Abstract
The complete structure and connectivity of the Caenorhabditis elegans nervous system (“mind of a worm”) was first published in 1986, representing a critical milestone in the field of connectomics. The reconstruction of the nervous system (connectome) at the level of synapses provided a unique perspective of understanding how behavior can be coded within the nervous system. The following decades have seen the development of technologies that help understand how neural activity patterns are connected to behavior and modulated by sensory input. Investigations on the developmental origins of the connectome highlight the importance of role of neuronal cell lineages in the final connectivity matrix of the nervous system. Computational modeling of neuronal dynamics not only helps reconstruct the biophysical properties of individual neurons but also allows for subsequent reconstruction of whole-organism neuronal network models. Hence, combining experimental datasets with theoretical modeling of neurons generates a better understanding of organismal behavior. This review discusses some recent technological advances used to analyze and perturb whole-organism neuronal function along with developments in computational modeling, which allows for interrogation of both local and global neural circuits, leading to different behaviors. Combining these approaches will shed light into how neural networks process sensory information to generate the appropriate behavioral output, providing a complete understanding of the worm nervous system.
1. Introduction
The field of connectomics attempts to link brain function with behavior by comprehensively mapping the anatomical links between all constituent neurons within different brain regions [1,2]. Caenorhabditis elegans, a microscopic roundworm, has served as a model for macroscopic research for over six decades (Figure 1). To date C. elegans remains the only organism to be fully mapped at the level of the nervous system [3,4,5]. The nervous system of both sexes; hermaphrodite (302 neurons) and male (385 neurons) have been completely mapped at the level of electron microscopy [4,6]. This has served as a prototype for analytical studies of larger scale connectome networks. However, can this complete mapping of synaptic connections in the brain (connectome) shape the understanding of the mechanistic basis of behavior? In other words, does structural connectivity define function within the nervous system? One mindset is that wiring diagrams can serve as a starting point for generating mechanistic hypotheses for the investigation of the neural basis of behavior. Therefore, knowledge of connectome structure enables the generation of testable hypotheses about the specific as well as general roles of individual neurons [7]. However, the physical connectivity patterns of the C. elegans nervous system, are unable to predict how the nervous system functions as a whole in order to enact behaviors [8,9,10]. Along with synapses, most nervous systems also contain gap junctions, which mediate fast, potentially bidirectional electrical coupling between cells [11,12]. In addition, extrasynaptic signaling between neurons within nervous system happens via monoamines and neuropeptides, occurring primarily outside the synaptic connectome [13]. These signaling systems act over both short and long ranges and are independent of synaptic connections, allowing them to shape behavioral responses to either the same or different stimuli [13,14]. Therefore, a complete characterization of a neuronal dynamics under different conditions is essential for generating a complete functional understanding of the nervous system. Since behavior is not a linear summation of sensory information, the same circuit can lead to different behavioral outputs. This could be a result of molecular events, stimuli concentration or physiological states—all of which are non-linear in nature—generating different behaviors via the same connectome. Hence, we define “functional connectomics” as the study of the relationship between a neuron’s function and its connections—both anatomical and extrasynaptic.
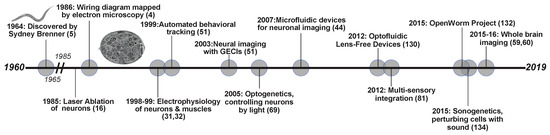
Figure 1.
Timeline of neuroscience related discoveries in C. elegans. The past 60 years have resulted in development in technologies and breakthroughs in understanding the neural connections and the nervous system.
2. Functional Characterization of Neural Circuits: Using Connectome to Generate Hypotheses
C. elegans neurons can be divided into three functional “classes” of neurons: sensory neurons, motor neurons, and interneurons or premotor neurons [7,9]. The sensory neurons have dendrites that extend to the tip of the nose and terminate into diverse ciliated structures to detect stimuli from the environment. These neurons account for a third of the neurons with more connections being pre-synaptic than post-synaptic. Conversely, motor neurons, another third of all neurons, have more post-synaptic connections. The remaining neurons are considered to be premotor interneurons, with large numbers of both pre- and post-synaptic connections [10]. Understanding the connections alone between these neuron classes helps increase our understanding of how a signal is transduced, processed, to ultimately produce behavioral outputs, as seen in previous work on C. elegans’ navigation [15].
