Recent Advances in the Genetic, Anatomical, and Environmental Regulation of the C. elegans Germ Line Progenitor Zone
Abstract
1. Introduction
2. Results
2.1. Anatomy
2.1.1. Hermaphrodite Gonad Structure
2.1.2. Gonad Development
2.1.3. Distal Tip Cell Structure
2.1.4. Open Questions about Somatic Gonad Structure and Function
2.2. Genetics
2.2.1. Genetic Regulation of the Progenitor Zone
2.2.2. Identification of the Link between Notch Signaling and PUF/FBF
2.2.3. Open Questions of Notch Signaling in the Germ Line
2.2.4. The Germ Stem Cells
2.2.5. Mitotically Dividing, non-Stem Progenitor Cells
2.2.6. Cells Undergoing Meiotic Entry
2.3. Topics Arising in Germ Line Regulation
2.3.1. Spatial Restriction of Cell Fate Determinants
2.3.2. A Role for Gonad Structure in Patterning the Germ Line
2.3.3. Testing the Model with Existing Data
2.3.4. Environmental Inputs into the Gonad and Germ Line
2.3.5. The Somatic Gonad as a Relay Point
Funding
Acknowledgments
Conflicts of Interest
References
- Kimble, J.E.; White, J.G. On the control of germ cell development in Caenorhabditis elegans. Dev. Biol. 1981, 81, 208–219. [Google Scholar] [CrossRef]
- Jane Albert Hubbard, E.; Greenstein, D. Introduction to the germ line. WormBook 2005. [Google Scholar] [CrossRef]
- Hubbard, E.J.A. Caenorhabditis elegans germ line: A model for stem cell biology. Dev. Dyn. 2007, 236, 3343–3357. [Google Scholar] [CrossRef]
- Kimble, J.; Seidel, H. C. Elegans Germline Stem Cells and Their Niche; Harvard Stem Cell Institute: Cambridge, MA, USA, 2013. [Google Scholar]
- Kershner, A.; Crittenden, S.L.; Friend, K.; Sorensen, E.B.; Porter, D.F.; Kimble, J. Germline Stem Cells and Their Regulation in the Nematode Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013, 786, 29–46. [Google Scholar] [PubMed]
- Albert Hubbard, E.J.; Schedl, T. Biology of the Caenorhabditis elegans Germline Stem Cell System. Genetics 2019, 213, 1145–1188. [Google Scholar] [CrossRef]
- Lints, R.; Hall, D.H. R. System. I.W. 2. 1. Reproductive System. Wormatlas 2018. [Google Scholar] [CrossRef]
- Brenner, J.L.; Schedl, T. Germline Stem Cell Differentiation Entails Regional Control of Cell Fate Regulator GLD-1 in Caenorhabditis elegans. Genetics 2016, 202, 1085–1103. [Google Scholar] [CrossRef]
- Cinquin, A.; Zheng, L.; Taylor, P.H.; Paz, A.; Zhang, L.; Chiang, M.; Snow, J.J.; Nie, Q.; Cinquin, O. Semi-permeable diffusion barriers enhance patterning robustness in the C. elegans germ line. Dev. Cell 2015, 35, 405–417. [Google Scholar] [CrossRef]
- Gordon, K.L.; Zussman, J.W.; Li, X.; Martin, C.M.; Sherwood, D.R. Stem cell niche exit in C. elegans via orientation and segregation of daughter cells by a cryptic cell outside the niche. Elife 2020. [Google Scholar] [CrossRef]
- Hall, D.H.; Winfrey, V.P.; Blaeuer, G.; Hoffman, L.H.; Furuta, T.; Rose, K.L.; Hobert, O.; Greenstein, D. Ultrastructural features of the adult hermaphrodite gonad of Caenorhabditis elegans: Relations between the germ line and soma. Dev. Biol. 1999, 212, 101–123. [Google Scholar] [CrossRef]
- Amini, R.; Goupil, E.; Labella, S.; Zetka, M.; Maddox, A.S.; Labbé, J.-C.; Chartier, N.T. C. elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J. Cell Biol. 2014, 206, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Maddox, A.S.; Habermann, B.; Desai, A.; Oegema, K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development 2005, 132, 2837–2848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Rolls, M.M.; Hanna-Rose, W. A postmitotic function and distinct localization mechanism for centralspindlin at a stable intercellular bridge. Dev. Biol. 2013, 376, 13–22. [Google Scholar] [CrossRef]
