crest cell pathfinding. Dev. Dyn. 2007, 236, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Krispin, S.; Nitzan, E.; Kassem, Y.; Kalcheim, C. Evidence for a dynamic spatiotemporal fate map and early fate restrictions of premigratory avian neural crest. Development 2010, 137, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Nitzan, E.; Kalcheim, C. Neural crest and somitic mesoderm as paradigms to investigate cell fate decisions during development. Dev. Growth Differ. 2013, 55, 60–78. [Google Scholar] [CrossRef]
- Lukoseviciute, M.; Gavriouchkina, D.; Williams, R.M.; Hochgreb-Hagele, T.; Senanayake, U.; Chong-Morrison, V.; Thongjuea, S.; Repapi, E.; Mead, A.; Sauka-Spengler, T. From pioneer to repressor: Bimodal foxd3 activity dynamically remodels neural crest regulatory landscape in vivo. Dev. Cell. 2018, 47, 608–628. [Google Scholar] [CrossRef]
- Knecht, A.K.; Bronner-Fraser, M. Induction of the neural crest: A multigene process. Nat. Rev. 2002, 3, 453–461. [Google Scholar] [CrossRef]
- Streit, A.; Berliner, A.J.; Papanayotou, C.; Sirulnik, A.; Stern, C.D. Initiation of neural induction by FGF signaling before gastrulation. Nature 2000, 406, 74–78. [Google Scholar] [CrossRef]
- Milet, C.; Monsoro-Burq, A.H. Neural crest induction at the neural plate border in vertebrates. Dev. Biol. 2012, 366, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Stuhlmiller, T.J.; García-Castro, M.I. Current perspectives on the signaling pathways directing neural crest induction. Cell Mol. Life Sci. 2012, 69, 3715–3737. [Google Scholar] [CrossRef] [PubMed]
- Pla, P.; Monsoro-Burq, A.H. The neural border: Induction, specification and maturation of the territory that generates neural crest cells. Dev. Biol. 2018, 444, S36–S46. [Google Scholar] [CrossRef] [PubMed]
- Meulemans, D.; Bronner-Fraser, M. Gene-regulatory interactions in neural crest evolution and development. Dev. Cell. 2004, 7, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Khudyakov, J.; Bronner-Fraser, M. Comprehensive spatiotemporal analysis of early chick neural crest network genes. Dev. Dyn. 2009, 238, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.S.; Sauka-Spengler, T.; LaBonne, C. Induction of the neural crest state: Control of stem cell attributes by gene regulatory, post-transcriptional and epigenetic interactions. Dev. Biol. 2012, 366, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, A.; Berx, G. Cadherins and epithelial-to-mesenchymal transition. Prog. Mol. Biol. Transl. Sci. 2013, 116, 317–336. [Google Scholar]
- Schulz, Y.; Wehner, P.; Opitz, L.; Salinas-Riester, G.; Bongers, E.M.; van Ravenswaaij-Arts, C.M.; Wincent, J.; Schoumans, J.; Kohlhase, J.; Borchers, A.; et al. CHD7, the gene mutated in CHARGE syndrome, regulates genes involved in neural crest cell guidance. Hum. Genet. 2014, 133, 997–1009. [Google Scholar] [CrossRef]
- Dady, A.; Duband, J.L. Cadherin interplay during neural crest segregation from the non-neural ectoderm and neural tube in the early chick embryo. Dev. Dyn. 2017, 246, 550–565. [Google Scholar] [CrossRef]
- Bérubé-Simard, F.A.; Pilon, N. Molecular dissection of CHARGE syndrome highlights the vulnerability of neural crest cells to problems with alternative splicing and other transcription-related processes. Transcription 2019, 10, 21–28. [Google Scholar] [CrossRef]
- Scarpa, E.; Szabó, A.; Bibonne, A.; Theveneau, E.; Parsons, M.; Mayor, R. Cadherin switch during EMT in neural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev. Cell 2015, 34, 421–434. [Google Scholar] [CrossRef] [PubMed]
- Okuno, H.; Mihara, F.R.; Ohta, S.; Fukuda, K.; Kurosawa, K.; Akamatsu, W.; Sanosaka, T.; Kohyama, J.; Hayashi, K.; Nakajima, K.; et al. CHARGE syndrome modeling using patient-iPSCs reveals defective migration of neural crest cells harboring CHD7 mutations. Elife 2017, 6, e21114. [Google Scholar] [CrossRef] [PubMed]
- Kunisada, T.; Tezulka, K.-I.; Aoki, H.; Motohashi, T. The stemness of neural crest cells and their derivatives. Birth. Defects Res. C Embryo. Today 2014, 102, 251–262. [Google Scholar] [CrossRef] [PubMed]
- Shellard, A.; Mayor, R. Integrating chemical and mechanical signals in neural crest cell migration. Curr. Opin. Genet. Dev. 2019, 57, 16–24. [Google Scholar] [CrossRef]
- Davidson, D. The function and evolution of Msx genes: Pointers and paradoxes. Trends Genet. 1995, 11, 405–411. [Google Scholar] [CrossRef]
- Alappat, S.; Zhang, Z.Y.; Chen, Y.P. Msx homeobox gene family and craniofacial development. Cell Res. 2003, 13, 429–442. [Google Scholar] [CrossRef]
- Catron, K.M.; Wang, H.; Hu, G.; Shen, M.M.; Abate-Shen, C. Comparison of Msx-1 and Msx-2 suggests a molecular basis for functional redundancy. Mech. Dev. 1996, 55, 185–199. [Google Scholar] [CrossRef]
- Tribulo, C.; Aybar, M.J.; Nguyen, V.H.; Mullins, M.C.; Mayor, R. Regulation of Msx genes by a Bmp gradient is essential for neural crest specification. Development 2003, 130, 6441–6452. [Google Scholar] [CrossRef]
- Ishii, M.; Han, J.; Yen, H.Y.; Sucov, H.M.; Chai, Y.; Maxson, R.E.J. Combined deficiencies of Msx1 and Msx2 cause impaired patterning and survival of the cranial neural crest. Development 2005, 132, 4937–4950. [Google Scholar] [CrossRef]