One of the first major findings of the worm connectome from the structural connectivity data was the characterization of the mechanosensory circuitry [16]. Using laser ablations, components of a mechanosensory circuit were identified, consisting of sensory neurons, premotor interneurons, and ventral cord motorneurons, responsible for escape behavior in response to body touch [16]. By testing each neuron’s function by laser microsurgery, [16,17], a set of premotor interneurons were identified that control the direction of locomotion; six neurons that promoted forward locomotion and four neurons promoting backward locomotion [16]. This work not only opened up the genetic and molecular studies of the C. elegans touch circuit, but also implicated these major premotor interneurons in several other behaviors [18,19,20,21]. This study was a landmark in understanding how structural changes within the connectome can impact function and signaling between neurons.
The compact nervous system of C. elegans allows for a single neuronal class to be involved in the sensation of diverse stimuli or elicitation of different behaviors [7,9,22]. From a functional connectomic perspective, this suggests that not all stimuli utilize the same pathways and connections. This suggests a non-linearity and complexity in the information processing of stimuli, for example, the polymodal, amphid, single-ciliated, nociceptive neuron, ASH, detects a myriad of different mechano, osmo, and chemo stimuli that result in aversive behaviors [23,24,25,26,27,28,29]. The diversity in neuronal circuitries may also be due to the intracellular machinery used within individual neurons. C. elegans are equipped with a large set of G protein subunits that exhibit overlapping expression, rendering particular intracellular pathways important in different behavioral circuits [30]. The nematodes genome codes for 21 Gα protein subunits, and 2 subunits each of both Gβ and Gγ proteins [30]. Of the 21 Gα subunits, 16 are expressed throughout the chemosensory neurons, and many overlap in their expression profiles [30]. For example, on its own, ASH expresses ten different Gα subunits, while a different amphid, single-ciliated, ASE, expresses only three [30].
3. Technologies Employed to Unravel the Functional Connectome
One of the earliest methods to monitor neuron function was patch clamp electrophysiology [31,32,33,34]. This technique lends itself to understanding functional connectomics as it monitors the flow of ions across neuronal membranes [33]. In C. elegans it was used to understand the role of graded potentials, in opposition to all or nothing action potentials observed in mammals [31,32,33,34]. This method is rather invasive, requiring fixed samples of individual neurons for testing [35].
3.1. Neuronal Imaging in C. elegans
Optical techniques are available in many experimental systems but are highly applied in C. elegans research as the nematodes are optically transparent and can be imaged while fully intact. In addition, a variety of Genetically Encoded Calcium Indicators (GECIs) are available targeting individual neuron/s of interest with specific promoters [36,37]. These GECIs can specifically target the neuronal cell body and/or distribute throughout the entire neuron. Fluorescent imaging with GECIs has achieved rapid progress in visualizing Ca2+ flux at the levels of cell populations [38], single cells [39], or even subcellular compartments [40]. Among available GECIs, Green fluorescent Calmodulin M13 fusion Protein (GCaMP) is one of the most successful and popular, due to its ability to convey Ca2+ levels with impressive signal-noise ratios (Figure 2A) [41,42,43].
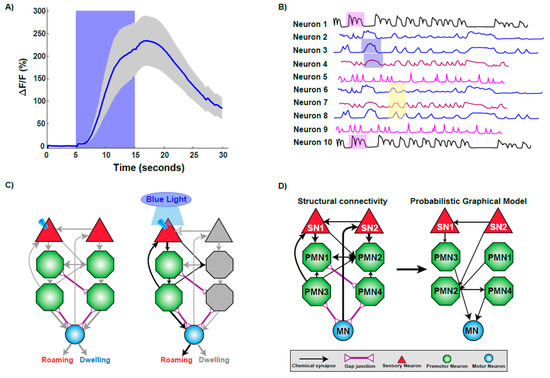
Figure 2.
