Abstract
The bone morphogenetic protein (BMP) pathway is best known for its role in promoting bone formation, however it has been shown to play important roles in both development and regeneration of many different tissues. Recent work has shown that the BMP proteins have a number of functions in skeletal muscle, from embryonic to postnatal development. Furthermore, complementary studies have recently demonstrated that specific components of the pathway are required for efficient muscle regeneration.
1. Introduction
As its name implies, the bone morphogenetic protein (BMP) signaling pathway was first identified as a key regulator of bone formation. Urist and colleagues isolated proteins from rabbit bone and then used them to induce bone formation in vitro or in vivo in the rat [1]. Later work showed that these proteins were highly conserved during evolution and most species had a number of paralogues [2,3,4]. Mapping of the mouse short ear gene, essential for skeletal patterning and bone repair, to the Bmp5 locus, was the first demonstration of the importance of this pathway in mice [5]. In contrast to the relatively mild phenotype of Bmp5 deletions in short-eared mice, deletion of either Bmp4 or Bmp2 and their receptors Bmpr1a and Bmpr2 caused early embryonic lethality, with severe defects in mesoderm formation, extraembryonic tissues and cardiac development [6,7,8,9] (Table A1). In addition to demonstrating that certain BMP genes are essential for development, this work implicated this signaling pathway to play important roles in multiple organ systems.
2. Signaling Mechanism
After decades of biochemical work, many components of the BMP signaling pathway and their mode of action have been described. Signaling activation requires two different receptors, first discovered by experiments with a human cell line [10]. Today the pathway consists of at least 20 different ligands, four type 1 receptors, and three type 2 receptors, varying combinations of which give distinct responses [11]. After binding of the BMP ligand, the receptors phosphorylate Smad cofactors, allowing an interaction with Smad4 and translocation of the Smad complex into the nucleus (Figure 1) [12,13,14]. The BMP-specific cofactors are Smad1, Smad5, and Smad9. Surprisingly, while single knockouts for Smad1 and Smad5 are embryonic lethal, Smad9 is dispensable for development [15,16,17,18,19]. The interaction between Smad1/5/8 and Smad4 is BMP-specific, while TGFβ signaling induces an interaction between Smad2/3 and Smad4 [20,21,22]. Furthermore, Smad1 and Smad4 have homologous transcriptional activation domains and their nuclear import leads to transcriptional competence [14,23]. While there is some overlap in the two pathways, the specific components, including extracellular and intracellular inhibitors, receptors, and Smads distinguish BMP from TGFβ signaling, leading to distinct outcomes and specific developmental consequences [24,25]. The regulation of skeletal muscle growth and regeneration by the TGFβ pathway falls beyond the scope of this review; readers may refer to previous works [26,27].
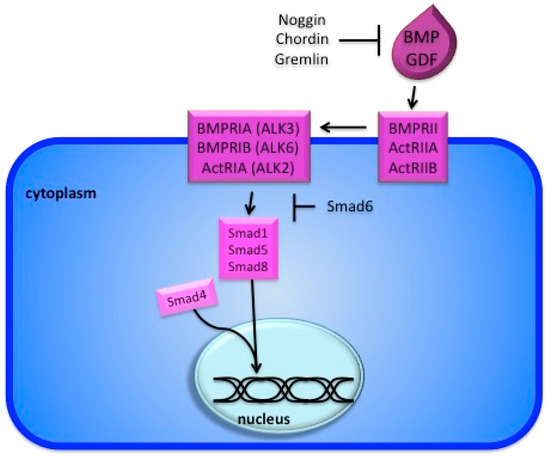
Figure 1.
The BMP signaling pathway. Binding of the ligand to the type II receptor recruits the type I receptor and activates Smad 1/5/8 by phosphorylation. The later engages Smad4, which is shared with the TGFb pathway, and the complex translocates to the nucleus to regulate transcription of target genes (e.g. Id). The pathway is blocked by several inhibitors (e.g. Noggin) as well as the inhibitory Smad6.
The most common BMP targets are Id and Msx genes. The canonical downstream target of BMP signaling is Id1, first identified in osteoblast-like cells [28]. Later work in embryonic stem cells and a number of different cell lines confirmed this target and also identified Id2, Id3, Msx1, and Msx2 as direct targets of the pathway [29]. The Drosophila homologue of the Smad proteins, Mad, binds to a GCCG motif in target genes [30]. When this site is present in a multimerized form, it is sufficient to bind Smad proteins and to confer BMP-specific activation in several mammalian cell lines [31].
Several inhibitors of the pathway have been identified. Work in Xenopus identified Smad6 as an inhibitor of the pathway, which functions by blocking Smad4 association with Smad1 (Figure 1) [32]. A number of secreted BMP inhibitors have also been described. Xenopus explant experiments demonstrated that Bmp4 and noggin have opposing effects on embryonic patterning [33]. Biochemical experiments demonstrated that noggin is able to bind to BMP ligands, rendering them incapable of binding to their receptors [34]. Related antagonists chordin and gremlin, function in the same way (Figure 1, Table A1) [35,36,37].
3. Role in Skeletal Muscle
Experiments with rodent myogenic cell lines were the first to demonstrate an inhibitory role for BMP signaling on muscle differentiation (Figure 2) [38,39]. The addition of BMP ligands to the myogenic L6 and C2C12 cell lines, as well as primary rat cell cultures, inhibited the formation of myotubes and the expression of the myogenic differentiation genes myogenin, MyoD, and Myf5, while it promoted a concomitant trans-differentiation into osteogenic fate [40,41,42,43,44,45,46,47,48,49,50]. Of note, Bmp2-activated muscle tissue was as efficient as autologous bone grafting in the treatment of large bone defects [51]. Furthermore, the implantation of BMP2-soaked beads in rat calf muscle resulted in osteogenic differentiation [52]. On the contrary, culture of myogenic cells in the presence of BMP inhibitors promoted myogenic differentiation (Figure 2, Table A1) [42,43,44,45,46,49,53,54]. However, the role of BMP signaling in muscle differentiation appears to be much more complex in embryonic development and adult muscle regeneration.
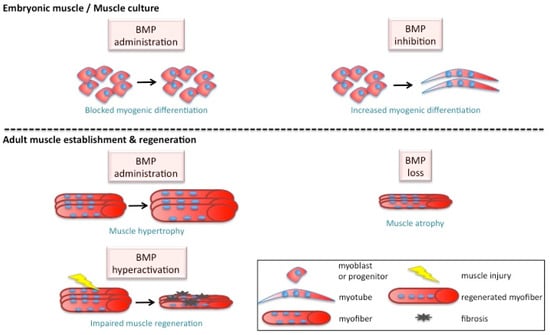
Figure 2.
BMP effect on myogenesis. Fine tuning of BMP is essential for normal muscle growth. On one hand, BMP ensures muscle hypertrophy and its loss leads to muscle atrophy and wasting. On the other hand, hyperactive BMP leads to impaired regeneration and reduced fiber size post-injury. Furthermore, in culture and during development, inhibiting BMP is associated with increased myogenic differentiation, while excess Bmp blocks myogenic differentiation.
4. Regulation of Prenatal Muscle Development
Skeletal muscle is of mesodermal origin, with paraxial mesoderm contributing to limb and trunk muscles, starting by a segmentation process called somitogenesis. As somites mature, dermomyotomal cells will either form the myotome by intercalating between the dorsal dermomyotome and ventral sclerotome or delaminate and migrate from the hypaxial domain specific somites to form the muscle masses of the limbs, diaphragm, and tongue [55,56,57,58]. Stem cells of the developing trunk muscle will then migrate from the dermomyotome into the myotome [59,60,61,62]. On the molecular level, Pax proteins are known to be involved in the control of many lineages during embryogenesis, with Pax3 and Pax7 acting as key regulators in the muscle lineage. Specifically, they orchestrate progenitor survival, proliferation, migration, self-renewal, and they trigger the myogenic program. At later stages, myogenic determination and differentiation are ensured by a family of bHLH transcription factors, collectively referred to as myogenic regulatory factors (family members: Myf5, MyoD, Mrf4/Myf6, myogenin) [63]. With the core myogenic program being very well described, several groups are trying to address impact of different factors or pathways, including the BMP pathway, on this program.