- Khadka, D.; Luo, T.; Sargent, T.D. Msx1 and Msx2 have shared essential functions in neural crest but may be dispensable in epidermis and axis formation in Xenopus. Int. J. Dev. Biol. 2006, 50, 499–502. [Google Scholar] [CrossRef]
- Phillips, B.T.; Kwon, H.J.; Melton, C.; Houghtaling, P.; Fritz, A.; Riley, B.B. Zebrafish msxB, msxC, and msxE function together to refine the neural-nonneural border and regulate cranial placodes and neural crest development. Dev. Biol. 2006, 294, 376–390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lincecum, J.M.; Fannon, A.; Song, K.; Wang, Y.; Sassoon, D.A. Msh homeobox genes regulate cadherin-mediated cell adhesion and cell-cell sorting. J. Cell Biochem. 1998, 70, 22–28. [Google Scholar] [CrossRef]
- Monaghan, A.P.; Davidson, D.R.; Sime, C.; Graham, E.; Baldock, R.; Bhattacharya, S.S.; Hill, R.E. The Msh-like homeobox genes define domains in the developing vertebrate eye. Development 1991, 112, 1053–1061. [Google Scholar] [PubMed]
- Suzuki, H.R.; Padanilam, B.J.; Vitale, E.; Ramirez, F.; Solursh, M. Repeating developmental expression of G-Hox7, a novel homeobox-containing gene in the chicken. Dev. Biol. 1991, 148, 375–388. [Google Scholar] [CrossRef]
- Foerst-Potts, L.; Sadler, T.W. Disruption of Msx-1 and Msx-2 reveals roles for these genes in craniofacial, eye, and axial development. Dev. Dyn. 1997, 209, 70–84. [Google Scholar] [CrossRef]
- Aruga, J.; Hatayama, M. Comparative genomics of the zic family genes. Adv. Exp. Med. Biol. 2018, 1046, 3–26. [Google Scholar]
- Diamond, K.E.M.; Barratt, K.S.; Arkell, R.M. Overview of rodent zic genes. Adv. Exp. Med. Biol. 2018, 1046, 179–207. [Google Scholar]
- Winata, C.L.; Korzh, V. Zebrafish zic genes mediate developmental signaling. Adv. Exp. Med. Biol. 2018, 1046, 157–177. [Google Scholar]
- Nakata, K.; Nagai, T.; Aruga, J.; Mikoshiba, K. Xenopus Zic3, a primary regulator both in neural and neural crest development. Proc. Natl. Acad. Sci. USA 1997, 94, 11980–11985. [Google Scholar] [CrossRef]
- Nakata, K.; Nagai, T.; Aruga, J.; Mikoshiba, K. Xenopus Zic family and its role in neural and neural crest development. Mech. Dev. 1998, 75, 43–51. [Google Scholar] [CrossRef]
- Marchal, L.; Luxardi, G.; Thomé, V.; Kodjabachian, L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc. Natl. Acad. Sci. USA 2009, 106, 17437–17442. [Google Scholar] [CrossRef] [PubMed]
- Aruga, J. The role of Zic genes in neural development. Mol. Cell Neurosci. 2004, 26, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Maurus, D.; Harris, W.A. Zic-associated holoprosencephaly: Zebrafish Zic1 controls midline formation and forebrain patterning by regulating Nodal, hedgehog, and retinoic acid signaling. Genes Dev. 2009, 23, 1461–1473. [Google Scholar] [CrossRef] [PubMed]
- McMahon, A.R.; Merzdorf, C.S. Expression of the zic1, zic2, zic3, and zic4 genes in early chick embryos. BMC Res. Notes 2010, 3, 167. [Google Scholar] [CrossRef] [PubMed]
- Aruga, J.; Millen, K.J. ZIC1 function in normal cerebellar development and human developmental pathology. Adv. Exp. Med. Biol. 2018, 1046, 249–268. [Google Scholar] [PubMed]
- Nyholm, M.K.; Abdelilah-Seyfried, S.; Grinblat, Y. A novel genetic mechanism regulates dorsolateral hinge-point formation during zebrafish cranial neurulation. J. Cell Sci. 2009, 122, 2137–2148. [Google Scholar] [CrossRef]
- Teslaa, J.J.; Keller, A.N.; Nyholm, M.K.; Grinblat, Y. Zebrafish zic2a and zic2b regulate neural crest and craniofacial development. Dev. Biol. 2013, 380, 73–86. [Google Scholar] [CrossRef]
- Elms, P.; Siggers, P.; Napper, D.; Greenfield, A.; Arkell, R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev. Biol. 2003, 264, 391–406. [Google Scholar] [CrossRef]
- Sedykh, I.; Yoon, B.; Roberson, L.; Moskvin, O.; Dewey, C.N.; Grinblat, Y. Zebrafish zic2 controls formation of periocular neural crest and choroid fissure morphogenesis. Dev. Biol. 2017, 429, 92–104. [Google Scholar] [CrossRef]
- Sanek, N.A.; Grinblat, Y. A novel role for zebrafish zic2a during forebrain development. Dev. Biol. 2008, 317, 325–335. [Google Scholar] [CrossRef][Green Version]
- Pillai-Kastoori, L.; Wen, W.; Wilson, S.G.; Strachan, E.; Lo-Castro, A.; Fichera, M.; Musumeci, S.A.; Lehmann, O.J.; Morris, A.C. Sox11 is required to maintain proper levels of hedgehog signaling during vertebrate ocular morphogenesis. PLoS Genet. 2014, 10, e1004491. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Pillai-Kastoori, L.; Wilson, S.G.; Morris, A.C. Sox4 regulates choroid fissure closure by limiting Hedgehog signaling during ocular morphogenesis. Dev. Biol. 2015, 399, 139–153. [Google Scholar] [CrossRef]
- Kuwajima, T.; Soares, C.A.; Sitko, A.A.; Lefebvre, V.; Mason, C. SoxC transcription factors promote contralateral retinal ganglion cell differentiation and axon guidance in the mouse visual system. Neuron 2017, 93, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, T.L.; Javier, A.L.; Campeau, S.A.; Knight, R.D.; Schilling, T.F. Tfap2 transcription factors in zebrafish neural crest development and ectodermal evolution. J. Exp. Zool. B Mol. Dev. Evol. 2007, 308, 679–691. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cornell, R.A. Redundant activities of Tfap2a and Tfap2c are required for neural crest induction and development of other non-neural ectoderm derivatives in zebrafish embryos. Dev. Biol. 2007, 304, 338–354. [Google Scholar] [CrossRef] [PubMed]