Technologies used to decipher the functional connectome. (A) Imaging calcium changes using GCaMP sensor. The trace plot shows the activation of a sensory neuron upon stimulus presentation by increase in calcium influx as measured by increase in fluorescence intensity. Increase in calcium is sustained while the worm experiences the stimulus, as fluorescence decreases upon stimulus removal. (B) A representative brain phase plot where neurons are activated in different phases (shaded regions) during exposure to a stimulus. (C) Optogenetic interrogation of the connectome. Expressing and activating Channelrhodopsin via blue light exposure in a particular sensory neuron activates a subset of downstream neurons, resulting in the elicitation of roaming behavior. (D) Computational modeling helps to unravel the functional connections within a structural framework of neurons.
Calcium imaging of neurons is widely employed in C. elegans neuroscience as it allows for the activity of a single or multiple neurons to be monitored over different time scales. Advances in microfabrication technology have permitted the construction of well-controllable microenvironments for monitoring neural function in C. elegans. One of the first microfluidic devices used to monitor neuronal calcium dynamics was termed the ‘olfactory chip’. This device was used to examine stimulus-response relationships in chemosensory neurons over a short temporal timescale [44]. Over the last decade, the applications of microfabrication techniques have exponentially increased in neuroscience with chips being designed for high-throughput and high resolution- based applications [44,45,46,47,48].
3.2. Whole-Brain Imaging in C. elegans
Measuring neural activity of a single neuron over a short time course is helpful in identifying neuronal dynamics upon stimuli exposure and characterizing circuits of activity [31,33,44,45,47,49,50,51]. However, it offers little insight into the processes at play during long-term behaviors. To execute motor commands during a particular behavior, more is occurring than cross-talk via the connection between neurons. Global-brain or whole-brain imaging enables characterization of behaviors with the integration of sensory neurons with motor neurons to be quantified [40,52]. While imaging a single neuron, sensory or premotor neurons are often the focus [45,53], whole-brain imaging enables elucidating on the role of multiple neurons within the connectome simultaneously. Studies in zebrafish have been accomplished on a global-brain scale with single-neuron resolution with specific regard to motor neurons [54,55].
The first whole-brain calcium imaging experiment in C. elegans was conducted in an immobilized setting, imaged neural activity across 100 neurons in a small channel. This technique relied on existing neural maps to match captured neural responses to individual neurons [4,38]. This study achieved single-neuron resolution across most of the brain and, when combined with the extensive existing knowledge of C. elegans neural anatomy, supported identification of most neurons. Using this technology, it is possible to track the calcium changes through circuits, generating a brain state phase plot (Figure 2B) [38]. This temporal experiment was key in identifying repeating patterns of stimulation similar to models of central pattern or rhythmic motor generation [38,56,57,58]. Tracking the whole-brain changes in calcium signals during this repetitive behavior allows for different classes of neurons as well as other movement variables, such as speed, to be incorporated into the understanding of this behavior [38]. While this information generated is sufficient to understand the culmination of a behavior, the neural information can also be parsed out to understand the signaling occurring in each part of the action, such as moving forward, slowing, reversing, or turning, on the scale of an individual neuron or across the brain [38]. This study suggested that high-level organization of behavior is encoded in the brain by globally distributed, continuous, and low-dimensional dynamics [38].
Recent developments have allowed researchers to develop two methods for imaging the neurons of C. elegans while roaming. These techniques rely on simultaneously recording several neurons expressing a calcium indicator using spinning disk confocal microscopy to ultimately produce volumetric imaging [59,60]. Both methods allow for the simultaneous recording of approximately 80 neurons, gathering and correlating information on body posture and location in a moving worm. One method expresses both the calcium indicator and another fluorescent protein, and also infers body posture and position via head ganglia orientation [59]. In contrast, the other method utilizes a custom software, which ensures that the head of the worm is always in the correct location of the microscope stage despite the worm’s movement, and actively records the body’s position [60]. The ability to generate multi-neuron data of freely roaming worms will be vital in moving towards a more thorough understanding of the functional connectome [61].