The first studies of BMP in muscle development were largely concerned with the role of the pathway in axis patterning and repression of the myogenic program. BMP diverts paraxial mesoderm fate towards lateral-plate fate, while BMP loss-of-function leads to expansion of the paraxial mesoderm domain [64]. Several groups showed that exogenous BMP ligands could skew Xenopus and avian mesoderm towards a ventral fate, at the expense of dorsal fates, including muscle [35,65,66,67,68], while these findings were more recently confirmed by Smad1 addition to Xenopus dorsal mesoderm [43]. In contrast, addition of the BMP inhibitors noggin or chordin to Xenopus embryo explants or chick embryos was able to drastically increase transcripts of muscle actin and MyoD, respectively (Table A1) [33,35,69,70]. In keeping with a muscle-repressive role, experiments in chick embryo explants demonstrated that BMP signaling favored the differentiation of cartilage over muscle [71]. Of note, co-injection of noggin and constitutively active BMP receptor led to a failure of noggin to dorsalize Xenopus embryos [34]. Thus, initial studies argued for a repressor role of BMP on myogenic fate.
The idea that BMP universally repressed muscle formation was soon challenged (Table A1) by explant experiments demonstrating that Bmp4 acted as a morphogen in Xenopus embryos [72]. A specific range of concentrations actually promoted the expression of Myf5 and formation of muscle. Furthermore the antagonizing function of noggin led to a gradient of Bmp4 activity distinct from the expression pattern of Bmp4. In chick embryo explants, exogenous Bmp4 had no effect on expression of the broad dermomyotomal marker Pax3, but repressed genes involved in myogenic differentiation including MyoD [70]. Further work in chick embryos implicated ectodermal and mesenchymal tissue Bmp2, Bmp4, and Bmp7 in sustaining Pax3 expression in myogenic progenitors of the limb and trunk as well as in proliferation of these progenitors [57,73,74,75]. This effect was shown to be dose-dependent in the limb [73]. Ectopic Bmp4 or constitutively active Bmpr1a during fetal myogenesis increased the number of Pax7+ and differentiating (MF20+) cells, while ectopic Bmp4 and ectopic noggin at E18 respectively increased and decreased the number of satellite cells, which are the stem cells of adult skeletal muscle [76]. Cranial myogenesis also relies upon a balance of ectodermal BMP ligands and the BMP antagonists noggin and gremlin produced in the cranial neural crest and other head tissues [77], while Bmp2 was found to negatively affect myogenic differentiation in embryonic tongues [78]. Specifically, in vivo transduction of cranial paraxial mesoderm with Bmp4 leads to loss of the branchial arch musculature, while activation of skeletal myogenesis correlates with the presence of noggin/gremlin-expressing cranial neural crest cells [77]. The above studies point to an essential contribution of BMP effectors and inhibitors on proper muscle formation.
While it is clear that BMP signaling plays a key role in myogenesis, it remains ambiguous exactly which cells of the muscle express BMP ligands and antagonists, and which cells transduce BMP signals in vivo. During zebrafish muscle development, BMP was specifically absent from the slow-twitch muscle pioneers and medial fast-twitch fibers (MFFs) of the central domain of the myotome, while BMP inhibition by dorsomorphin or Bmpr1a morpholino treatment led to premature activation of the pioneer marker Engrailed and subsequent increase in the numbers of pioneers and MFFs [79,80,81]. Analysis of the BMP expression pattern in chick embryos revealed Bmp2 and Bmp4 presence in the posterior margin and apical ectodermal ridge (Bmp2) or anterior mesenchyme (Bmp4) [81], while Bmp4-expressing cells were flanking Pax3-expressing cells in the trunk [73,74]. In line with these findings, Pax7+ dermomyotomal cells in zebrafish embryos co-expressed pSmad1/5/8 and there was an increase in Pax7+ cells in response to Bmp2b overexpression [82]. On the contrary, noggin was expressed in the mesenchyme core of the developing limb bud, in close proximity to MyoD-expressing cells [73] and in the dorsomedial lip of the dermomyotome of more rostral somites [70]. Work in fetal human muscle showed that most Bmp4- and Bmpr1a-expressing cells were located in between developing fibers, while gremlin was expressed in interstitial progenitor cells and fibers [83]. Ex vivo, sorted Bmp4-expressing cells could give rise to muscle, while they were able to inhibit the differentiation of gremlin-expressing cells. Immunostaining for phosphorylated Smad1/5/8 in chick embryos showed that muscle progenitors at the tips of developing muscles adjacent to tendon were transducing BMP signals [76]. Implantation of Bmp2- or noggin-soaked beads changed the domains of Tcf4-expressing cells [81], a population that has been shown to pre-pattern limb muscles [84]. Furthermore, increasing and decreasing the levels of BMP signaling with viruses led to an expansion or reduction, respectively, of muscle progenitors and muscle size. Genetically increasing BMP signaling by knocking out noggin in mice led to lack or malformation of many muscles, decrease in thick fibers and reduction in Pax7+ cells [85], confirming previous findings on Pax7+ cells by ectopic noggin expression [76]. Analysis of noggin KO in different backgrounds showed that even though the early specification (either Pax3+ progenitor cells or MyoD+ precursor cells) was unaffected, the terminal stages of myogenesis were delayed, with mutant E14.5 embryos and newborns having less fusion and disorganized muscles, respectively [86]. Chordino mutant zebrafish embryos, which lack the chordino BMP inhibitor, exhibit upregulated BMP leading to a dorsally expanded Bmp4 expression domain and enlarged tail somites [87,88].
Three groups tried to address when BMP has a stronger impact on myogenesis, analyzing more or less advanced stages of myogenic progression during prenatal development. In an ex vivo system to develop Pax7+ myogenic progenitors from Xenopus embryos, it was shown that Bmp4 overexpression increased Pax7+ cells and Pax7 expression, an effect counteracted by induction of heat-shock-inducible noggin. Bmp4 treatment even induced paraxial explants to develop Pax7+ cells, although such cells are not formed normally from paraxial tissue. However, this was only observed when the treatment was done at stage 13, but not 18 or 22. The findings were confirmed in vivo, with Bmp4 and the BMP downstream target Msx1 increasing the number of Pax7+ cells, when injected in Xenopus embryos [89]. Transcriptional analysis of Myf5-expressing cells from embryonic and fetal mouse muscle revealed a higher expression of the BMP target genes Id2 and Smad6 in embryonic myoblasts [90]. Bmp4 addition to cultured embryonic myoblasts had no effect, while it inhibited myotube formation in fetal cultures [90]. Together these results suggested that BMP signaling supports embryonic myogenesis but represses muscle differentiation in fetal stages. In keeping with a potential role of BMP in earlier stages of the myogenic differentiation, Bmpr1a ablation in Myf5+ cells led to smaller muscles comprised of less and smaller myofibers and fat accumulation, while Bmpr1a ablation in MyoD+ cells did not affect muscle mass [91]. Overall, it appears that BMP signaling plays a more important role in early prenatal stages of myogenic differentiation.