- Luo, T.; Zhang, Y.; Khadka, D.; Rangarajan, J.; Cho, K.W.Y.; Sargent, T.D. Regulatory targets for transcription factor AP2 in Xenopus embryos. Dev. Growth Differ. 2005, 47, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Luo, T.; Sargent, T.D. Expression of TFAP2beta and TFAP2gamma genes in Xenopus laevis. Gene Exp. Patterns 2006, 6, 589–595. [Google Scholar] [CrossRef]
- Van Otterloo, E.; Li, H.; Jones, K.L.; Williams, T. AP-2α and AP-2ß cooperatively orchestrate homeobox gene expression during branchial arch patterning. Development 2018, 145, dev157438. [Google Scholar] [CrossRef]
- Milunsky, J.M.; Maher, T.M.; Zhao, G.; Roberts, A.E.; Stalker, H.J.; Zori, R.T.; Burch, M.N.; Clemens, M.; Mulliken, J.B.; Smith, R.; et al. TFAP2A mutations result in branchio-oculo-facial syndrome. Am. J. Hum. Genet. 2008, 82, 1171–1177. [Google Scholar] [CrossRef]
- Milunsky, J.M.; Maher, T.M.; Zhao, G.; Wang, Z.; Mulliken, J.B.; Chitayat, D.; Clemens, M.; Stalker, H.J.; Bauer, M.; Burch, M.; et al. Genotype-phenotype analysis of the branchio-oculo-facial syndrome. Am. J. Med. Genet. A 2011, 144A, 22–32. [Google Scholar] [CrossRef]
- Kwon, H.J.; Bhat, N.; Sweet, E.M.; Cornell, R.A.; Riley, B.B. Identification of early requirements for preplacodal ectoderm and sensory organ development. PLoS Genet. 2010, 6, e1001133. [Google Scholar] [CrossRef] [PubMed]
- Stewart, R.A.; Arduini, B.L.; Berghmans, S.; George, R.E.; Kanki, J.P.; Henion, P.D.; Look, A.T. Zebrafish foxd3 is selectively required for neural crest specification, migration and survival. Dev. Biol. 2006, 292, 174–188. [Google Scholar] [CrossRef] [PubMed]
- Dooley, C.M.; Wali, N.; Sealy, I.M.; White, R.J.; Stemple, D.L.; Collins, J.E.; Busch-Nentwich, E.M. The gene regulatory basis of genetic compensation during neural crest induction. PLoS Genet. 2019, 15, e1008213. [Google Scholar] [CrossRef] [PubMed]
- Gestri, G.; Osborne, R.J.; Wyatt, A.W.; Gerrelli, D.; Gribble, S.; Stewart, H.; Fryer, A.; Bunyan, D.J.; Prescott, K.; Collin, J.R.O.; et al. Reduced TFAP2A function causes variable optic fissure closure and retinal defects and sensitizes eye development to mutations in other morphogenetic regulators. Hum. Genet. 2009, 126, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Bassett, E.A.; Pontoriero, G.F.; Feng, W.; Marquardt, T.; Fini, M.E.; Williams, T.; West-Mays, J.A. Conditional deletion of activating protein 2alpha (AP-2alpha) in the developing retina demonstrates non-cell-autonomous roles for AP-2alpha in optic cup development. Mol. Cell. Biol. 2007, 27, 7497–7510. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.M. Epigenetic developmental disorders: CHARGE syndrome, a case study. Curr. Genet. Med. Rep. 2015, 3, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Baralena, L.; Marcocci, C.; Tanda, M.L.; Manetti, L.; Dell’Unto, E.; Bartolomei, M.P.; Nardi, M.; Martino, E.; Pinchera, A. Cigarette smoking and treatment outcomes in Graves ophthalmopathy. Ann. Intern. Med. 1998, 129, 632–635. [Google Scholar] [CrossRef] [PubMed]
- Bergman, J.E.; Janssen, N.; Hoefsloot, L.H.; Jongmans, M.C.; Hofstra, R.M.; van Ravenswaaij-Arts, C.M. CHD7 mutations in CHARGE syndrome: The clinical implications of an expanding phenotype. J. Med. Genet. 2011, 48, 334–342. [Google Scholar] [CrossRef]
- Hsu, P.; Ma, A.; Wilson, M.; Williams, G.; Curotta, J.; Munns, C.F.; Mehr, S. CHARGE syndrome: A review. J. Paediatr. Child. Health 2014, 50, 504–511. [Google Scholar] [CrossRef]
- Bajpai, R.; Chen, D.A.; Rada-Iglesias, A.; Zhang, J.; Xiong, Y.; Helms, J.; Chang, C.-P.; Zhao, Y.; Swigut, T.; Wysocka, J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 2010, 463, 958–962. [Google Scholar] [CrossRef]
- Ufartes, R.; Schwenty-Lara, J.; Freese, L.; Neuhofer, C.; Möller, J.; Wehner, P.; van Ravenswaaij-Arts, C.M.A.; Wong, M.T.Y.; Schanze, I.; Tzschach, A.; et al. Sema3a plays a role in the pathogenesis of CHARGE syndrome. Hum. Mol. Genet. 2018, 27, 1343–1352. [Google Scholar] [CrossRef] [PubMed]
- Sperry, E.D.; Hurd, E.A.; Durham, M.A.; Reamer, E.N.; Stein, A.B.; Martin, D.M. The chromatin remodeling protein CHD7, mutated in CHARGE syndrome, is necessary for proper craniofacial and tracheal development. Dev. Dyn. 2014, 243, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Gage, P.J.; Hurd, E.A.; Martin, D.M. Mouse models for the dissection of CHD7 functions in eye development and the molecular basis for ocular defects in CHARGE syndrome. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7923–7930. [Google Scholar] [CrossRef] [PubMed]
- Shpargel, K.B.; Starmer, J.; Wang, C.; Ge, K.; Magnuson, T. UTX-guided neural crest function underlies craniofacial features of Kabuki syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, E9046–E9055. [Google Scholar] [CrossRef]
- Schwenty-Lara, J.; Nehl, D.; Borchers, A. The histone methyltransferase KMT2D, mutated in Kabuki syndrome patients, is required for neural crest cell formation and migration. Hum. Mol. Genet. 2020, 29, 305–319. [Google Scholar] [CrossRef]
- Shpargel, K.B.; Mangini, C.L.; Xie, G.; Ge, K.; Magnuson, T. The KMT2D Kabuki syndrome histone methylase controls neural crest cell differentiation and facial morphology. Development 2020, 147, dev187997. [Google Scholar] [CrossRef]