3.3. Optogenetics and the Worm Connectome
Optogenetics offers temporal control of activity of individual neurons utilizing light-controlled ion channels [62]. This unique optical control of neurons allows researchers to probe neural circuits and investigate neuronal function in a highly specific and controllable fashion [63,64,65,66,67,68,69]. C. elegans is a popular platform for probing the nervous system at length, with scales spanning from synapse to whole circuit [63,64,67]. The initial studies on optogenetic control of C. elegans neurons involved using whole-field illumination together with specific genetic mutations, involving activation of Channelrhodopsin (ChR2) in excitable motor neurons [68,69]. Specific neurons or muscles expressing ChR2 can be quickly and reversibly activated by light in both live and behaving animals [68,69]. Expressing a light-sensitive protein in specific neurons using specific promoters highlighted that functional neuronal circuits during behaviors can be elucidated in C. elegans (Figure 2C).
A drawback of whole-field illumination was lack of cellular specificity as the illumination occurred over a region of the worm’s body. Newer technologies have been generated wherein targeted illumination of multiple fluorophores expressing optically sensitive proteins, and extensively employed to observe behavior [67,70,71,72]. Using these illumination systems, it is now possible to track a freely moving C. elegans and spatiotemporally excite and/or inhibit specific nodes of neural networks to probe for function across several types of locomotory behaviors. In addition, this technology enables the use of combinations of optogenetic tools and fluorescent GECIs with high reproducibility and light intensity control. In addition to development of illumination systems, progress has been made in terms of development of variants of the optogenetic proteins that both depolarize and hyperpolarize neurons over longer temporal scales [64]. These highly light-sensitive optogenetic tools, are highly effective and display fast kinetics, allowing better investigation prolonged neuronal activity states in C. elegans.
3.4. Computational Strategies
C. elegans is a powerful biophysical system that can be understood at different levels such as sensory stimulation and motor output. Computational models serve as an excellent platform to implicate how dynamics of different neurons can affect network connectivity [73]. The development of neuroimaging tools and technologies enable us to record neural dynamics over a longer timescale, which help generating dynamic models of synaptic connectivity. Newer computational approaches help elucidate the dynamic connectome by comparing longer timescale studies containing higher-dimensions of data [73].
An interesting application of computational strategies is the characterization of sensory-motor integration [74]. Sensory-motor integration attempts to understand the neural pathways from stimuli input to motor-neuron driven behavioral responses. Given the connectome data, physical circuits can be identified, and then simulated to understand responses [74]. Modeling the dynamics of the neural network is possible by combining the known structural connectome data of C. elegans with a physiologically model of a neuron [74]. Models such as probabilistic graphical models (PGMs), use known circuits of repetitive behavior, (Figure 2D), to predict responses to novel situations [75,76].
A recent study has developed the “dynome” of the worm nervous system [77]. This predictive system relies on the simulation of neural dynamics, or temporal experiments on multiple neurons, to allow application of stimuli to neurons to examine network properties. Such a model enables the user to apply or modify stimuli to the network, observe the neural dynamics on various time and population scales, and allow for network structural changes. Changing these parameters allows for calculation of neural response patterns associated with different stimuli. The strength of this approach lies in the ability of removing neurons from a network and studying the dynamics of the circuit [77,78]. This unique computational approach is a first step in making any nervous system’s architecture being able to predict a behavior based on network properties.
4. Analysis of the Functional Connectome
The development of the technologies discussed above, allow us to investigate specific aspects of the connectome, such as sensation or processing of diverse multisensory stimuli. Sensory systems continuously receive complex types of environmental input, which are processed by the nervous system to identify relevant facts about the surroundings and internally represent that information to help the animal successfully navigate its environment. This complex neural function is further complicated when combined with the internal physiological state of the animal. In Drosophila, recent work on the multilevel multimodal convergence circuit, which relies on multisensory integration, was limited by the lack of connectomic data unveiling multisensory neuronal convergence [79].
Multisensory integration in C. elegans falls into two broad categories: co-exposure to two distinct stimuli, aversive and attractive, or exposure to one stimuli in conjunction with an environmental indicator, both of which can be tested via avoidance assays [80,81,82]. Furthermore, with only sixteen pairs of chemosensory neurons, neuronal ablation techniques such as the laser ablation method as well as genetic ablations have become effective tools for understanding the diverse roles of individual sensory neurons within the realm of the connectome [83,84].