- Rehain-Bell, K.; Love, A.; Werner, M.E.; Macleod, I.; Yates Iii, J.R.; Shaub, A.; Correspondence, M. A Sterile 20 Family Kinase and Its Co-factor CCM-3 Regulate Contractile Ring Proteins on Germline Intercellular Bridges. Curr. Biol. 2017, 27, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Dé, F.; Landmann, R.; Bain, O.; Martin, C.; Uni, S.; Taylor, M.J.; Sullivan, W. Both asymmetric mitotic segregation and cell-to-cell invasion are required for stable germline transmission of Wolbachia in filarial nematodes. Biol. Open 2012, 1, 536–547. [Google Scholar] [CrossRef]
- Foray, V.; Pérez-Jiménez, M.M.; Fattouh, N.; Landmann, F. Wolbachia Control Stem Cell Behavior and Stimulate Germline Proliferation in Filarial Nematodes. Dev. Cell 2018, 45, 198–211.e3. [Google Scholar] [CrossRef]
- Kelley, C.A.; Cram, E.J. Regulation of actin dynamics in the C. elegans somatic gonad. J. Dev. Biol. 2019, 7, 6. [Google Scholar] [CrossRef]
- Cecchetelli, A.D.; Hugunin, J.; Tannoury, H.; Cram, E.J. CACN-1 is required in the Caenorhabditis elegans somatic gonad for proper oocyte development. Dev. Biol. 2016, 414, 58–71. [Google Scholar] [CrossRef]
- Jayadev, R.; Chi, Q.; Keeley, D.P.; Hastie, E.L.; Kelley, L.C.; Sherwood, D.R. α-Integrins dictate distinct modes of type IV collagen recruitment to basement membranes. J. Cell Biol. 2019, 218, 3098–3116. [Google Scholar] [CrossRef]
- Kramer, J.M. Basement membrane. WormBook 2005. [Google Scholar] [CrossRef]
- Kimble, J.; Hirsh, D. The Postembryonic Cell Lineages of the Hermaphrodite and Male Gonads in Caenorhabditis Elegans; Academic Press: Cambridge, MA, USA, 1979; Volume 70, pp. 396–417. [Google Scholar]
- Cecchetelli, A.D.; Cram, E.J. Regulating distal tip cell migration in space and time. Mech. Dev. 2017, 148, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Sherwood, D.R.; Plastino, J. Invading, Leading and Navigating Cells in Caenorhabditis elegans: Insights into Cell Movement in Vivo. Genetics 2018, 208, 53–78. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.-C.; Schwarzbauer, J.E. Gonad morphogenesis and distal tip cell migration in the Caenorhabditis elegans hermaphrodite. Wiley Interdiscip. Rev. Dev. Biol. 2012, 1, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Byrd, D.T.; Knobel, K.; Affeldt, K.; Crittenden, S.L.; Kimble, J. A DTC niche plexus surrounds the germline stem cell pool in Caenorhabditis elegans. PLoS ONE 2014, 9, e88372. [Google Scholar] [CrossRef]
- Linden, L.M.; Gordon, K.L.; Pani, A.M.; Payne, S.G.; Garde, A.; Burkholder, D.; Chi, Q.; Goldstein, B.; Sherwood, D.R. Identification of regulators of germ stem cell enwrapment by its niche in C. elegans. Dev. Biol. 2017, 429, 271–284. [Google Scholar] [CrossRef]
- Crittenden, S.L.; Lee, C.; Mohanty, I.; Battula, S.; Knobel, K.; Kimble, J. Sexual dimorphism of niche architecture and regulation of the Caenorhabditis elegans germline stem cell pool. Mol. Biol. Cell 2019, 30, 1757–1769. [Google Scholar] [CrossRef]
- Wong, B.G.; Paz, A.; Corrado, M.A.; Ramos, B.R.; Cinquin, A.; Cinquin, O.; Hui, E.E. Live imaging reveals active infiltration of mitotic zone by its stem cell niche. Integr. Biol. 2013, 5, 976–982. [Google Scholar] [CrossRef]