5. Regulation of Early Postnatal Muscle Growth
The passage from small, proliferative prenatal myogenic populations to a large, post-mitotic adult tissue requires a critical early postnatal period with major changes in terms of hyperplasia, hypertrophy, cell-cycle status and acquisition of quiescence. In the late fetal stage, muscle progenitors are progressively embedded under the basal lamina [59,60,61,62]. At that point, they are described as muscle satellite cells, providing the pool of muscle stem cells required for postnatal growth and muscle repair after achieving quiescence at adult stage [92]. Only a limited number of studies investigated the role of BMP in the crucial early postnatal phase of muscle formation. Several components of the pathway were shown to be expressed both in whole muscle extracts and in satellite cells at different time-points from P3 to P28 in mice and rats (Table A1) [93,94]. Using the muscle fiber-specific myosin light chain 1 fast Cre, Sartori and colleagues found that deletion of Smad4 in muscle fibers led to decreased muscle size (Table A1) [95]. However, in that study it was not investigated whether smaller muscles were the result of defects in embryonic, fetal or postnatal myogenesis. Moreover, the deletion of Smad4 blocks both TGFβ and BMP signaling, making result interpretation difficult. For instance, Myf5-Cre:Smad4f/f mice have small tongues prenatally, which was attributed to the loss of TGFβ signaling rather than BMP, as revealed by subsequent analysis [96]. Hence, BMP is likely to play a role in postnatal myogenesis, but the negative impact of Smad4 blockade on muscle formation should be cautiously interpreted, as Smad4 is in the interconnection of the TGFβ and BMP pathways.
Specific blockade of the BMP pathway with multiple genetic and pharmacological studies in neonatal mice helped clarify the situation [94]. Stantzou and colleagues showed that the loss of BMP signaling in satellite cells led to decreased satellite cell numbers, reduced muscle weight and fiber length. Thus BMP signaling appeared to play a similar role as in embryogenesis by maintaining proliferation of activated satellite cells. Furthermore, introduction of exogenous BMPs into postnatal muscle led to pronounced hypertrophy [97]. Upstream of BMP, miR-26a seems to target Smad1, Smad4, and the BMP target Id3 [98]. Blocking miR-26a in neonatal mice increased proliferation and decreased myogenic differentiation [98]. An unexpected role for the BMP pathway in muscle was found when deleting the receptor Bmpr1a in MyoD- and Myf5-expressing cells [91]. The epaxial muscles of mutants contained smaller myofibers and increased fat infiltration, potentially due to the role of this pathway in endothelial progenitors, some of which also underwent Cre recombination [91].
6. Regulation of Adult Muscle Homeostasis and Regeneration
Once muscle growth ceases, adult muscle is comprised of multinuclear, post-mitotic myofibers, and mononuclear quiescent muscle stem cells. In response to injuries or homeostatic needs, satellite cells become activated and support the formation of new myofibers, while a subpopulation will renew the quiescent pool for future needs. The skeletal muscle niche also includes a microvascular network, neurons, immune cells, and interstitial populations of mesenchymal progenitor cells, all of which participate in different stages of muscle maintenance and regeneration (for a comprehensive review see [99]). The role of BMP on adult muscle maintenance and repair has been investigated by analysis of homeostatic and regenerating muscle, respectively, by overexpression or deletion of different components of the pathway.
Two studies reported the effect of BMP administration on homeostatic (resting) muscle and both groups demonstrated that BMP promotes hypertrophy under these conditions (Figure 2). In the absence of injury, administration of Bmp7, Follistatin, or Bmpr1a led to increased Smad1/5 phosphorylation and subsequently fiber hypertrophy, mediated by the IGF and AKT/mTOR axes [95,97]. Hypertrophy was counteracted by simultaneous injection of Smad6 and Bmp7 or Smad6 and Follistatin [97]. Noggin administration or Smad1/5 knock-down also led to muscle wasting due to atrophy [95]. On the contrary, loss of the downstream Id1 and Id3 was not sufficient to induce a muscle phenotype [100]. Thus, in the absence of muscle injury (i.e., homeostatic muscle), exogenous BMP promotes muscle hypertrophy, raising the question of whether BMP is also involved in the generation of new muscle fibers following muscle injury and the associated muscle fiber loss and satellite cell activation.
Work from several groups established that quiescent satellite cells are pSmad-negative, but BMP signaling is active in adult regenerating muscle, in which satellite cells are activated in response to injury (Table A1) [44,45,76,91,100]. Of note, over a third of satellite cells co-express Smad1/5/8 and the Smad targets Id1 and Id3 at day 3 post-injury [100], while pSmad was also found in myonuclei and connective tissue cells [76]. BMP signaling through Acvr1, Bmpr1a, and Bmpr1b was identified as an important cue to stimulate cell cycle entry of differentiated myotubes and muscle fibers during newt limb regeneration [101]. Loss of Id1 and Id3 delays regeneration by reduction of satellite cell numbers and satellite cell proliferation at day 3 after injury [100]. Of note, Pax7 has been found to block premature differentiation of quiescent satellite cells by inducing the expression of Id2 and Id3, with quiescent satellite cells being Pax7+Id3+ [102]. Ono and colleagues used single fiber cultures to examine the involvement of BMP signaling in satellite cell activation. They first showed that Bmpr1a, pSmad1/5/8, Smad4, and noggin are all absent from the quiescent population and induced by culture, with noggin expressed in differentiating myogenin+ cells [45]. Furthermore, they found that exogenous BMP treatment inhibited differentiation of multiple cultures, and blockade of BMP signaling during cardiotoxin-induced muscle regeneration delayed the repair process [45]. Regeneration was also delayed when BMP was overactivated by relieving the miR-26a block on Smad1/Smad4/Id3 during muscle repair [98]. Similarly, hyperactivation of BMP signaling through the receptor Acvr1 led to a decrease in fiber size and an increase in fibrosis post-injury [103], showing that fine tuning of BMP levels is essential during the regeneration process (Figure 2). The effects of hyperactive Acvr1 were counteracted by rapamycin, which improved myofiber size and dissolved fibrosis [103]. Finally, BMP electroporation into regenerating rat muscles led to differentiation into osteogenic cells [104], corroborating in vitro results of BMP driving muscle cell lines into a bone fate [40,41,42,43,44,45,46,47,48,49,50].
Apart from satellite cells, BMP has been implicated in the regulation of additional cell populations during muscle regeneration. BMP inhibition in mesangioblasts contributed to enhanced grafting potential and muscle recovery [105]. Similarly, noggin treatment of bone marrow-derived mesenchymal stromal cells led to more dystrophin expression after engraftment into mice that lack dystrophin, a structural protein that maintains myofiber stability. On the contrary, Bmp4 treatment resulted in reduced dystrophin restoration [106]. Huang and colleagues found increased fat deposition in cardiotoxin-induced regenerating muscle of MyoD-Cre:Bmpr1af/f and Myf5-Cre:Bmpr1af/f mice. Their analysis suggested that BMP signaling in myo-endothelial progenitors repressed an adipogenic program both in developing and regenerating muscle [91].
Several studies also investigated the effects of conditional ablation of Smad4 on muscle regeneration (Table A1). Analysis of Myl1-Cre; Smad4f/f mice showed that denervation injury significantly increased the level of atrophy in mutant mice, due to an increase in transcription of ubiquitin ligases, including Musa1 [95,97]. Later work demonstrated that inducible deletion of Smad4 with the use of a Pax7-CreERT2 allele led to significant impairment of muscle regeneration following cardiotoxin injection, with decreased fiber size and increased fibrosis [48]. On the contrary, knock-down of Smad4 in all muscle populations during regeneration (siRNA or miR-431 administration at D2 and D5 post-injury) leads to fiber hypertrophy [107]. However, these findings must be interpreted carefully, as the deletion of Smad4 abrogates TGF-β signaling, in addition to the BMP pathway.