4.1. Divergent Functions within a Single Neuronal Class
Modifications in functional connections downstream of sensory neurons can be achieved by differential cellular signaling mechanisms both within sensory neurons and between sensory and premotor interneurons. The nociceptive sensory neuron ASH detects various stimuli; chemical, mechanical, osmotic, all of which result in the same behavioral outcome: avoidance [25,28,29,85,86,87]. Studies have characterized differential use of both intra- and inter-signaling molecules by ASH sensory neuron leading to the same behavior (Figure 3A) [85]. For example, nose touch avoidance, requires expression of itr-1 in ASH neurons and is not required for osmotic aversive responses [85]. Conversely, specific genes within ASH (such as osm-10) are specific for detecting osmotic stress, and not required for tactile response. This implies that in addition to expression of specific G protein-coupled subunits, distinct downstream effectors within the signaling pathway are specifically recruited to initiate functional connections. [85].
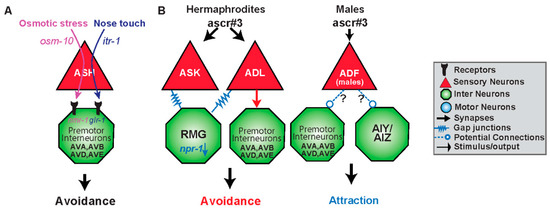
Figure 3.
Functional circuits are created by differential use of neurons and distinct intra- and intercellular signaling pathways. (A) The sensory neuron ASH responds to two different stimuli resulting in an avoidance response via the premotor interneurons AVA, AVB, AVD and AVE. Osmotic stress is mediated via osm-10 whereas nose touch utilizes itr-1 within ASH. Downstream of ASH, osmotic stress targets the N-methyl-D-aspartate (NMDA)-type receptor nmr-1, whereas nose touch activates the glutamate receptor glr-1 in the premotor interneurons [86,88]. (B) Gender and type of synapse govern behavioral output to the same stimulus. ADL senses the ascaroside, ascr#3, and in solitary hermaphrodites (high npr-1) results in avoidance via electrical synapses. However, hermaphroditic animals with low npr-1 activity dampen or even reverse the valence of response to ascr#3 by the hub-and-spoke gap junction circuit, specifically ASK and RMG. Gender also shapes the response to ascr#3. Males are also able to detect ascr#3 via the masculine mab-3 expressing state of ADF and are attracted to the compound. It is likely that ADF is opposing the ADL promoted avoidance response either via input to command interneurons or the first layer amphid interneurons it synapses with.
ASH has synaptic connections with AVA, AVB, and AVD, AVE, the forward and backward premotor interneurons [4]. Differential release of glutamate from ASH neurons may activate different types of glutamate receptors on these premotor interneurons mediating the nociceptive escape response (Figure 3A) [25,28,87]. The glutamate receptor, glr-1, is utilized primarily in nose touch avoidance, in downstream, premotor neurons (Figure 3A) [29,86,87]. Weak activation of ASH, elicited by nose touch, activates non-NMDA ionotropic glutamate receptor (iGluR) subunits GLR-1 (Figure 3A). Hyperosmolarity evokes higher levels of Ca2+ release, activating the NMDA ionotropic glutamate receptor NMR-1 along with GLR-1 [87]. Therefore, differential activation of inter/intra-signaling pathways leads to specific downstream neurons establishing different functional connections within the structural connectome.
While certain intracellular components and synaptic connections are vital in some behaviors, they may be irrelevant in other behavioral circuits. One example of this is the amphid sensory neuron, ADL, and its involvement in the response to ascaroside #3 (ascr#3) (Figure 3B). Ascarosides (ascr) are small-molecule signals which serve diverse functions in inter-organismal chemical signaling [88,89,90,91]. Ascr#3 is a small-molecule pheromone that causes different behavioral responses in males and hermaphrodites. Males are attracted by ascr#3, hermaphrodites are repelled by the cue [89,92,93]. Hermaphrodites avoid ascr#3 via ADL chemical synaptic transmission, presumably, to the backward command interneurons AVA and AVD [4,92]. ADL neuron’s avoidance to ascr#3 is regulated via the gap junction hub-and-spoke RMG circuit, whereas the interneuron RMG serves as a hub to modulate sensory neuron responses [92,93]. RMG, through the activity level of the neuropeptide receptor npr-1, and input from the sensory neuron ASK, can inhibit ADL triggered avoidance by altering gap junction properties [92,93]. Therefore, chemical synapses are involved in the avoidance to ascr#3, whereas gap junctions are necessary for modulating the response in an npr-1 dependent manner to elicit aggregation or attraction (Figure 3B).