- Seidel, H.S.; Smith, T.A.; Evans, J.K.; Stamper, J.Q.; Mast, T.G.; Kimble, J. C. elegans germ cells divide and differentiate in a folded tissue. Dev. Biol. 2018, 442, 173–187. [Google Scholar] [CrossRef]
- Lee, C.H.; Shin, H.; Kimble, J. Dynamics of Notch-Dependent Transcriptional Bursting in Its Native Context. Dev. Cell 2019, 50, 426–435.e4. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Yadav, S.; DeVault, L.; Jan, Y.N.; Sherwood, D.R. RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization. PLoS Genet. 2015, 11, e1005484. [Google Scholar] [CrossRef] [PubMed]
- Gordon, K.L.; Payne, S.G.; Linden-High, L.M.; Pani, A.M.; Goldstein, B.; Hubbard, E.J.A.; Sherwood, D.R. Ectopic Germ Cells Can Induce Niche-like Enwrapment by Neighboring Body Wall Muscle. Curr. Biol. 2019, 29, 823–833.e5. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Pani, A.M.; Heppert, J.K.; Higgins, C.D.; Goldstein, B. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 2015, 200, 1035–1049. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Ward, J.D.; Reiner, D.J.; Goldstein, B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods 2013, 10, 1028–1034. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, D.J.; Goldstein, B. CRISPR-based methods for caenorhabditis elegans genome engineering. Genetics 2016, 202, 885–901. [Google Scholar] [CrossRef] [PubMed]
- Paix, A.; Folkmann, A.; Seydoux, G. Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 2017, 121–122, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Dokshin, G.A.; Ghanta, K.S.; Piscopo, K.M.; Mello, C.C. Robust genome editing with short single-stranded and long, partially single-stranded DNA donors in caenorhabditis elegans. Genetics 2018, 210, 781–787. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Colaiácovo, M.P. CRISPR-Cas9-Guided Genome Engineering in Caenorhabditis elegans. Curr. Protoc. Mol. Biol. 2019, 129, e106. [Google Scholar] [CrossRef] [PubMed]
- Au, V.; Li-Leger, E.; Raymant, G.; Flibotte, S.; Chen, G.; Martin, K.; Fernando, L.; Doell, C.; Rosell, F.I.; Wang, S.; et al. CRISPR/Cas9 methodology for the generation of knockout deletions in caenorhabditis elegans. G3 Genes Genom. Genet. 2019, 9, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Gerhold, A.R.; Ryan, J.; Vallée-Trudeau, J.-N.; Dorn, J.F.; Labbé, J.-C.; Maddox, P.S. Investigating the Regulation of Stem and Progenitor Cell Mitotic Progression by In Situ Imaging. Curr. Biol. 2015, 25, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Gritti, N.; Kienle, S.; Filina, O.; van Zon, J.S. Long-term time-lapse microscopy of C. elegans post-embryonic development. Nat. Commun. 2016, 7, 12500. [Google Scholar] [CrossRef]
- Rog, O.; Dernburg, A.F. Direct visualization reveals kinetics of meiotic chromosome synapsis. Cell Rep. 2015, 10, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Austin, J.; Kimble, J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell 1987, 51, 589–599. [Google Scholar] [CrossRef]
- Pepper, A.S.R.; Killian, D.J.; Hubbard, E.J.A. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics 2003, 163, 115–132. [Google Scholar]
- Berry, L.W.; Westlund, B.; Schedl, T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Development 1997, 124, 925–936. [Google Scholar] [PubMed]
- Henderson, S.T.; Gao, D.; Lambie, E.J.; Kimble, J. lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development 1994, 120, 2913–2924. [Google Scholar] [PubMed]