The role of BMP in aging or pathological muscles is less characterized. One study has shown that late middle-aged rats have lower BMP (measured by Bmp2, Bmp7, and pSmad1/5/8 levels) than young controls, while BMP levels can increase with exercise [108]. On the contrary, muscles exhibiting wasting associated with motor nerve transaction, degeneration or disuse were shown to have increased Smad1/5 phosphorylation and suppressed Smad6 and noggin, while administration of Bmp7 or Bmpr1a prior to denervation partially rescued the muscle wasting [97]. Deficiency in Fibrillin2, which is inversely correlated with BMP signaling, led to myopathic features in mice, including poor muscle architecture, reduction in muscle mass, and increase in centronucleated fibers, which are indicative of ongoing damage and regeneration [109]. The myopathic phenotype was exacerbated by Bmp7 overexpression but rescued by Bmp7 partial ablation or noggin overexpression [109]. In Duchenne muscular dystrophy (DMD), one of the most prevalent myopathies, Bmp4 was shown to be higher than in control samples and this increase was associated with the defective differentiation of myoblasts into myotubes [110]. Increased BMP was also observed in DMD iPSC-derived myoblasts [111]. Expectedly, noggin administration to an animal model of the disease induced the expression of myogenic markers (MyoD, myogenin) and mildly alleviated the disease phenotype [63]. Furthermore, Bmp4 was increased in patient myoblasts of the LMNA myopathy, while Bmp4 downregulation or Smad6 overexpression in mutant myoblast cultures promoted myogenic differentiation in vitro [112]. In Myhre syndrome, which is characterized by generalized muscular hypertrophy, there are Smad4 mutations associated with 8-fold increase in pSmad2/3 and 11-fold increase in pSmad1/5/8 [113,114]. Fibrodysplasia ossificans progressiva (FOP) is an extremely rare condition in which muscle and other soft tissues are spontaneously transformed into bone, and it is caused by a specific activating mutation in Acvr1 [115].
7. Perspectives
After several decades of work on BMP signaling in myogenic cells, it is clear that there is significant overlap in the function of this pathway during development and regeneration (Figure 2). In both scenarios, activation of the pathway seems crucial to maintaining expression of satellite cell master regulators like Pax3, while its timely repression is likewise required for differentiation and formation of muscle fibers. The main difference in BMP requirements between developmental and adult myogenesis seems to be that in the prenatal stage BMP has been shown to act as morphogen. However, several questions remain open regarding the levels and dynamics of BMP signaling in regeneration. Many groups have shown that a precise level of BMP signaling, tempered by the presence of inhibitory proteins, is crucial for initial stages of myogenic specification. We do not have the same detailed understanding of BMP levels in regenerating muscle, necessitating the development of new functional approaches, for instance that could modify the output of the BMP signaling pathway. It is also worthwhile to investigate the role of this pathway in other cell types that make up the skeletal muscle niche, including endothelial cells, immune cells, or mesenchymal progenitors, both during development and regeneration.
Funding
The FR laboratory receives funding from Association Française contre les Myopathies (AFM) via TRANSLAMUSCLE (project 19507), Labex REVIVE (ANR-10-LABX-73), Agence Nationale pour la Recherche (ANR) grants BMP-myomass (ANR-12-BSV1-0038- 04) and BMP-MyoStem (ANR-16-CE14-0002-03).
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A. Skeletal Muscle Glossary
Appendix A.1. Core Muscle Cells
Muscle progenitor cells: the stem cells building the skeletal muscle tissue during development.
Myoblasts: myogenic precursor cells.
Myocytes: mononuclear cells, engaged to myogenic differentiation.
Myotubes: multinucleated cells emanating from fusion of myogenic cells.
Myofibers: long syncytial multinucleated post-mitotic cells that comprise the adult skeletal muscle.
Satellite cells: the skeletal muscle stem cells, located on the periphery of the myofibers under the basal lamina. Quiescent in the adult muscle, but capable of re-entering the cell cycle, activating the myogenic program, forming new myofibers, and renewing the quiescent pool.
Appendix A.2. Rough Molecular Regulation
Pax3/7: paired box transcription factors that regulate myogenic progenitors and stem cells.
Myf5, MyoD, Mrf4/Myf6, myogenin: a family of basic helix-loop-helix (bHLH) factors that ensure progenitor (during development) or stem cell (postnatal satellite cells) commitment and differentiation into the myogenic lineage. Collectively referred to as myogenic regulatory factors (MRFs).
Appendix A.3. Muscle Origin and Regeneration
Somites: segmented units of paraxial mesoderm that give rise, among other tissues, to muscle.
Developmental myogenesis: embryonic and fetal waves of myogenesis, including progressive differentiation and fusion to form myofibers.
Early postnatal myogenesis: critical period during which (a) muscle size increases by hyperplasia (nuclei number increase in myofibers through fusion of postnatal satellite cells) and hypertrophy (fiber size increase), and (b) satellite cell quiescence is established.
Muscle regeneration: injury-induced reestablishment of the structural and functional properties of the skeletal muscle. Its phases include necrosis, inflammation, myogenesis/fusion, remodeling/maturation.
Appendix B. Studying the Skeletal Muscle (Methodologies)
Appendix B.1. In Vitro
- (a)
- Murine myogenic cell line C2C12,
- (b)
- Primary myoblasts extracted by enzymatic dissociation of muscles and FACS isolation of their myoblasts.
Both cultures are proliferative under high serum conditions, but serum starvation induces differentiation into myogenin + myocytes and eventually MyHC + myotubes.
Appendix B.2. Ex Vivo
- (a)
- Explant cultures (mostly for developmental studies),
- (b)
- Single myofiber cultures (isolation and culture of single fibers from early postnatal and adult animals).
Appendix B.3. In Vivo
- (a)
- Gross macroscopic analysis or cryosection analysis of resting (uninjured) muscles,
- (b)
- Lesions with physical (e.g., cryoinjury) or chemical (e.g., BaCl2) techniques for induction of regeneration and study of muscle repair.
Appendix C

Table A1.
Major Findings on BMP Effects on Myogenesis.
Table A1.
Major Findings on BMP Effects on Myogenesis.