4.2. Developmental Connectomics: Rewiring of the Connectome during Larval Development
Throughout development, the nervous system undergoes drastic changes in neuron number and neural connectivity. From sensory systems to neuromuscular junctions, new cellular components expand neural circuits as they differentiate from progenitors [4,94,95]. These structural changes at each larval stage update sensorimotor responses and adapt to changing body plans. Though the adult wiring diagram has long been completed, no other developmental stage or other organism has been mapped with such synaptic resolution [4,96]. However, emerging technologies has allowed neurogenesis of the connectome to be followed throughout the development of C. elegans, utilizing the well-established electron miscopy data of the adult worm and embryonic cellular information [97].
From the first cell division to hatching, the C. elegans connectivity matrix [6,96] can be followed throughout the development of the C. elegans [97]. A fertilized C. elegans undergoes the first round of divisions to form two cells called AB and P1, named for their anterior and posterior locations, respectively [98]. The AB cell gives rise to the neurons [95]. The larval connectome is a dynamic structure with connections changing very quickly over a given period of development. For instance, the first neuronal cell RMEV, is born just shy of 5 hours post fertilization. This neuron has no connections to the four other neurons present, ADF and AWB right and left. However, ten minutes later, RMEV temporarily becomes hub of connection within the group of >30 neurons [97]. At the 5 hours mark, connectivity is more complex with neurons forming feedback loops among other large scale networking features [99]. After 10 hours, the embryo hatches, entering the L1 stage of development [100].
Neuronal cells migrate from their point of origin during development and rewiring occurs throughout the development of the worm [97]. At the L1 larval stage, the worm has 222 neurons [94], 22 of which are motor neurons [101,102]. Of the 80 neurons added into adulthood, the majority of these neurons belong to the motor class [94]. The 22 motor neurons of the L1 worm migrate to innervate the dorsal region in adulthood with the addition of 36 ventral motor neurons throughout the larval stages [102]. The point of genesis of a neuron in development does not have a great influence on its level of connectivity, with no clear connection between cells having greater connections in adulthood with neurons that have a large number of connections during the development of the worm [97]. However, two studies which investigated the origins of the C. elegans connectome in the embryo suggest that there are some relationships between neurons that are born early versus those that are born later in development [97,103]. Both studies used a complex network approach combining elements of the published connectome with the birth times and spatial locations of neurons. Using this approach, the researchers found that as the C. elegans embryo develops, a neural network emerges that is shaped by their ancestral developmental cell lineages and proximal relationships between these cells [97]. The growth of the network transitions from an accelerated to a constant increase in the number of synaptic connections as a function of the neuronal number [103]. These investigations highlight the fact that a full understanding of the interplay between anatomical, functional, and behavioral changes across development, requires dynamic and structural models of complete neural circuits at different stages of development.
4.3. Sex Differences in the Functional Connectome
Interestingly, the sex of the animal can establish the synaptic connection and function of a neuron. One example of a sex-specific circuit change is in the sensation of ascr#3 (Figure 3B). Ascr#3 is also sensed by ADF, but only in males, and hermaphrodites which have been masculinized through expression of fem-3, a sex-determination protein which inhibits the sexual regulator gene, tra-1 [104,105]. Neuronal activation of ADF by ascr#3 also requires mab-3, which is naturally inhibited in hermaphroditic animals [104]. As ADL activation in males, results in attraction, masculinized ADF in hermaphrodites inhibits the aversive response to ascr#3. This inhibition may be taking place via extrasynaptic connections, or through serotonin signaling on downstream neuronal target of ADL (Figure 3B). Sex can also result in different physical circuits, where synapses between certain neurons are only present in males, and pruned in hermaphrodites [106].