- Nadarajan, S.; Govindan, J.A.; McGovern, M.; Hubbard, E.J.A.; Greenstein, D. MSP and GLP-1/Notch signaling coordinately regulate actomyosin-dependent cytoplasmic streaming and oocyte growth in C. elegans. Development 2009, 136, 2223–2234. [Google Scholar] [CrossRef] [PubMed]
- Tax, F.E.; Yeargers, J.J.; Thomas, J.H. Sequence of C. elegans lag-2 reveals a cell-signalling domain shared with Delta and Serrate of Drosophila. Nature 1994, 368, 150–154. [Google Scholar] [CrossRef]
- Haupt, K.A.; Law, K.T.; Enright, A.L.; Kanzler, C.R.; Shin, H.; Wickens, M.; Kimble, J. A PUF hub drives self-renewal in caenorhabditis elegans Germline stem cells. Genetics 2020, 214, 147–161. [Google Scholar] [CrossRef]
- Lamont, L.B.; Crittenden, S.L.; Bernstein, D.; Wickens, M.; Kimble, J. FBF-1 and FBF-2 Regulate the Size of the Mitotic Region in the C. elegans Germline. Dev. Cell 2004, 7, 697–707. [Google Scholar] [CrossRef]
- Kershner, A.M.; Shin, H.; Hansen, T.J.; Kimble, J. Discovery of two GLP-1/Notch target genes that account for the role of GLP-1/Notch signaling in stem cell maintenance. Proc. Natl. Acad. Sci. USA 2014, 111, 3739–3744. [Google Scholar] [CrossRef]
- Shin, H.; Haupt, K.A.; Kershner, A.M.; Kroll-Conner, P.; Wickens, M.; Kimble, J. SYGL-1 and LST-1 link niche signaling to PUF RNA repression for stem cell maintenance in Caenorhabditis elegans. PLoS Genet. 2017, 13, e1007121. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.A.; Enright, A.L.; Ferdous, A.S.; Kershner, A.M.; Shin, H.; Wickens, M.; Kimble, J. The molecular basis of LST-1 self-renewal activity and its control of stem cell pool size. Development 2019, 146, dev181644. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, S.L.; Bernstein, D.S.; Bachorik, J.L.; Thompson, B.E.; Gallegos, M.; Petcherski, A.G.; Moulder, G.; Barstead, R.; Wickens, M.; Kimble, J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 2002, 417, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Kershner, A.M.; Kimble, J. Genome-wide analysis of mRNA targets for Caenorhabditis elegans FBF, a conserved stem cell regulator. Proc. Natl. Acad. Sci. USA 2010, 107, 3936–3941. [Google Scholar] [CrossRef] [PubMed]
- Kadyk, L.C.; Kimble, J. Genetic regulation of entry into meiosis in Caenorhabditis elegans. Development 1998, 125, 1803–1813. [Google Scholar]
- Eckmann, C.R.; Crittenden, S.L.; Suh, N.; Kimble, J. GLD-3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans. Genetics 2004, 168, 147–160. [Google Scholar] [CrossRef]
- Hansen, D.; Wilson-Berry, L.; Dang, T.; Schedl, T. Control of the proliferation versus meiotic development decision in the C. elegans germline through regulation of GLD-1 protein accumulation. Development 2004, 131, 93–104. [Google Scholar] [CrossRef]
- Hansen, D.; Hubbard, E.J.A.; Schedl, T.; Albert Hubbard, E.J.; Schedl, T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 2004, 268, 342–357. [Google Scholar] [CrossRef]
- Mohammad, A.; Vanden Broek, K.; Wang, C.; Daryabeigi, A.; Jantsch, V.; Hansen, D.; Schedl, T. Initiation of meiotic development is controlled by three post-transcriptional pathways in caenorhabditis elegans. Genetics 2018, 209, 1197–1224. [Google Scholar] [CrossRef]
- Fox, P.M.; Vought, V.E.; Hanazawa, M.; Lee, M.H.; Maine, E.M.; Sched, T. Cyclin e and CDK-2 regulate proliferative cell fate and cell cycle progression in the C. elegans germline. Development 2011, 138, 2223–2234. [Google Scholar] [CrossRef]
- Jeong, J.; Verheyden, J.M.; Kimble, J. Cyclin E and Cdk2 Control GLD-1, the Mitosis/Meiosis Decision, and Germline Stem Cells in Caenorhabditis elegans. PLoS Genet. 2011, 7, e1001348. [Google Scholar] [CrossRef] [PubMed]