| Finding [Reference] | Stage |
|---|---|
| Bmp signaling is necessary for mesoderm formation [6,7,8,9] | development |
| In vitro inhibition of Bmp signaling promotes muscle differentiation [42,43,44,45,46,49,53,54] | in vitro evaluation |
| In vivo inhibition of Bmp signaling promotes muscle differentiation [33,35,69,70] | development |
| Specific Bmp ligand concentrations promote embryonic muscle differentiation in vivo [57,72,73,74,75,76,77] | development |
| Bmp signaling is required for postnatal muscle growth [93,94,95] | early postnatal growth |
| Bmp signaling is activated during muscle regeneration [44,45,76,91,100] | adulthood (regeneration assay) |
| Smad4 deletion is detrimental to muscle regeneration [48,95,97] | adulthood (regeneration assay) |
References
- Urist, M.R.; Mikulski, A.; Lietze, A. Solubilized and insolubilized bone morphogenetic protein. Proc. Natl. Acad. Sci. USA 1979, 76, 1828–1832. [Google Scholar] [CrossRef] [PubMed]
- Celeste, A.J.; Iannazzi, J.A.; Taylor, R.C.; Hewick, R.M.; Rosen, V.; Wang, E.A.; Wozney, J.M. Identification of transforming growth factor beta family members present in bone-inductive protein purified from bovine bone. Proc. Natl. Acad. Sci. USA 1990, 87, 9843–9847. [Google Scholar] [CrossRef] [PubMed]
- Ozkaynak, E.; Rueger, D.C.; Drier, E.A.; Corbett, C.; Ridge, R.J.; Sampath, T.K.; Oppermann, H. OP-1 cDNA encodes an osteogenic protein in the TGF-beta family. EMBO J. 1990, 9, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Wozney, J.M.; Rosen, V.; Celeste, A.J.; Mitsock, L.M.; Whitters, M.J.; Kriz, R.W.; Hewick, R.M.; Wang, E.A. Novel regulators of bone formation: Molecular clones and activities. Science 1988, 242, 1528–1534. [Google Scholar] [CrossRef]
- Kingsley, D.M.; Bland, A.E.; Grubber, J.M.; Marker, P.C.; Russell, L.B.; Copeland, N.G.; Jenkins, N.A. The mouse short ear skeletal morphogenesis locus is associated with defects in a bone morphogenetic member of the TGF beta superfamily. Cell 1992, 71, 399–410. [Google Scholar] [CrossRef]
- Mishina, Y.; Suzuki, A.; Ueno, N.; Behringer, R.R. Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 1995, 9, 3027–3037. [Google Scholar] [CrossRef]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, X.; We, R.; Derynck, R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature 1996, 383, 168–172. [Google Scholar] [CrossRef]
- Beppu, H.; Kawabata, M.; Hamamoto, T.; Chytil, A.; Minowa, O.; Noda, T.; Miyazono, K. BMP type II receptor is required for gastrulation and early development of mouse embryos. Dev. Biol. 2000, 221, 249–258. [Google Scholar] [CrossRef]
- Paralkar, V.M.; Hammonds, R.G.; Reddi, A.H. Identification and characterization of cellular binding proteins (receptors) for recombinant human bone morphogenetic protein 2B, an initiator of bone differentiation cascade. Proc. Natl. Acad. Sci. USA 1991, 88, 3397–3401. [Google Scholar] [CrossRef]
- Antebi, Y.E.; Linton, J.M.; Klumpe, H.; Bintu, B.; Gong, M.; Su, C.; McCardell, R.; Elowitz, M.B. Combinatorial signal perception in the BMP pathway. Cell 2017, 170, 1184–1196. [Google Scholar] [CrossRef] [PubMed]
- Hoodless, P.A.; Haerry, T.; Abdollah, S.; Stapleton, M.; O’Connor, M.B.; Attisano, L.; Wrana, J.L. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell 1996, 85, 489–500. [Google Scholar] [CrossRef]
- Thomsen, G.H. Xenopus mothers against decapentaplegic is an embryonic ventralizing agent that acts downstream of the BMP-2/4 receptor. Development 1996, 122, 2359–2366. [Google Scholar] [PubMed]
- Kretzschmar, M.; Liu, F.; Hata, A.; Doody, J.; Massagué, J. The TGF-beta family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997, 11, 984–995. [Google Scholar] [CrossRef]
- Chang, H.; Huylebroeck, D.; Verschueren, K.; Guo, Q.; Matzuk, M.M.; Zwijsen, A. Smad5 knockout mice die at mid-gestation due to multiple embryonic and extraembryonic defects. Development 1999, 126, 1631–1642. [Google Scholar]
- Yang, X.; Castilla, L.H.; Xu, X.; Li, C.; Gotay, J.; Weinstein, M.; Liu, P.P.; Deng, C.X. Angiogenesis defects and mesenchymal apoptosis in mice lacking SMAD5. Development 1999, 126, 1571–1580. [Google Scholar]
- Lechleider, R.J.; Ryan, J.L.; Garrett, L.; Eng, C.; Deng, C.; Wynshaw-Boris, A.; Roberts, A.B. Targeted mutagenesis of Smad1 reveals an essential role in chorioallantoic fusion. Dev. Biol. 2001, 240, 157–167. [Google Scholar] [CrossRef]
- Tremblay, K.D.; Dunn, N.R.; Robertson, E.J. Mouse embryos lacking Smad1 signals display defects in extra-embryonic tissues and germ cell formation. Development 2001, 128, 3609–3621. [Google Scholar]
- Hester, M.; Thompson, J.C.; Mills, J.; Liu, Y.; El-Hodiri, H.M.; Weinstein, M. Smad1 and Smad8 function similarly in mammalian central nervous system development. Mol Cell Biol. 2005, 25, 4683–4692. [Google Scholar] [CrossRef][Green Version]
- Lagna, G.; Hata, A.; Hemmati-Brivanlou, A.; Massagué, J. Partnership between DPC4 and SMAD proteins in TGF-beta signalling pathways. Nature 1996, 383, 832–836. [Google Scholar] [CrossRef]
- Chen, Y.; Bhushan, A.; Vale, W. Smad8 mediates the signaling of the ALK-2 [corrected] receptor serine kinase. Proc. Natl. Acad. Sci. USA 1997, 94, 12938–12943. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Kato, Y.; Chen, D.; Harris, S.E.; Mundy, G.R.; Yoneda, T. Smad5 and DPC4 are key molecules in mediating BMP-2-induced osteoblastic differentiation of the pluripotent mesenchymal precursor cell line C2C12. J. Biol. Chem. 1998, 273, 1872–1879. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hata, A.; Baker, J.C.; Doody, J.; Cárcamo, J.; Harland, R.M.; Massagué, J. A human Mad protein acting as a BMP-regulated transcriptional activator. Nature 1996, 381, 620–623. [Google Scholar] [CrossRef] [PubMed]
- Graff, J.M.; Bansal, A.; Melton, D.A. Xenopus Mad proteins transduce distinct subsets of signals for the TGF beta superfamily. Cell 1996, 85, 479–487. [Google Scholar] [CrossRef]
- Yingling, J.M.; Das, P.; Savage, C.; Zhang, M.; Padgett, R.W.; Wang, X.F. Mammalian dwarfins are phosphorylated in response to transforming growth factor beta and are implicated in control of cell growth. Proc. Natl. Acad. Sci. USA 1996, 93, 8940–8944. [Google Scholar] [CrossRef]
- Sartori, R.; Gregorevic, P.; Sandri, M. TGFβ and BMP signaling in skeletal muscle: Potential significance for muscle-related disease. Trends Endocrinol. Metab. 2014, 25, 464–471. [Google Scholar] [CrossRef]
- Kim, J.; Lee, J. Role of transforming growth factor-β in muscle damage and regeneration: Focused on eccentric muscle contraction. J. Exerc. Rehabil. 2017, 13, 621–626. [Google Scholar] [CrossRef]
- Ogata, T.; Wozney, J.M.; Benezra, R.; Noda, M. Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc. Natl. Acad. Sci. USA 1993, 90, 9219–9222. [Google Scholar] [CrossRef]