There are 294 neurons that are present in both the hermaphrodite and male worm [104,107,108,109,110,111,112]. These common neurons constitute a significant portion of the 302 hermaphrodite neurons [4] but the male has an additional 83 neurons primarily localized in the nose and tail [113]. In embryonic development, only two sets of sex specific cells develop in both sexes, HSN and CEM; the other cells develop throughout the larval stages [95]. In hermaphrodites, HSN cells are motor neurons involved in egg laying. The male-specific CEMs are involved with detecting hermaphrodite pheromones and help innervate cephalic sensilla [108,114]. Programed apoptosis eliminates the unnecessary category of neuron in each sex [115,116]. Complete differentiation doesn’t occur embryonically but in larval stages when it is important to complete sexually different neural circuits prior to their use in adulthood [94,113]. Sexually dimorphic neuronal connectivity comes about primarily in the L4 stage, when sexual maturity is reached [4,6,117,118]. At this stage of development, pruning occurs in particular neurons which later have an impact of sex specific behaviors especially those related to mate finding. Beyond physical changes, pruning of cells impacts sensory circuits leading to sex-specific reception of chemosensory information [104,119,120,121,122].
The importance in pruning of connections is prevalent in the PHB and AVA connection. In worms with this connection, hermaphrodites and young worms, there is an avoidance to noxious chemicals, e.g. SDS, [82,117] closely related to kairomone secreted their predator Pristionchus pacificus [123]. This connection is pruned in L4 males and they do not avoid noxious chemicals [82,117,118]. This difference in behavior is necessary to alter the way that males seek mates. Males must actively seek a mate to reproduce and may perform more ecologically dangerous behaviors to pass genetic information. Hermaphrodites do not need to do this and take action to preserve life. Etiological studies [124] focusing on this valence have shown that which changes in food availability effects this connection. When there is a lack of food as a juvenile L1, the male neuronal pruning is altered, affecting reproduction efficacy as the male does not maintain contact with the hermaphrodite [125]. This behavior is rescued with the addition of food prior to the L3 state [118].
Sex-specific circuits have been identified that govern the male response to sex pheromones, demonstrating the importance of fully mapping neural circuits in both hermaphrodites and males [122]. This highlights the necessity of investigating how specific connections underlying a behavioral circuit is regulated by sex of the organism, not merely the requisite neuron, in order to generate a more complete functional connectome.
4.4. Modulation of Neural Circuits
Behavioral circuits are dependent on the state of the animal. While receptor expression profiles and the sex of the animal are set variables, more flexible states—such as the physiological state of the animal—shape and modulate these circuits. Sensory networks are altered by neuromodulators (neurotransmitters and neuropeptides) in a context specific manner; over varying distances and timescales. The effect of these modulations varies based on site of release and local concentration as governed by release, degradation, and reuptake of neuromodulators [7,10].
The neurotransmitter serotonin (5-HT) has been shown to have a large role in behaviors related to foraging, egg laying, and locomotion, dependent on the presence or absence of food, as expression levels are correlated with being either fed or starved [126]. Interestingly, it was found that the site of release is important, able to generate opposing effects of 5-HT-mediated locomotion [127]. These findings highlight how a single neurotransmitter, within the same circuit, can give rise to different synaptic strengths and fine-tuned behavioral outputs. Moreover, the same stimulus does not necessarily utilize the same circuit at different concentrations [126]. Furthermore, the duration of stimulus detection is coded into neural circuits suggesting a role of temporal activity in shaping functional circuits. For instance, avoidance to copper, is a short-term behavioral state mediated by a cross-talk ASI and ASH inhibition circuit that fine tunes the behavioral response. ASH neurons respond quickly and robustly in comparison to a slower, weaker response by ASI, which inhibits further ASH activation [128].