- Lui, D.Y.; Colaiácovo, M.P. Meiotic Development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013, 757, 133–170. [Google Scholar] [PubMed]
- Ellis, R.E.; Stanfield, G.M. The regulation of spermatogenesis and sperm function in nematodes. Semin. Cell Dev. Biol. 2014, 29, 17–30. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Spike, C.; Greenstein, D. Control of Oocyte Growth and Meiotic Maturation in Caenorhabditis elegans. Adv. Exp. Med. Biol. 2013, 757, 277–320. [Google Scholar] [PubMed]
- Lee, M.-H.; Hook, B.; Lamont, L.B.; Wickens, M.; Kimble, J. LIP-1 phosphatase controls the extent of germline proliferation in Caenorhabditis elegans. EMBO J. 2006, 25, 88–96. [Google Scholar] [CrossRef]
- Chen, J.; Mohammad, A.; Pazdernik, N.; Huang, H.; Bowman, B.; Tycksen, E.; Schedl, T. GLP-1 Notch-LAG-1 CSL control of the germline stem cell fate is mediated by transcriptional targets lst-1 and sygl-1. PLoS Genet. 2020, 16, e1008650. [Google Scholar] [CrossRef]
- Michaelson, D.; Korta, D.Z.; Capua, Y.; Hubbard, E.J.A. Insulin signaling promotes germline proliferation in C. elegans. Development 2010, 137, 671–680. [Google Scholar] [CrossRef]
- Biedermann, B.; Wright, J.; Senften, M.; Kalchhauser, I.; Sarathy, G.; Lee, M.-H.; Ciosk, R. Developmental Cell Translational Repression of Cyclin E Prevents Precocious Mitosis and Embryonic Gene Activation during C. elegans Meiosis. Dev. Cell 2009, 17, 355–364. [Google Scholar] [CrossRef]
- Chi, C.; Ronai, D.; Than, M.T.; Walker, C.J.; Sewell, A.K.; Han, M. Nucleotide levels regulate germline proliferation through modulating GLP-1/Notch signaling in C. elegans. Genes Dev. 2016, 30, 307–320. [Google Scholar] [CrossRef]
- Marsac, R.; Pinson, B.; Saint-Marc, C.; Olmedo, M.; Artal-Sanz, M.; Daignan-Fornier, B.; Gomes, J.E. Purine homeostasis is necessary for developmental timing, germline maintenance and muscle integrity in caenorhabditis elegans. Genetics 2019, 211, 1297–1313. [Google Scholar] [CrossRef]
- Li, X.; Johnson, R.; Park, D.; Chin-Sang, I.; Chamberlin, H. Somatic gonad sheath cells and Eph receptor signaling promote germ-cell death in C. elegans. Cell Death Differ. 2012, 19, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Starich, T.A.; Hall, D.H.; Greenstein, D. Two classes of gap junction channels mediate soma-germline interactions essential for germline proliferation and gametogenesis in Caenorhabditis elegans. Genetics 2014, 198, 1127–1153. [Google Scholar] [CrossRef] [PubMed]
- Aprison, E.Z.; Ruvinsky, I. Coordinated Behavioral and Physiological Responses to a Social Signal Are Regulated by a Shared Neuronal Circuit. Curr. Biol. 2019, 29, 4108–4115.e4. [Google Scholar] [CrossRef] [PubMed]
- Aprison, E.Z.; Ruvinsky, I. Counteracting Ascarosides Act through Distinct Neurons to Determine the Sexual Identity of C. elegans Pheromones. Curr. Biol. 2017, 27, 2589–2599.e3. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Yoon, D.S. A phenotype-based RNAi screening for Ras-ERK/MAPK signaling-associated stem cell regulators in C. elegans. Methods Mol. Biol. 2017, 1622, 207–221. [Google Scholar] [PubMed]
- Killian, D.J.; Hubbard, E.J.A. Caenorhabditis elegans germline patterning requires coordinated development of the somatic gonadal sheath and the germ line. Dev. Biol. 2005, 279, 322–335. [Google Scholar] [CrossRef] [PubMed]