- Hollnagel, A.; Oehlmann, V.; Heymer, J.; Rüther, U.; Nordheim, A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J. Biol. Chem. 1999, 274, 19838–19845. [Google Scholar] [CrossRef]
- Kim, J.; Johnson, K.; Chen, H.J.; Carroll, S.; Laughon, A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature 1997, 388, 304–308. [Google Scholar] [CrossRef]
- Kusanagi, K.; Inoue, H.; Ishidou, Y.; Mishima, H.K.; Kawabata, M.; Miyazono, K. Characterization of a bone morphogenetic protein-responsive Smad-binding element. Mol. Biol. Cell 2000, 11, 555–565. [Google Scholar] [CrossRef]
- Hata, A.; Lagna, G.; Massagué, J.; Hemmati-Brivanlou, A. Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev. 1998, 12, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Re’em-Kalma, Y.; Lamb, T.; Frank, D. Competition between noggin and bone morphogenetic protein 4 activities may regulate dorsalization during Xenopus development. Proc. Natl. Acad. Sci. USA 1995, 92, 12141–12145. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, L.B.; De Jesús-Escobar, J.M.; Harland, R.M. The Spemann organizer signal noggin binds and inactivates bone morphogenetic protein 4. Cell 1996, 86, 599–606. [Google Scholar] [CrossRef]
- Piccolo, S.; Sasai, Y.; Lu, B.; De Robertis, E.M. Dorsoventral patterning in Xenopus: Inhibition of ventral signals by direct binding of chordin to BMP-4. Cell 1996, 86, 589–598. [Google Scholar] [CrossRef]
- Hsu, D.R.; Economides, A.N.; Wang, X.; Eimon, P.M.; Harland, R.M. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol. Cell 1998, 1, 673–683. [Google Scholar] [CrossRef]
- Larraín, J.; Bachiller, D.; Lu, B.; Agius, E.; Piccolo, S.; De Robertis, E.M. BMP-binding modules in chordin: A model for signalling regulation in the extracellular space. Development 2000, 127, 821–830. [Google Scholar]
- Yamaguchi, A.; Katagiri, T.; Ikeda, T.; Wozney, J.M.; Rosen, V.; Wang, E.A.; Kahn, A.J.; Suda, T.; Yoshiki, S. Recombinant human bone morphogenetic protein-2 stimulates osteoblastic maturation and inhibits myogenic differentiation in vitro. J. Cell Biol. 1991, 113, 681–687. [Google Scholar] [CrossRef]
- Murray, S.S.; Murray, E.J.; Glackin, C.A.; Urist, M.R. Bone morphogenetic protein inhibits differentiation and affects expression of helix-loop-helix regulatory molecules in myoblastic cells. J. Cell Biochem. 1993, 53, 51–60. [Google Scholar] [CrossRef]
- Katagiri, T.; Yamaguchi, A.; Komaki, M.; Abe, E.; Takahashi, N.; Ikeda, T.; Rosen, V.; Wozney, J.M.; Fujisawa-Sehara, A.; Suda, T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J. Cell Biol. 1994, 127, 1755–1766. [Google Scholar] [CrossRef]
- Chalaux, E.; López-Rovira, T.; Rosa, J.L.; Bartrons, R.; Ventura, F. JunB is involved in the inhibition of myogenic differentiation by bone morphogenetic protein-2. J. Biol. Chem. 1998, 273, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Ginn, S.L.; Lek, M.; North, K.N.; Alexander, I.E.; Little, D.G.; Schindeler, A. Myoblast sensitivity and fibroblast insensitivity to osteogenic conversion by BMP-2 correlates with the expression of Bmpr-1a. BMC Musculoskelet. Disord. 2009, 10, 51. [Google Scholar] [CrossRef] [PubMed]
- Nojima, J.; Kanomata, K.; Takada, Y.; Fukuda, T.; Kokabu, S.; Ohte, S.; Takada, T.; Tsukui, T.; Yamamoto, T.S.; Sasanuma, H.; et al. Dual roles of smad proteins in the conversion from myoblasts to osteoblastic cells by bone morphogenetic proteins. J. Biol. Chem. 2010, 285, 15577–15586. [Google Scholar] [CrossRef] [PubMed]
- Friedrichs, M.; Wirsdöerfer, F.; Flohé, S.B.; Schneider, S.; Wuelling, M.; Vortkamp, A. BMP signaling balances proliferation and differentiation of muscle satellite cell descendants. BMC Cell Biol. 2011, 12, 26. [Google Scholar] [CrossRef] [PubMed]
- Ono, Y.; Calhabeu, F.; Morgan, J.E.; Katagiri, T.; Amthor, H.; Zammit, P.S. BMP signalling permits population expansion by preventing premature myogenic differentiation in muscle satellite cells. Cell Death Differ. 2011, 18, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Terada, K.; Misao, S.; Katase, N.; Nishimatsu, S.; Nohno, T. Interaction of Wnt signaling with BMP/Smad signaling during the transition from cell proliferation to myogenic differentiation in mouse myoblast-derived cells. Int. J. Cell Biol. 2013, 2013, 616294. [Google Scholar] [CrossRef] [PubMed]
- Almodóvar, J.; Guillot, R.; Monge, C.; Vollaire, J.; Selimović, S.; Coll, J.L.; Khademhosseini, A.; Picart, C. Spatial patterning of BMP-2 and BMP-7 on biopolymeric films and the guidance of muscle cell fate. Biomaterials 2014, 35, 3975–3985. [Google Scholar] [CrossRef]
- Paris, N.D.; Soroka, A.; Klose, A.; Liu, W.; Chakkalakal, J.V. Smad4 restricts differentiation to promote expansion of satellite cell derived progenitors during skeletal muscle regeneration. Elife 2016, 5, e19484. [Google Scholar] [CrossRef]
- Dörpholz, G.; Murgai, A.; Jatzlau, J.; Horbelt, D.; Belverdi, M.P.; Heroven, C.; Schreiber, I.; Wendel, G.; Ruschke, K.; Stricker, S.; et al. IRS4, a novel modulator of BMP/Smad and Akt signalling during early muscle differentiation. Sci. Rep. 2017, 7, 8778. [Google Scholar] [CrossRef]
- Ferrazzo, P.C.; Niccoli, S.; Khaper, N.; Rathbone, C.R.; Lees, S.J. Ascorbic acid diminishes bone morphogenetic protein 2-induced osteogenic differentiation of muscle precursor cells. Muscle Nerve 2019, 59, 501. [Google Scholar] [CrossRef]
- Betz, O.B.; Betz, V.M.; Schröder, C.; Penzkofer, R.; Göttlinger, M.; Mayer-Wagner, S.; Augat, P.; Jansson, V.; Müller, P.E. Repair of large segmental bone defects: BMP-2 gene activated muscle grafts vs. autologous bone grafting. BMC Biotechnol. 2013, 13, 65. [Google Scholar] [CrossRef] [PubMed]
- Okubo, Y.; Bessho, K.; Fujimura, K.; Kusumoto, K.; Ogawa, Y.; Iizuka, T. Expression of bone morphogenetic protein in the course of osteoinduction by recombinant human bone morphogenetic protein-2. Clin. Oral. Implant. Res. 2002, 13, 80. [Google Scholar] [CrossRef] [PubMed]
- Shi, S.; Hoogaars, W.M.; de Gorter, D.J.; van Heiningen, S.H.; Lin, H.Y.; Hong, C.C.; Kemaladewi, D.U.; Aartsma-Rus, A.; ten Dijke, P.; Hoen, P.A. BMP antagonists enhance myogenic differentiation and ameliorate the dystrophic phenotype in a DMD mouse model. NeuroBiol. Dis. 2011, 41, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Horbelt, D.; Boergermann, J.H.; Chaikuad, A.; Alfano, I.; Williams, E.; Lukonin, I.; Timmel, T.; Bullock, A.N.; Knaus, P. Small molecules dorsomorphin and LDN-193189 inhibit myostatin/GDF8 signaling and promote functional myoblast differentiation. J. Biol. Chem. 2015, 290, 3390–3404. [Google Scholar] [CrossRef]
- Brohmann, H.; Jagla, K.; Birchmeier, C. The role of Lbx1 in migration of muscle precursor cells. Development 2000, 127, 437–445. [Google Scholar]
- Gros, J.; Scaal, M.; Marcelle, C. A two-Step mechanism for myotome formation in chick. Dev. Cell 2004, 6, 875–882. [Google Scholar] [CrossRef]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. Divergent functions of murine Pax3 and Pax7 in limb muscle development. Genes Dev. 2004, 18, 1088–1105. [Google Scholar] [CrossRef]