- Cocciadiferro, D.; Augello, B.; De Nittis, P.; Zhang, J.; Mandriani, B.; Malerba, N.; Squeo, G.M.; Romano, A.; Piccinni, B.; Verri, T.; et al. Dissecting KMT2D missense mutations in Kabuki syndrome patients. Hum. Mol. Genet. 2018, 27, 3651–3668. [Google Scholar] [CrossRef]
- Shangguan, H.; Su, C.; Ouyang, Q.; Cao, B.; Wang, J.; Gong, C.; Chen, R. Kabuki syndrome: Novel pathogenic variants, new phenotypes and review of literature. Orphanet. J. Rare Dis. 2019, 14, 255. [Google Scholar] [CrossRef]
- Wang, Y.-R.; Xu, N.-X.; Wang, J.; Wang, X.-M. Kabuki syndrome: Review of the clinical features, diagnosis and epigenetic mechanisms. World J. Pediatr. 2019, 15, 528–535. [Google Scholar] [CrossRef]
- Schwarz, Q.; Vieira, J.M.; Howard, B.; Eickholt, B.J.; Ruhrberg, C. Neuropilin 1 and 2 control cranial gangliogenesis and axon guidance through neural crest cells. Development 2008, 135, 1605–1613. [Google Scholar] [CrossRef]
- Nakayama, H.; Bruneau, S.; Kochupurakkal, N.; Coma, S.; Briscoe, D.M.; Klagsbrun, M. Regulation of mTOR signaling by semaphorin 3F-neuropilin 2 interactions in vitro and in vivo. Sci. Rep. 2015, 5, 11789. [Google Scholar] [CrossRef] [PubMed]
- Van Nostrand, J.L.; Brady, C.A.; Jung, H.; Fuentes, D.R.; Kozak, M.M.; Johnson, T.M.; Lin, C.-Y.; Lin, C.-J.; Swiderski, D.L.; Vogel, H.; et al. Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 2014, 514, 228–232. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, W.A.; Trainor, P.A. Neural crest cell evolution: How and when did a neural crest cell become a neural crest cell. Curr. Top. Dev. Biol. 2015, 111, 3–26. [Google Scholar] [PubMed]
- Higashi, Y.; Maruhashi, M.; Nelles, L.; Van de Putte, T.; Verschueren, K.; Miyoshi, T.; Yoshimoto, A.; Kondoh, H.; Huylebroeck, D. Generation of the floxed allele of the SIP1 (Smad-interacting protein 1) gene for Cre-mediated conditional knockout in the mouse. Genesis 2002, 33, 82–84. [Google Scholar] [CrossRef] [PubMed]
- Van de Putte, T.; Maruhashi, M.; Francis, A.; Nelles, L.; Kondoh, H.; Huylebroeck, D.; Higashi, Y. Mice lacking ZFHX1B, the gene that codes for Smad-interacting protein-1, reveal a role for multiple neural crest defects in the etiology of Hirschsprung disease-mental retardation syndrome. Am. J. Hum. Genet. 2003, 72, 465–470. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, M.; Bessonova, M.; Gu, H.F.; Groop, L.C.; Jönsson, J.I. Characterization, chromosomal localization, and expression during hematopoietic differentiation of the gene encoding Arl6ip, ADP-ribosylation-like factor-6 interacting protin (ARL6). Genomics 2000, 68, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Esmail, M.A.; Ansley, S.J.; Blacque, O.E.; Boroevich, K.; Ross, A.J.; Moore, S.J.; Badano, J.L.; May-Simera, H.; Compton, D.S.; et al. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedel syndrome. Nat. Genet. 2004, 36, 989–993. [Google Scholar] [CrossRef]
- Kobayashi, T.; Hori, Y.; Ueda, N.; Kajiho, H.; Muraoka, S.; Shima, F.; Kataoka, T.; Kontani, K.; Katada, T. Biochemical characterization of missense mutations in the Arf/Arl-family small GTPase Arl6 causing Bardet-Biedl syndrome. Biochem. Biophys. Res. Commun. 2009, 381, 439–442. [Google Scholar] [CrossRef]
- Tu, C.-T.; Yang, T.-C.; Huang, H.-Y.; Tsai, H.-J. Zebrafish arl6ip1 is required for neural crest development during embryogenesis. PLoS ONE 2012, 7, e32899. [Google Scholar] [CrossRef]
- Barembaum, M.; Bronner, M.E. Identification and dissection of a key enhancer mediating cranial neural crest specific expression of transcription factor, Ets-1. Dev. Biol. 2013, 382, 567–575. [Google Scholar] [CrossRef][Green Version]
- Sieweke, M.H.; Tekotte, H.; Frampton, J.; Graf, T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 1996, 85, 49–60. [Google Scholar] [CrossRef]
- Javadiyan, S.; Craig, J.E.; Sharma, S.; Lower, K.M.; Casey, T.; Haan, E.; Souzeau, E.; Burdon, K.P. Novel missense mutation in the bZIP transcription factor, MAF, associated with congenital cataract, developmental delay, seizures, and hearing loss (Aymé-Gripp syndrome). BMC Med. Genet. 2017, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Niceta, M.; Barbuti, D.; Gupta, N.; Ruggiero, C.; Tizzano, E.F.; Graul-Neumann, L.; Barresi, S.; Nishimura, G.; Valenzuela, I.; López-Grondona, F.; et al. Skeletal abnormalities are common features in Aymé-Gripp syndrome. Clin. Genet. 2020, 97, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Lu, N.; Eberspaecher, H.; De Crombrugghe, B. A new long form of c-Maf cooperates with Sox9 to activate the type II collagen gene. J. Biol. Chem. 2002, 277, 50668–50675. [Google Scholar] [CrossRef]
- Rogers, C.D.; Phillips, J.L.; Bronner, M.E. Elk3 is essential for the progression from progenitor to definitive neural crest cell. Dev. Biol. 2013, 374, 225–263. [Google Scholar] [CrossRef]
- Delalande, J.-M.; Guyote, M.E.; Smith, C.M.; Shepherd, I.T. Zebrafish sip1a and sip1b are essential for normal axial and neural patterning. Dev. Dyn. 2008, 237, 1060–1069. [Google Scholar] [CrossRef]
- Mowat, D.R.; Wilson, M.J.; Goossens, M. Mowat-Wilson syndrome. J. Med. Genet. 2003, 40, 305–310. [Google Scholar] [CrossRef]