- Chiang, M.; Cinquin, A.; Paz, A.; Meeds, E.; Price, C.A.; Welling, M.; Cinquin, O. Control of Caenorhabditis elegans germ-line stem-cell cycling speed meets requirements of design to minimize mutation accumulation. BMC Biol. 2015, 13, 51. [Google Scholar] [CrossRef]
- Cinquin, A.; Chiang, M.; Paz, A.; Hallman, S.; Yuan, O.; Vysniauskaite, I.; Fowlkes, C.C.; Cinquin, O. Intermittent Stem Cell Cycling Balances Self-Renewal and Senescence of the C. elegans Germ Line. PLoS Genet. 2016, 12, e1005985. [Google Scholar] [CrossRef]
- Kocsisova, Z.; Kornfeld, K.; Schedl, T. Rapid population-wide declines in stem cell number and activity during reproductive aging in C. elegans. Development 2019, 146. [Google Scholar] [CrossRef]
- Roy, D.; Michaelson, D.; Hochman, T.; Santella, A.; Bao, Z.; Goldberg, J.D.; Jane, E.; Hubbard, A. Cell cycle features of C. elegans germline stem/progenitor cells vary temporally and spatially. Dev Biol. 2016, 1, 261–271. [Google Scholar] [CrossRef]
- Christensen, S.; Kodoyianni, V.; Bosenberg, M.; Friedman, L.; Kimble, J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su (H). Development 1996, 122, 1373–1383. [Google Scholar] [PubMed]
- Lee, C.H.; Sorensen, E.B.; Lynch, T.R.; Kimble, J. C. elegans GLP-1/Notch activates transcription in a probability gradient across the germline stem cell pool. Elife 2016, 5, e18370. [Google Scholar] [CrossRef] [PubMed]
- Porter, D.F.; Prasad, A.; Carrick, B.H.; Kroll-Connor, P.; Wickens, M.; Kimble, J. Toward identifying subnetworks from FBF binding landscapes in caenorhabditis spermatogenic or oogenic germlines. G3 Genes Genom. Genet. 2019, 9, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Lambie, E.J.; Kimble, J. Two homologous regulatory genes, lin-12 and glp-1, have overlapping functions. Development 1991, 112, 231–240. [Google Scholar]
- Wickens, M.; Bernstein, D.S.; Kimble, J.; Parker, R. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 2002, 18, 150–157. [Google Scholar] [CrossRef]
- Beadell, A.V.; Liu, Q.; Johnson, D.M.; Haag, E.S. Independent recruitments of a translational regulator in the evolution of self-fertile nematodes. Proc. Natl. Acad. Sci. USA 2011, 108, 19672–19677. [Google Scholar] [CrossRef]
- Beadell, A.V.; Haag, E.S. Evolutionary dynamics of GLD-1-mRNA complexes in Caenorhabditis nematodes. Genome Biol. Evol. 2014, 7, 314–335. [Google Scholar] [CrossRef]
- Thomas, C.G.; Li, R.; Smith, H.E.; Woodruff, G.C.; Oliver, B.; Haag, E.S. Report Simplification and Desexualization of Gene Expression in Self-Fertile Nematodes. Curr. Biol. 2012, 22, 2167–2172. [Google Scholar] [CrossRef]
- Yin, D.; Schwarz, E.M.; Thomas, C.G.; Felde, R.L.; Korf, I.F.; Cutter, A.D.; Schartner, C.M.; Ralston, E.J.; Meyer, B.J.; Haag, E.S. Rapid genome shrinkage in a self-fertile nematode reveals sperm competition proteins. Science 2018, 359, 55–61. [Google Scholar] [CrossRef]
- Yin, D.; Haag, E.S. Evolution of sex ratio through gene loss. Proc. Natl. Acad. Sci. USA 2019, 116, 12919–12924. [Google Scholar] [CrossRef]
- Chen, Y.-Z.; Mapes, J.; Lee, E.-S.; Skeen-Gaar, R.R.; Xue, D. Caspase-mediated activation of Caenorhabditis elegans CED-8 promotes apoptosis and phosphatidylserine externalization. Nat. Commun. 2013, 4, 2726. [Google Scholar] [CrossRef] [PubMed]
- Fox, P.M.; Schedl, T. Analysis of Germline Stem Cell Differentiation Following Loss of GLP-1 Notch Activity in Caenorhabditis elegans. Genetics 2015, 201, 167–184. [Google Scholar] [CrossRef] [PubMed]