- Vasyutina, E.; Stebler, J.; Brand-Saberi, B.; Schulz, S.; Raz, E.; Birchmeier, C. CXCR4 and Gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev. 2005, 19, 2187–2198. [Google Scholar] [CrossRef]
- Ben-Yair, R.; Kalcheim, C. Lineage analysis of the avian dermomyotome sheet reveals the existence of single cells with both dermal and muscle progenitor fates. Development 2005, 132, 689–701. [Google Scholar] [CrossRef]
- Gros, J.; Manceau, M.; Thomé, V.; Marcelle, C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 2005, 435, 954–958. [Google Scholar] [CrossRef]
- Kassar-Duchossoy, L.; Giacone, E.; Gayraud-Morel, B.; Jory, A.; Gomès, D.; Tajbakhsh, S. Pax3/Pax7 mark a novel population of primitive myogenic cells during development. Genes Dev. 2005, 19, 1426–1431. [Google Scholar] [CrossRef] [PubMed]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nature 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed]
- Bismuth, K.; Relaix, F. Genetic regulation of skeletal muscle development. Exp. Cell Res. 2010, 316, 3081–3086. [Google Scholar] [CrossRef] [PubMed]
- Tonegawa, A.; Funayama, N.; Ueno, N.; Takahashi, Y. Mesodermal subdivision along the mediolateral axis in chicken controlled by different concentrations of BMP-4. Development 1997, 124, 1975–1984. [Google Scholar]
- Dale, L.; Howes, G.; Price, B.M.; Smith, J.C. Bone morphogenetic protein 4: A ventralizing factor in early Xenopus development. Development 1992, 115, 573–585. [Google Scholar]
- Jones, C.M.; Lyons, K.M.; Lapan, P.M.; Wright, C.V.; Hogan, B.L. DVR-4 (bone morphogenetic protein-4) as a posterior-ventralizing factor in Xenopus mesoderm induction. Development 1992, 115, 639–647. [Google Scholar]
- Pourquié, O.; Fan, C.M.; Coltey, M.; Hirsinger, E.; Watanabe, Y.; Bréant, C.; Francis-West, P.; Brickell, P.; Tessier-Lavigne, M.; Le Douarin, N.M. Lateral and axial signals involved in avian somite patterning: A role for BMP4. Cell 1996, 84, 461–471. [Google Scholar] [CrossRef]
- Nikaido, M.; Tada, M.; Takeda, H.; Kuroiwa, A.; Ueno, N. In vivo analysis using variants of zebrafish BMPR-IA: Range of action and involvement of BMP in ectoderm patterning. Development 1999, 126, 181–190. [Google Scholar]
- Hirsinger, E.; Duprez, D.; Jouve, C.; Malapert, P.; Cooke, J.; Pourquié, O. Noggin acts downstream of Wnt and Sonic Hedgehog to antagonize BMP4 in avian somite patterning. Development 1997, 124, 4605–4614. [Google Scholar]
- Reshef, R.; Maroto, M.; Lassar, A.B. Regulation of dorsal somitic cell fates: BMPs and Noggin control the timing and pattern of myogenic regulator expression. Genes Dev. 1998, 12, 290–303. [Google Scholar] [CrossRef]
- Duprez, D.M.; Coltey, M.; Amthor, H.; Brickell, P.M.; Tickle, C. Bone morphogenetic protein-2 (BMP-2) inhibits muscle development and promotes cartilage formation in chick limb bud cultures. Dev. Biol. 1996, 174, 448–452. [Google Scholar] [CrossRef] [PubMed]
- Dosch, R.; Gawantka, V.; Delius, H.; Blumenstock, C.; Niehrs, C. Bmp-4 acts as a morphogen in dorsoventral mesoderm patterning in Xenopus. Development 1997, 124, 2325–2334. [Google Scholar] [PubMed]
- Amthor, H.; Christ, B.; Weil, M.; Patel, K. The importance of timing differentiation during limb muscle development. Curr. Biol. 1998, 8, 642–652. [Google Scholar] [CrossRef]
- Amthor, H.; Christ, B.; Patel, K. A molecular mechanism enabling continuous embryonic muscle growth - a balance between proliferation and differentiation. Development 1999, 126, 1041–1053. [Google Scholar]
- Amthor, H.; Christ, B.; Rashid-Doubell, F.; Kemp, C.F.; Lang, E.; Patel, K. Follistatin regulates bone morphogenetic protein-7 (BMP-7) activity to stimulate embryonic muscle growth. Dev. Biol. 2002, 243, 115–127. [Google Scholar] [CrossRef]
- Wang, H.; Noulet, F.; Edom-Vovard, F.; Tozer, S.; Le Grand, F.; Duprez, D. Bmp signaling at the tips of skeletal muscles regulates the number of fetal muscle progenitors and satellite cells during development. Dev. Cell 2010, 18, 643–654. [Google Scholar] [CrossRef]
- Tzahor, E.; Kempf, H.; Mootoosamy, R.C.; Poon, A.C.; Abzhanov, A.; Tabin, C.J.; Dietrich, S.; Lassar, A.B. Antagonists of Wnt and BMP signaling promote the formation of vertebrate head muscle. Genes Dev. 2003, 17, 3087–3099. [Google Scholar] [CrossRef]
- Aoyama, K.; Yamane, A.; Suga, T.; Suzuki, E.; Fukui, T.; Nakamura, Y. Bone morphogenetic protein-2 functions as a negative regulator in the differentiation of myoblasts, but not as an inducer for the formations of cartilage and bone in mouse embryonic tongue. BMC Dev. Biol. 2011, 11, 44. [Google Scholar] [CrossRef]
- Dolez, M.; Nicolas, J.F.; Hirsinger, E. Laminins, via heparan sulfate proteoglycans, participate in zebrafish myotome morphogenesis by modulating the pattern of Bmp responsiveness. Development 2011, 138, 97–106. [Google Scholar] [CrossRef]
- Maurya, A.K.; Tan, H.; Souren, M.; Wang, X.; Wittbrodt, J.; Ingham, P.W. Integration of Hedgehog and BMP signalling by the engrailed2a gene in the zebrafish myotome. Development 2011, 138, 755–765. [Google Scholar] [CrossRef]
- Bonafede, A.; Köhler, T.; Rodriguez-Niedenführ, M.; Brand-Saberi, B. BMPs restrict the position of premuscle masses in the limb buds by influencing Tcf4 expression. Dev. Biol. 2006, 299, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.E.; Bird, N.C.; Devoto, S.H. BMP regulation of myogenesis in zebrafish. Dev. Dyn. 2010, 239, 806–817. [Google Scholar] [CrossRef] [PubMed]
- Frank, N.Y.; Kho, A.T.; Schatton, T.; Murphy, G.F.; Molloy, M.J.; Zhan, Q.; Ramoni, M.F.; Frank, M.H.; Kohane, I.S.; Gussoni, E. Regulation of myogenic progenitor proliferation in human fetal skeletal muscle by BMP4 and its antagonist Gremlin. J. Cell Biol. 2006, 175, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Kardon, G.; Harfe, B.D.; Tabin, C.J. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Dev. Cell 2003, 5, 937–944. [Google Scholar] [CrossRef]
- Costamagna, D.; Mommaerts, H.; Sampaolesi, M.; Tylzanowski, P. Noggin inactivation affects the number and differentiation potential of muscle progenitor cells in vivo. Sci. Rep. 2016, 6, 31949. [Google Scholar] [CrossRef]
- Tylzanowski, P.; Mebis, L.; Luyten, F.P. The Noggin null mouse phenotype is strain dependent and haploinsufficiency leads to skeletal defects. Dev. Dyn. 2006, 235, 1599–1607. [Google Scholar] [CrossRef]
- Schulte-Merker, S.; Lee, K.J.; McMahon, A.P.; Hammerschmidt, M. The zebrafish organizer requires chordino. Nature 1997, 387, 862–863. [Google Scholar] [CrossRef]
- O’Neill, K.; Thorpe, C. BMP signaling and spadetail regulate exit of muscle precursors from the zebrafish tailbud. Dev. Biol. 2013, 375, 117–127. [Google Scholar] [CrossRef]