- Garavelli, L.; Mainardi, P.C. Mowat-Wilson syndrome. Orphanet. J. Rare Dis. 2007, 2, 42. [Google Scholar] [CrossRef]
- Garcia-Castro, M.I.; Marcelle, C.; Bronner-Fraser, M. Ectodermal Wnt function as a neural crest inducer. Science 2002, 297, 848–851. [Google Scholar]
- Chen, Y.; Gridley, T. The SNAI1 and SNAI2 proteins occupy their own and each other’s promoter during chondrogenesis. Biochem. Biophys. Res. Commun. 2013, 435, 356–360. [Google Scholar] [CrossRef]
- Plouhinec, J.-L.; Roche, D.D.; Pegoraro, C.; Figueiredo, A.L.; Maczkowiak, F.; Brunet, L.J.; Milet, C.; Vert, J.-P.; Pollet, N.; Harland, R.M.; et al. Pax3 and Zic1 trigger the early neural crest gene regulatory network by the direct activation of multiple key neural crest specifiers. Dev. Biol. 2014, 386, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.J. Treacher Collins syndrome. J. Med. Genet. 1995, 32, 806–808. [Google Scholar] [CrossRef]
- Valdez, B.C.; Henning, C.; So, R.B.; Dixon, J.; Dixon, M.J. The Treacher Collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. USA 2004, 101, 10709–10714. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.; Jones, N.C.; Sandell, L.L.; Jayasinghe, S.M.; Crane, J.; Rey, J.P.; Dixon, M.J.; Trainor, P.A. Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl. Acad. Sci. USA 2006, 103, 13403–13408. [Google Scholar] [CrossRef] [PubMed]
- Hong, C.-S.; Saint-Jeannet, J.-P. Znf703, a novel target of Pax3 and Zic1, regulates hindbrain and neural crest development in Xenopus. Genesis 2017, 55, 10. [Google Scholar] [CrossRef]
- Janesick, A.; Tang, W.; Ampig, K.; Blumberg, B. Znf703 is a novel RA target in the neural plate border. Sci. Rep. 2019, 9, 8275. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Locascio, R.A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Taneyhill, L.A.; Coles, E.G.; Bronner-Fraser, M. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development 2007, 134, 1481–1490. [Google Scholar] [CrossRef]
- Coles, E.G.; Taneyhill, L.A.; Bronner-Fraser, M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev. Biol. 2007, 312, 533–544. [Google Scholar] [CrossRef]
- Blanco, M.J.; Barrallo-Gimeno, A.; Acloque, H.; Reyes, A.E.; Tada, M.; Allende, M.L.; Mayor, R.; Nieto, M.A. Snail1a and Snail1b cooperate in the anterior migration of the axial mesendoderm in the zebrafish embryo. Development 2007, 134, 4073–4081. [Google Scholar] [CrossRef]
- Qiao, L.; Guo, H.; Zhang, T.; Jing, L.; Xiao, C.; Xiao, Y.; Luo, N.; Zhu, H.; Meng, W.; Xu, H.; et al. Snail modulates the assembly of fibronectin via α5 integrin for myocardial migration in zebrafish embryos. Sci. Rep. 2014, 26, 4470. [Google Scholar] [CrossRef] [PubMed]
- Le Douarin, N.M.; Dupin, E. The pluripotency of neural crest cells and their role in brain development. Curr. Top. Dev. Biol. 2016, 116, 659–678. [Google Scholar] [PubMed]
- Sarkar, A.; Hochedlinger, K. The sox family of transcription factors: Versatile regulators of stem and progenitor cell fate. Cell Stem. Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [PubMed]
- Julian, L.M.; McDonald, A.C.; Stanford, W.L. Direct reprogramming with SOX factors: Masters of cell fate. Curr. Opin. Genet. Dev. 2017, 46, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Southard-Smith, E.M.; Kos, L.; Pavan, W.J. Sox10 mutation disrupts neural crest development in Dom Hirschsprung mouse model. Nat. Genet. 1998, 18, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Wegner, M.; Stolt, C.C. From stem cells to neurons and glia: A Soxist’s view of neural development. Trends Neurosci. 2005, 28, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Dutton, K.A.; Pauliny, A.; Lopes, S.S.; Elworthy, S.; Carney, T.J.; Rauch, J.; Geisler, R.; Haffter, P.; Kelsh, R.N. Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 2001, 128, 4113–4125. [Google Scholar] [PubMed]
- Matsuoka, T.; Ahlberg, P.; Kessaris, N.; Iannarelli, P.; Dennehy, U.; Richardson, W.D.; McMahon, A.P.; Koentges, G. Neural crest origins of the neck and shoulder. Nature 2005, 436, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, B.L.; Gallina, D.; Kahana, A. Phenothiourea sensitizes zebrafish cranial neural crest and extraocular muscle development to changes in retinoic acid and insulin-like growth factor signaling. PLoS ONE 2011, 6, e22991. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, M.; Kamel, G.; Shubinets, V.; Hickey, G.; Grimaldi, M.; Liao, E.C. Embryonic fate map of first pharyngeal arch structures in the sox10:kaede zebrafish transgenic model. J. Craniofac. Surg. 2012, 23, 1333–1337. [Google Scholar] [CrossRef] [PubMed]
- Read, A.P.; Newton, V.E. Waardenburg syndrome. J. Med. Genet. 1997, 34, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Pingault, V.; Ente, D.; Dastot-Le Moal, F.; Goossens, M.; Marlin, S.; Bondurand, N. Review and update of mutations causing Waardenburg syndrome. Hum. Mutat. 2010, 31, 391–406. [Google Scholar] [CrossRef] [PubMed]
- Britsch, S.; Goerich, D.E.; Riethmacher, D.; Peirano, R.I.; Rossner, M.; Nave, K.A. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 2001, 15, 66–78. [Google Scholar] [CrossRef] [PubMed]