- Cinquin, O.; Crittenden, S.L.; Morgan, D.E.; Kimble, J. Progression from a stem cell-like state to early differentiation in the C. elegans germ line. Proc. Natl. Acad. Sci. USA 2010, 107, 2048–2053. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.R.; Francis, R.; Schedl, T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- ans sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996, 180, 165–183. [Google Scholar] [CrossRef]
- Achache, H.; Laurent, L.; Hecker-Mimoun, Y.; Ishtayeh, H.; Rappaport, Y.; Kroizer, E.; Colaiácovo, M.P.; Tzur, Y.B. Progression of meiosis is coordinated by the level and location of MAPK activation via OGR-2 in Caenorhabditis elegans. Genetics 2019, 212, 213–229. [Google Scholar] [CrossRef]
- Lee, M.H.; Hook, B.; Pan, G.; Kershner, A.M.; Merritt, C.; Seydoux, G.; Thomson, J.A.; Wickens, M.; Kimble, J. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 2007, 3, 2540–2550. [Google Scholar] [CrossRef]
- McCarter, J.; Bartlett, B.; Dang, T.; Schedl, T. On the Control of Oocyte Meiotic Maturation and Ovulation inCaenorhabditis elegans. Dev. Biol. 1999, 205, 111–128. [Google Scholar] [CrossRef]
- Marin, V.A.; Evans, T.C. Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 2003, 130, 2623–2632. [Google Scholar] [CrossRef]
- Maciejowski, J.; Ugel, N.; Mishra, B.; Isopi, M.; Hubbard, E.J.A. Quantitative analysis of germline mitosis in adult C. elegans. Dev. Biol. 2006, 292, 142–151. [Google Scholar] [CrossRef]
- Raiders, S.A.; Eastwood, M.D.; Bacher, M.; Priess, J.R. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLoS Genet. 2018, 14, e1007417. [Google Scholar] [CrossRef]
- Crittenden, S.L.; Leonhard, K.A.; Byrd, D.T.; Kimble, J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 2006, 17, 3051–3061. [Google Scholar] [CrossRef] [PubMed]
- Rosu, S.; Cohen-Fix, O. Live-imaging analysis of germ cell proliferation in the C. elegans adult supports a stochastic model for stem cell proliferation. Dev. Biol. 2017, 423, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Seidel, H.S.; Kimble, J. Cell-cycle quiescence maintains Caenorhabditis elegans germline stem cells independent of GLP-1/Notch. Elife 2015, 4, e10832. [Google Scholar] [CrossRef] [PubMed]
- Rangel-Huerta, E.; Maldonado, E. Transit-Amplifying Cells in the Fast Lane from Stem Cells towards Differentiation. Stem Cells Int. 2017, 2017, 7602951. [Google Scholar] [CrossRef]
- Zhang, B.; Hsu, Y.-C. Emerging roles of transit-amplifying cells in tissue regeneration and cancer. Wiley Interdiscip. Rev. Dev. Biol. 2017, 6, e282. [Google Scholar] [CrossRef]
- Templeman, N.M.; Cota, V.; Keyes, W.; Kaletsky, R.; Murphy, C.T.; Templeman, N.M.; Cota, V.; Keyes, W.; Kaletsky, R.; Murphy, C.T. CREB Non-autonomously Controls Reproductive Aging through Hedgehog/Patched Signaling. Dev. Cell 2020, 54, 92–105.e5. [Google Scholar] [CrossRef]
- Aprison, E.Z.; Ruvinsky, I. Sexually Antagonistic Male Signals Manipulate Germline and Soma of C. elegans Hermaphrodites. Curr. Biol. 2016, 26, 2827–2833. [Google Scholar] [CrossRef]
- Dalfó, D.; Michaelson, D.; Hubbard, E.J.A. Sensory regulation of the C. elegans germline through TGF-β-dependent signaling in the niche. Curr. Biol. 2012, 22, 712–719. [Google Scholar] [CrossRef]
- Pekar, O.; Ow, M.C.; Hui, K.Y.; Noyes, M.B.; Hall, S.E.; Jane Albert Hubbard, E. Linking the environment, DAF-7/TGFβ signaling and LAG-2/DSL ligand expression in the germline stem cell niche. Development 2017, 144, 2896–2906. [Google Scholar] [CrossRef]