- Daughters, R.S.; Chen, Y.; Slack, J.M. Origin of muscle satellite cells in the Xenopus embryo. Development 2011, 138, 821–830. [Google Scholar] [CrossRef]
- Biressi, S.; Tagliafico, E.; Lamorte, G.; Monteverde, S.; Tenedini, E.; Roncaglia, E.; Ferrari, S.; Ferrari, S.; Cusella-De Angelis, M.G.; Tajbakhsh, S.; et al. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev. Biol. 2007, 304, 633–651. [Google Scholar] [CrossRef]
- Huang, P.; Schulz, T.J.; Beauvais, A.; Tseng, Y.H.; Gussoni, E. Intramuscular adipogenesis is inhibited by myo-endothelial progenitors with functioning Bmpr1a signalling. Nat. Commun. 2014, 5, 4063. [Google Scholar] [CrossRef] [PubMed]
- Relaix, R.; Zammit, P.S. Satellite cells are essential for skeletal muscle regeneration: The cell on the edge returns centre stage. Development 2012, 139, 2845–2856. [Google Scholar] [CrossRef] [PubMed]
- Suryawan, A.; Frank, J.W.; Nguyen, H.V.; Davis, T.A. Expression of the TGF-beta family of ligands is developmentally regulated in skeletal muscle of neonatal rats. Pediatr. Res. 2006, 59, 175–179. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stantzou, A.; Schirwis, E.; Swist, S.; Alonso-Martin, S.; Polydorou, I.; Zarrouki, F.; Mouisel, E.; Beley, C.; Julien, A.; Le Grand, F.; et al. BMP signaling regulates satellite cell-dependent postnatal muscle growth. Development 2017, 144, 2737–2747. [Google Scholar] [CrossRef]
- Sartori, R.; Schirwis, E.; Blaauw, B.; Bortolanza, S.; Zhao, J.; Enzo, E.; Stantzou, A.; Mouisel, E.; Toniolo, L.; Ferry, A.; et al. BMP signaling controls muscle mass. Nat. Genet. 2013, 45, 1309–1318. [Google Scholar] [CrossRef]
- Han, D.; Zhao, H.; Parada, C.; Hacia, J.G.; Bringas, P., Jr.; Chai, Y. A TGFβ-Smad4-Fgf6 signaling cascade controls myogenic differentiation and myoblast fusion during tongue development. Development 2012, 139, 1640–1650. [Google Scholar] [CrossRef]
- Winbanks, C.E.; Chen, J.L.; Qian, H.; Liu, Y.; Bernardo, B.C.; Beyer, C.; Watt, K.I.; Thomson, R.E.; Connor, T.; Turner, B.J.; et al. The bone morphogenetic protein axis is a positive regulator of skeletal muscle mass. J. Cell Biol. 2013, 203, 345–357. [Google Scholar] [CrossRef]
- Dey, B.K.; Gagan, J.; Yan, Z.; Dutta, A. miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012, 26, 2180–2191. [Google Scholar] [CrossRef]
- Evano, B.; Tajbakhsh, S. Skeletal muscle stem cells in comfort and stress. NPJ Regen Med. 2018, 3, 24. [Google Scholar] [CrossRef]
- Clever, J.L.; Sakai, Y.; Wang, R.A.; Schneider, D.B. Inefficient skeletal muscle repair in inhibitor of differentiation knockout mice suggests a crucial role for BMP signaling during adult muscle regeneration. Am. J. Physiol. Cell Physiol. 2010, 298, C1087–C1099. [Google Scholar] [CrossRef]
- Wagner, I.; Wang, H.; Weissert, P.M.; Straube, W.L.; Shevchenko, A.; Gentzel, M.; Brito, G.; Tazaki, A.; Oliveira, C.; Sugiura, T.; et al. Serum proteases potentiate BMP-induced cell cycle re-entry of dedifferentiating muscle cells during newt limb regeneration. Dev. Cell 2017, 40, 608–617. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Shadrach, J.L.; Wagers, A.J.; Lassar, A.B. Id3 is a direct transcriptional target of Pax7 in quiescent satellite cells. Mol. Biol. Cell 2009, 20, 3170–3177. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Cholok, D.; Loder, S.; Li, J.; Breuler, C.; Chung, M.T.; Sung, H.H.; Ranganathan, K.; Habbouche, J.; Drake, J.; et al. mTOR inhibition and BMP signaling act synergistically to reduce muscle fibrosis and improve myofiber regeneration. JCI Insight 2016, 1, e89805. [Google Scholar] [CrossRef] [PubMed]
- Kawai, M.; Ohmori, Y.K.; Nishino, M.; Yoshida, M.; Tabata, K.; Hirota, D.S.; Ryu-Mon, A.; Yamamoto, H.; Sonobe, J.; Kataoka, Y.H.; et al. Determination of cell fate in skeletal muscle following BMP gene transfer by in vivo electroporation. Eur. J. Histochem. 2017, 61, 2772. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Costamagna, D.; Quattrocelli, M.; van Tienen, F.; Umans, L.; de Coo, I.F.; Zwijsen, A.; Huylebroeck, D.; Sampaolesi, M. Smad1/5/8 are myogenic regulators of murine and human mesoangioblasts. J. Mol. Cell Biol. 2016, 8, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Jiqing, C.; Yaqin, L.; Yingyin, L.; Fei, C.; Huili, Z.; Yuling, Z.; Juan, Y.; Shanwei, F.; Cheng, Z. BMP4 inhibits myogenic differentiation of bone marrow-derived mesenchymal stromal cells in mdx mice. Cytotherapy 2015, 17, 1213–1219. [Google Scholar] [CrossRef][Green Version]
- Lee, K.P.; Shin, Y.J.; Panda, A.C.; Abdelmohsen, K.; Kim, J.Y.; Lee, S.M.; Bahn, Y.J.; Choi, J.Y.; Kwon, E.S.; Baek, S.J.; et al. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015, 29, 1605–1617. [Google Scholar] [CrossRef]
- Kim, J.S.; Lee, Y.H.; Yi, H.K. Gradual downhill running improves age-related skeletal muscle and bone weakness: Implication of autophagy and bone morphogenetic proteins. Exp. Physiol. 2016, 101, 1528–1540. [Google Scholar] [CrossRef]
- Sengle, G.; Carlberg, V.; Tufa, S.F.; Charbonneau, N.L.; Smaldone, S.; Carlson, E.J.; Ramirez, F.; Keene, D.R.; Sakai, L.Y. Abnormal activation of BMP signaling causes myopathy in Fbn2 null mice. PLoS Genet. 2015, 11, e1005340. [Google Scholar] [CrossRef]
- Sterrenburg, E.; van der Wees, C.G.; White, S.J.; Turk, R.; de Menezes, R.X.; van Ommen, G.J.; den Dunnen, J.T.; Hoen, P.A. Gene expression profiling highlights defective myogenesis in DMD patients and a possible role for bone morphogenetic protein 4. NeuroBiol. Dis. 2006, 23, 228–236. [Google Scholar] [CrossRef]
- Choi, I.Y.; Lim, H.; Estrellas, K.; Mula, J.; Cohen, T.V.; Zhang, Y.; Donnelly, C.J.; Richard, J.P.; Kim, Y.J.; Kim, H.; et al. Concordant but varied phenotypes among Duchenne Muscular Dystrophy patient-specific myoblasts derived using a human iPSC-based model. Cell Rep. 2016, 15, 2301–2312. [Google Scholar] [CrossRef]
- Janin, A.; Bauer, D.; Ratti, F.; Valla, C.; Bertrand, A.; Christin, E.; Chopin, E.; Streichenberger, N.; Bonne, G.; Gache, V.; et al. SMAD6 overexpression leads to accelerated myogenic differentiation of LMNA mutated cells. Sci. Rep. 2018, 8, 5618. [Google Scholar] [CrossRef] [PubMed]
- Myhre, S.A.; Ruvalcaba, R.H.; Graham, C.B. A new growth deficiency syndrome. Clin Genet. 1981, 20, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Le Goff, C.; Mahaut, C.; Abhyankar, A.; Le Goff, W.; Serre, V.; Afenjar, A.; Destrée, A.; di Rocco, M.; Héron, D.; Jacquemont, S.; et al. Mutations at a single codon in Mad homology 2 domain of SMAD4 cause Myhre syndrome. Nat. Genet. 2011, 44, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Shore, E.M.; Xu, M.; Feldman, G.J.; Fenstermacher, D.A.; Cho, T.J.; Choi, I.H.; Connor, J.M.; Delai, P.; Glaser, D.L.; LeMerrer, M.; et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat. Genet. 2006, 38, 525. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).