- Truch, K.; Arter, J.; Turnescu, T.; Weider, M.; Hartwig, A.C.; Tamm, E.R.; Sock, E.; Wegner, M. Analysis of the human SOX10 mutation Q377X in mice and its implications for genotype-phenotype correlation in SOX10-related human disease. Hum. Mol. Genet. 2018, 27, 1078–1092. [Google Scholar] [CrossRef]
- Teng, L.; Mundell, N.A.; Frist, A.Y.; Wang, Q.; Labosky, P.A. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 2008, 135, 1615–1624. [Google Scholar] [CrossRef]
- Mundell, N.A.; Labosky, P.A. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development 2011, 138, 641–652. [Google Scholar] [CrossRef]
- Schunter, J.A.; Löffler, D.; Wiesner, T.; Kovacs, P.; Badenhoop, K.; Aust, G.; Tönjes, A.; Müller, P.; Baber, R.; Simon, J.C.; et al. A novel FoxD3 variant is associated with vitiligo and elevated thyroid auto-antibodies. J. Clin. Endocrinol. Metab. 2015, 100, E1335–E1342. [Google Scholar] [CrossRef]
- Lee, H.-C.; Huang, H.-Y.; Lin, C.-Y.; Chen, Y.-H.; Tsai, H.-J. Foxd3 mediates zebrafish myf5 expression during early somitogenesis. Dev. Biol. 2006, 290, 359–372. [Google Scholar] [CrossRef]
- Curran, K.; Raible, D.W.; Lister, J.A. Foxd3 controls melanophore specification in the zebrafish neual crest by regulation of Mitf. Dev. Biol. 2009, 332, 408–417. [Google Scholar] [CrossRef]
- Wang, W.D.; Melville, D.B.; Montero-Balaguer, M.; Hatzopoulos, A.K.; Knapik, E.W. Tfap2a and Foxd3 regulate early steps in the development of the neural crest progenitor population. Dev. Biol. 2011, 360, 173–185. [Google Scholar] [CrossRef]
- Patel, S.; Rauf, A.; Khan, H.; Meher, B.R.; Hassan, S.S.U. A holistic review on the autoimmune disease vitiligo with emphasis on the causal factors. Biomed. Pharmacother. 2017, 92, 501–508. [Google Scholar] [CrossRef] [PubMed]
- Kloss, B.A.; Reis, L.M.; Brémond-Gignac, D.; Glaser, T.; Semina, E.V. Analysis of FOXD3 sequence variation in human ocular disease. Mol. Vis. 2012, 18, 1740–1749. [Google Scholar] [PubMed]
- McGonnell, I.M.; Graham, A.; Richardson, J.; Fish, J.L.; Depew, M.J.; Dee, C.T.; Holland, P.W.H.; Takahashi, T. Evolution of the Alx homeobox gene family: Parallel retention and independent loss of the vertebrate Alx3 gene. Evol. Dev. 2011, 13, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Dee, C.T.; Szymoniuk, C.R.; Mills, P.E.D.; Takahashi, T. Defective neural crest migration revealed by a zebrafish model of Alx1-related frontonasasl dysplasia. Hum. Mol. Genet. 2013, 22, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Uz, E.; Alanay, Y.; Aktas, D.; Vargel, I.; Gucer, S.; Tuncbilek, G.; von Eggeling, F.; Yilmaz, E.; Deren, O.; Posorski, N.; et al. Disruption of ALX1 causes extreme microphthalmia and severe facial clefting: Expandig the spectrum of autosomal-recessive ALX-related frontonasasl dysplasia. Am. J. Hum. Genet. 2010, 86, 789–796. [Google Scholar] [CrossRef]
- Pini, J.; Kueper, J.; Hu, Y.D.; Kawasaki, K.; Yeung, P.; Tsimbal, C.; Yoon, B.; Carmichael, N.; Maas, R.L.; Cotney, J.; et al. ALX-1 related frontonasal dysplasia results from defective neural crest cell development and migration. EMBO Mol. Med. 2020, 12, e12013. [Google Scholar] [CrossRef]
- Twigg, S.R.; Versnel, S.L.; Nürnberg, G.; Lees, M.M.; Bhat, M.; Hammond, P.; Hennekam, R.C.; Hoogeboom, A.J.; Hurst, J.A.; Johnson, D.; et al. Frontorhiny, a distinctive presentation of frontonasal dysplasia caused by recessive mutations in the ALX3 homeobox gene. Am. J. Hum. Genet. 2009, 84, 698–705. [Google Scholar] [CrossRef]
- Bertola, D.R.; Rodrigues, M.G.; Quaio, C.R.; Kim, C.A.; Passos-Bueno, M.R. Vertical transmission of a frontonasal phenotype caused by a novel ALX4 mutation. Am. J. Med. Genet. A 2013, 161A, 600–604. [Google Scholar] [CrossRef]
- Trainor, P.A.; Tam, P.P. Cranial paraxial mesoderm and neural crest of the mouse embryo-codistribution in the craniofacial mesenchyme but distinct segreation in the branchial arches. Development 1995, 229, 14–29. [Google Scholar]
- Trainor, P.A.; Krumlauf, R. Patterning the cranial neural crest: Hindbrain segmentation and Hox gene plasticity. Nat. Rev. Neurosci. 2000, 1, 116–124. [Google Scholar] [CrossRef]
- Trainor, P.A. Specification of neural crest cell formation and migration in mouse embryos. Sem. Cell Dev. Biol. 2005, 16, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Bohnsack, B.L.; Kahana, A. Thyroid hormone and retinoic acid interact to regulate zebrafish craniofacial neural crest development. Dev. Biol. 2013, 373, 300–309. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; An, M.; Arduini, B.L.; Henion, P.D. Specific pan-neural crest expression of zebrafish crestin throughout embryonic development. Dev. Dyn. 2001, 220, 169–174. [Google Scholar] [CrossRef]
- Van Der Meulen, K.L.; Vöcking, O.; Weaver, M.L.; Meshram, N.N.; Famulski, J.K. Spatiotemporal characterization of anterior segment mesenchyme heterogeneity during zebrafish ocular anterior segment development. Front. Cell Dev. Biol. 2020, 8, 379. [Google Scholar] [CrossRef] [PubMed]
- Gage, P.J.; Suh, H.; Camper, S.A. Dosage requirement of Pitx2 for development of multiple organs. Development 1999, 126, 4643–4651. [Google Scholar] [PubMed]
- Bohnsack, B.L.; Kasprick, D.; Kish, P.E.; Goldman, D.; Kahana, A. A zebrafish model of Axenfeld-Rieger Syndrome reveals that pitx2 regulation by retinoic acid is essential for ocular and craniofacial development. Invest. Ophthalmol. Vis. Sci. 2012, 53, 7–22. [Google Scholar] [CrossRef]