- Aprison, E.Z.; Ruvinsky, I. Dynamic Regulation of Adult-Specific Functions of the Nervous System by Signaling from the Reproductive System. Curr. Biol. 2019, 29, 4116–4123.e3. [Google Scholar] [CrossRef]
- Leighton, D.H.W.; Choe, A.; Wu, S.Y.; Sternberg, P.W. Communication between oocytes and somatic cells regulates volatile pheromone production in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2014, 111, 17905–17910. [Google Scholar] [CrossRef] [PubMed]
- Aprison, E.Z.; Ruvinsky, I. Sex Pheromones of C. elegans Males Prime the Female Reproductive System and Ameliorate the Effects of Heat Stress. PLoS Genet. 2015, 11, e1005729. [Google Scholar] [CrossRef] [PubMed]
- Jayadev, R.; Sherwood, D.R. Basement membranes. Curr. Biol. 2017, 27, R207–R211. [Google Scholar] [CrossRef] [PubMed]
- McGovern, M.; Voutev, R.; Maciejowski, J.; Corsi, A.K.; Hubbard, E.J.A. A “latent niche” mechanism for tumor initiation. Proc. Natl. Acad. Sci. USA 2009, 106, 11617–11622. [Google Scholar] [CrossRef]
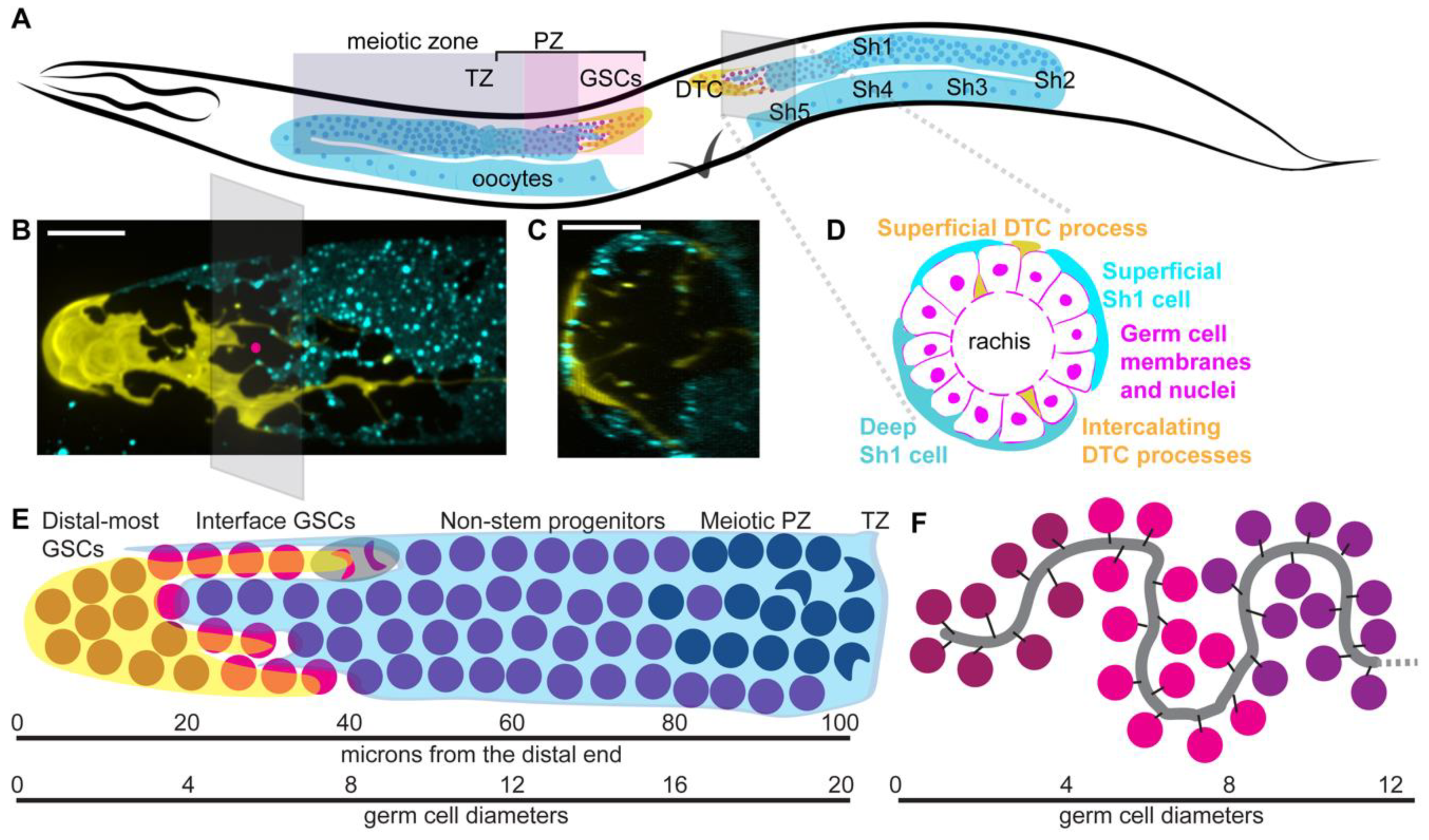
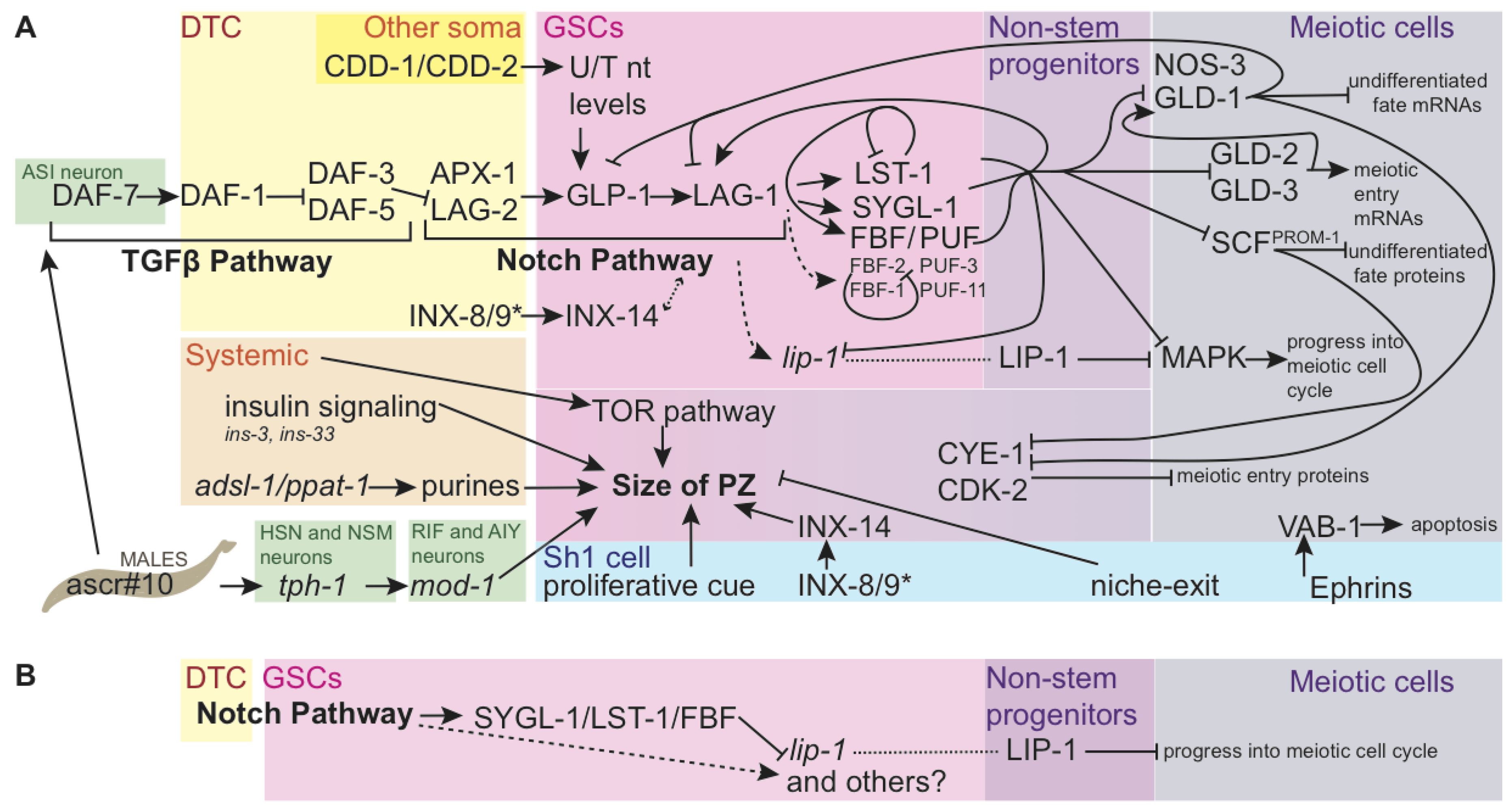
| Attribute | Distal-Most GSCs | Interface GSCs | Non-Stem Progenitors | Source |
|---|---|---|---|---|
| Average position | Distal 2–4 rows, ~0–20 μm | Rows 5–9, ~20–45 μm | Rows 10–16, ~45–100 μm | [6,10] |
| Contact with DTC | Enwrapped by plexus | Major branches | Longest branches only | [26,27] |
| Contact with Sh1 | None | Contact with distal edges | Uniformly covered | [10] |
| Division rate | Lowest | Highest | Lower | [101,103] |
| Division type | Symmetric | Asymmetric | Symmetric | [10] |
| DNA content | Lowest | Higher | Highest | [82] |
| GLD-1 abundance | Lowest | Faint | Highest of non-meiotic cells | [8,30] |
| lst-1 expression | Actively transcribed | Some transcript/protein | Less transcript/protein | [31,84] |
| sygl-1 expression | Actively transcribed | Actively transcribed | Less transcript/protein | [31,84] |
| emb-30(ts) result | Blocks at metaphase | Blocks at metaphase | Differentiates in place | [95] |
| glp-1(ts) result | Differentiates last | Differentiates second | Differentiates first in wave | [94,95] |
| Response to environment | Shrinks during aging; glp-1 independent during and resumes mitotic cycle after starvation | Shrinks during aging; glp-1 independent during and resumes mitotic cycle after starvation | Shrinks during aging; glp-1 independent during and differentiates after starvation | [81,105] |
| Additional notes | Retains position 8+ hours | Descendants leave niche | Accumulates LIP-1 protein | [67,104] |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gordon, K. Recent Advances in the Genetic, Anatomical, and Environmental Regulation of the C. elegans Germ Line Progenitor Zone. J. Dev. Biol. 2020, 8, 14. https://doi.org/10.3390/jdb8030014
Gordon K. Recent Advances in the Genetic, Anatomical, and Environmental Regulation of the C. elegans Germ Line Progenitor Zone. Journal of Developmental Biology. 2020; 8(3):14. https://doi.org/10.3390/jdb8030014
Chicago/Turabian StyleGordon, Kacy. 2020. "Recent Advances in the Genetic, Anatomical, and Environmental Regulation of the C. elegans Germ Line Progenitor Zone" Journal of Developmental Biology 8, no. 3: 14. https://doi.org/10.3390/jdb8030014
APA StyleGordon, K. (2020). Recent Advances in the Genetic, Anatomical, and Environmental Regulation of the C. elegans Germ Line Progenitor Zone. Journal of Developmental Biology, 8(3), 14. https://doi.org/10.3390/jdb8030014