- Hendee, K.E.; Sorokina, E.A.; Muheisen, S.S.; Reis, L.M.; Tyler, R.C.; Markovic, V.; Cuturilo, G.; Link, B.A.; Semina, E.V. PITX2 deficiency and associated human disease: Insights from the zebrafish model. Hum. Mol. Genet. 2018, 27, 1675–1695. [Google Scholar] [CrossRef]
- Evans, A.L.; Gage, P.J. Expression of the homeobox gene Pitx2 in neural crest is required for optic stalk and ocular anterior segment development. Hum. Mol. Genet. 2005, 14, 3347–3359. [Google Scholar] [CrossRef]
- Bhate, M.; Martin, F.J. Unilateral inferior rectus hypoplasia in a child with Axenfeld-Rieger syndrome. J. AAPOS 2012, 16, 304–306. [Google Scholar] [CrossRef]
- Shah, B.M.; Dada, T.; Panda, T.; Tanwar, M.; Bhartiya, S.; Dada, R. Novel occurence of Axenfeld-Rieger syndrome in a patient with blepharophimosis ptosis epicanthus inversus syndrome. Indian J. Ophthalmol. 2014, 62, 358–360. [Google Scholar]
- Semina, E.V.; Reiter, R.; Leysens, N.J.; Alward, W.L.; Small, K.W.; Datson, N.A.; Siegel-Barelt, J.; Bierke-Nelson, D.; Bitou, P.; Zabel, B.U.; et al. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat. Genet. 1996, 14, 392–399. [Google Scholar] [CrossRef] [PubMed]
- Kulak, S.C.; Kozlowski, K.; Semina, E.V.; Pearce, W.G.; Walter, M.A. Mutation in the RIEG1 gene in patients with iridogoniodysgenesis syndrome. Hum. Mol. Genet. 1998, 7, 1113–1117. [Google Scholar] [CrossRef] [PubMed]
- Saadi, I.; Semina, E.V.; Amendt, B.A.; Harris, D.J.; Murphy, K.P.; Murray, J.C.; Russo, A.F. Identification of a dominant negative homeodomain mutation in Rieger syndrome. J. Biol. Chem. 2001, 276, 23034–23041. [Google Scholar] [CrossRef] [PubMed]
- Saadi, I.; Toro, R.; Kuburas, A.; Semina, E.V.; Murray, J.C.; Russo, A.F. An unusual class of PITX2 mutations in Axenfeld-Rieger syndrome. Birth. Defects Res. A Clin. Mol. Teratol. 2006, 76, 175–181. [Google Scholar] [CrossRef]
- Zepeda, E.M.; Branham, K.; Moroi, S.E.; Bohnsack, B.L. Surgical outcomes of glaucoma associated with Axenfeld-Rieger syndrome. BMC Ophthalmol. 2020, 20, 172. [Google Scholar] [CrossRef]
- Arikawa, A.; Yoshida, S.; Yoshikawa, H.; Ishikawa, K.; Yamaji, Y.; Arita, R.-I.; Ueno, A.; Ishibashi, T. Case of novel PITX2 gene mutation associated with Peters’ anomaly and persistent hyperplastic primary vitreous. Eye (Lond.) 2009, 24, 391–393. [Google Scholar] [CrossRef][Green Version]
- Kumar, S.; Duester, G. Retinoic acid signaling in perioptic mesenchyme represses Wnt signaling via induction of Pitx2 and Dkk2. Dev. Biol. 2010, 340, 67–74. [Google Scholar] [CrossRef]
- Kitamura, K.; Miura, H.; Miyagawa-Tomita, S.; Yanazawa, M.; Katoh-Fukui, Y.; Suzuki, R.; Ohuchi, H.; Suehiro, A.; Motegi, Y.; Nakahara, Y.; et al. Mouse Pitx2 deficiency leads to anomalies of the ventral body wall, heart, extra- and periocular mesoderm and right pulmonary isomerism. Development 1999, 126, 5749–5758. [Google Scholar]
- Asai-Coakwell, M.; Backhouse, C.; Casey, R.J.; Gage, P.J.; Lehmann, O.J. Reduced human and murine corneal thickness in an Axenfeld-Rieger Syndrome subtype. Invest. Ophthalmol. Vis. Sci. 2006, 47, 4905–4909. [Google Scholar] [CrossRef]
- Matt, N.; Ghyselinck, N.B.; Wendling, O.; Chambon, P.; Mark, M. Retinoic acid-induced developmental defects are mediated by RARbeta/RXR heterodimers in the pharyngeal endoderm. Development 2003, 130, 2083–2093. [Google Scholar] [CrossRef]
- Molotkov, A.; Molotkova, N.; Duester, G. Retinoic acid guides eye morphogenetic movements via paracrine signaling but is unnecessary for retinal dorsoventral patterning. Development 2006, 133, 1901–1910. [Google Scholar] [CrossRef] [PubMed]
- Matt, N.; Ghyselinck, N.B.; Pellerin, I.; Dupe, V. Impairing retinoic acid signaling in the neural crest cells is sufficient to alter entire eye morphogenesis. Dev. Biol. 2008, 320, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Berry, F.B.; Lines, M.A.; Ooas, J.M.; Footz, T.; Underhill, D.A.; Gage, P.J.; Walter, M.A. Functional interactions between FOXC1 and PITX2 underlie the sensitifvity to FOXC! gene dose in Axenfeld=Rieger syndrome and anterior segment dysgenesis. Hum. Mol. Genet. 2006, 15, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Gage, P.J.; Qian, M.; Wu, D.; Rosenberg, K.I. The canonical Wnt signaling antagonist DKK2 is an essential effector of PITX2 function during normal eye development. Dev. Biol. 2008, 317, 310–324. [Google Scholar] [CrossRef]
- Pressman, C.L.; Chen, H.; Johnson, R.L. LMX1B, a LIM homeodomain class transcription factor, is necessary for normal development of multiple tissues in the anterior segment of the murine eye. Genesis 2000, 26, 15–25. [Google Scholar] [CrossRef]
- Vollrath, D.; Jaramillo-Babb, V.L.; Clough, M.V.; McIntosh, I.; Scott, K.M.; Lichter, P.R.; Richards, J.E. Loss-of-function mutations in the LIM-homeodomain gene, LMX1B in nail-patella syndrome. Hum. Mol. Genet. 1998, 7, 1091–1098. [Google Scholar] [CrossRef]
- McMahon, C.; Gestri, G.; Wilson, S.W.; Link, B.A. Lmx1b is essentail for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish. Dev. Biol. 2009, 332, 287–298. [Google Scholar] [CrossRef]
- Chen, L.; Martino, V.; Dombkowski, A.; Williams, T.; West-Mays, J.; Gage, P.J. AP-2ß is a downstream effector of PITX2 required to specify endothelium and establish angiogenic privilege during corneal development. Invest. Ophthalmol. Vis. Sci. 2016, 57, 1072–1081. [Google Scholar] [CrossRef]
- Martino, V.B.; Sabljic, T.; Deschamps, P.; Green, R.M.; Akula, M.; Peacock, E.; Ball, A.; Williams, T.; West-Mays, J.A. Conditional deletion of AP-2ß in mouse cranial neural crest results in anterior segment dysgenesis and early-onset glaucoma. Dis. Model Mech. 2016, 9, 849–961. [Google Scholar] [CrossRef]
- Bohnsack, B.L.; Gallina, D.; Thompson, H.; Kasprick, D.; Lucarelli, M.J.; Dootz, G.; Nelson, C.; McGonnell, I.M.; Kahana, A. Development of extraocular muscles require early signals from periocular neural crest and the developing eye. Arch. Ophthalmol. 2011, 129, 1030–1041. [Google Scholar] [CrossRef]
- Diehl, A.G.; Zareparst, S.; Qian, M.; Khanna, R.; Angeles, R.; Gage, P.J. Extraocular muscle morphogenesis and gene expression are regulated by Pitx 2 gene dose. Invest. Ophthalmol. Vis. Sci. 2006, 47, 1785–1793. [Google Scholar] [CrossRef] [PubMed]
- Zacharias, A.L.; Lewandoski, M.; Rudnicki, M.A.; Gage, P.J. Pitx2 is an upstream activator of extraocular myogenesis and survival. Dev. Biol. 2011, 349, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Langenberg, T.; Kahana, A.; Wszalek, J.A.; Halloran, M.C. The Eye Organizes Neural Crest Cell Migration. Dev. Dyn. 2008, 237, 1645–16521. [Google Scholar] [CrossRef] [PubMed]
- Smith, R.S.; Zabealeta, A.; Kume, T.; Savinova, O.V.; Kidson, S.H.; Martin, J.E.; Nishimura, D.Y.; Alward, W.L.; Hogan, B.L.; John, S.W.M. Haploinsufficiency of the transcription factors FOXC1 and FOXC2 results in aberrant ocular development. Hum. Mol. Genet. 2000, 9, 1021–1032. [Google Scholar] [CrossRef]
- French, C.R.; Seshadri, S.; Destafano, A.L.; Fornage, M.; Arnold, C.R.; Gage, P.J.; Skarie, J.M.; Dobyns, W.B.; Millen, K.J.; Liu, T.; et al. Mutation of FOXC1 and PITX2 induces cerebral small-vessel disease. J. Clin. Investig. 2014, 124, 4877–4881. [Google Scholar] [CrossRef]
- Tumer, Z.; Bach-Holm, D. Axenfeld-Rieger syndrome and spectrum of Pitx2 and Foxc1 mutations. Eur. J. Hum. Genet. 2009, 17, 1527–1539. [Google Scholar] [CrossRef]
- Saleem, R.A.; Banerjee-Basu, S.; Murphy, T.C.; Baxevanis, A.D.; Walter, M.A. Essential structural and functional determinants within the forkhead domain of FOXC1. Nucleic Acids Res. 2004, 32, 4182–4193. [Google Scholar] [CrossRef]
- David, D.; Cardoso, J.; Marques, B.; Marques, R.; Silva, E.D.; Santos, H.; Boavida, M.G. Molecular characterization of a familial translocation implicates disruption of HDAC9 and possible position effect on TGFbeta2 in the pathogenesis of Peters’ anomaly. Genomics 2003, 81, 489–503. [Google Scholar] [CrossRef]
- Iwao, K.; Inatani, M.; Matsumoto, Y.; Ogata-Iwao, M.; Takihara, Y.; Irie, F.; Yamaguchi, Y.; Okinami, S.; Tanihara, H. Heparan sulfate deficiency leads to Peters anomaly in mice by disturbing neural crest TGF-β2 signaling. J. Clin. Investig. 2009, 119, 997–1008. [Google Scholar] [CrossRef]
- Silla, Z.T.; Naidoo, J.; Kidson, S.H.; Sommer, P. Signals from the lens and Foxc1 regulate the expression of key genes during the onset of corneal endothelial development. Exp. Cell Res. 2014, 322, 381–388. [Google Scholar] [CrossRef]
- Matt, N.; Dupe, V.; Garnier, J.-M.; Dennefeld, C.; Chambon, P.; Mark, M.; Ghyselinck, N.B. Retinoic acid-dependent eye morphogenesis is orchestrated by neural crest cells. Development 2005, 132, 4789–4800. [Google Scholar] [CrossRef] [PubMed]
- Sommer, P.; Napier, H.R.; Hogan, B.L.; Kidson, S.H. Identification of Tgfbeta1i4 as a downstream target of Foxc1. Dev. Growth Differ. 2006, 48, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Lupo, G.; Gestri, G.; O’Brien, M.; Denton, R.M.; Chandraratna, R.A.S.; Ley, S.V.; Harris, W.A.; Wilson, S.W. Retinoic acid receptor signaling regulates choroid fissure closure through independent mechanisms in the ventral optic cup and periocular mesenchyme. Proc. Natl. Acad. Sci. USA 2011, 108, 8698–8703. [Google Scholar] [CrossRef]
- Tamimi, Y.; Skarie, J.M.; Footz, T.; Berry, F.B.; Link, B.A.; Walter, M.A. FGF19 is a target for FOXC1 regulation in ciliary body-derived cells. Hum. Mol. Genet. 2006, 15, 3229–3240. [Google Scholar] [CrossRef]
- Berry, F.B.; Skarie, J.M.; Mirzayans, F.; Fortin, Y.; Hudson, T.J.; Raymond, V.; Link, B.A.; Walter, M.A. FOXC1 is required for cell viability and resistance to oxidative stress in the eye through the transcriptional regulation of FOXO1A. Hum. Mol. Genet. 2008, 17, 490–505. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Chen, L.; Liu, W.; Zhao, D.; Schultz, K.M.; Sasman, A.; Liu, T.; Zhang, H.F.; Gage, P.J.; Kume, T. Foxc1 and Foxc2 in the neural crest are required for ocular anterior segment development. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1368–1377. [Google Scholar] [CrossRef] [PubMed]
- Schimmenti, L.A.; de la Cruz, J.; Lewis, R.A.; Karkera, J.D.; Manligas, G.S.; Ressler, E.; Muenke, M. Novel mutation in sonic hedgehog in non-syndrome colobomatous microphthalmia. Am. J. Med. Genet. 2003, 116A, 215–221. [Google Scholar] [CrossRef]
- Solebo, A.L.; Teoh, L.; Rahi, J. Epidemiology of blindness in children. Arch. Dis. Child. 2017, 102, 853–857. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).