Worms also exhibit long-term behavioral states for example, roaming and dwelling states in the presence of food alternate, and last for minutes at a time (Figure 4). This switch is achieved via two opposing neuromodulators: serotonergic signaling promotes dwelling, whereas the neuropeptide PDF-1, pigment dispersing factor, promotes the roaming state [129]. The neurons that produce and respond to each neuromodulator form a distributed circuit independent to the classical wiring diagram, with several essential neurons that express each molecule (Figure 4). Serotonergic signaling through mod-1 initiates and extends dwelling states by inhibiting the neurons that promote roaming, whereas PDF signaling through pdfr-1 initiates and extends roaming states (Figure 4). Despite the compact size of the C. elegans nervous system, the serotonin and PDF that regulate roaming and dwelling each have several important sources, and their receptors each act in several target neurons. Strikingly, this functional circuit defies classical circuit logic of sensory to motor organization: motor and interneurons modulate the activity of sensory neurons [129]. This largely extrasynaptic, long-term timescale circuit has many potential inputs that can bias signaling of one state over the other.
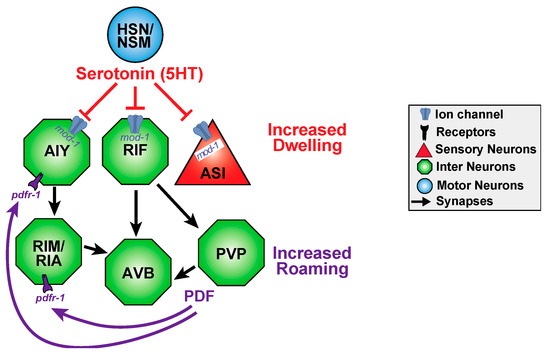
Figure 4.
Functional circuits may be shaped by both neurotransmitters and neuropeptides and by short and long timescales. C. elegans display long-term behavioral dwelling or roaming states which are triggered by serotonin and PDF, respectively. PDF prolongs roaming and shortens dwelling states, whereas serotonin has reciprocal effects. PVP has been hypothesized to secrete PDF. Roaming and dwelling behavior seems to be modulated by a distributed circuit with the switch between dwelling and roaming is seemingly spontaneous. NSM neurons are implicated in feeding, and HSN neurons are implicated in egg laying. AIY and RIM neurons regulate reversal frequencies. ASI neurons are sensory neurons (triangles) that sense food, pheromones, to regulate dauer larva development. RIA interneurons regulate head curving during locomotion. AVB interneurons are forward command neurons in the motor circuit. Interestingly, functional connections can be extrasynaptic and defy sensory to motor circuit logic as HSN/NSM serotonin inhibits ASI in this behavior. Circuit adapted from Flavell et al. [129].
Together, functional connectomes often take shape in drastically different ways than wiring diagrams suggest, with particular synaptic importance being dictated by physiological states and timescales. Additionally, functional circuits do not work in isolation, the final behavioral output is a readout of the fine tuning of multiple functional circuits converging to create a functional connectome.
Just as the internal state modulates the response to a particular cue, the presence of multiple stimuli is integrated into larger networks. How integration of cues allows for the modulation of circuits can further be exemplified by looking at threat tolerance. Well-fed C. elegans do not cross a high osmotic barrier to chemotax the food-odor attractant, diacetyl: the risk is not worth reward. However, animals which are deprived of food will cross the same osmotic barrier, presumably weighing that the risk no longer outweighs the reward [81]. This modulation requires slow accumulation of tyramine—the expression of which increases during extended times of starvation, thereby desensitizing ASH to the osmotic stressor—and requires a few hours of starvation to reach a level of tyramine which allows for the switch to decide to cross the osmotic barrier [81].
The aforementioned examples showcase the complexity underlying functional circuits, as there seem to be multiple levels of neuronal processing acting in parallel to finely adjust how the animal responds, including specific intercellular machinery allows for rapid adjustment of neuronal responses, thereby affecting the output, and these modulations can also take place over long time scales—not merely minutes, but instead hours. Therefore, to truly decipher a functional circuit, many different time points and stimulus concentrations must be investigated, as at one concentration and time scale there is likely hidden information acting at a deeper level.
5. Discussion and Future Directions
Nervous systems are comprised of structurally interconnected neuronal networks and brain regions with complex connectivity patterns [78]. As mapping and recording techniques become increasingly capable of capturing neural structure and activity across widely distributed circuits and systems, there is a growing need for new analytical tools and modeling approaches interpret these rich sets of “big data”. C. elegans, with its well characterized physical connections, provides a strong platform for functional connectomic analysis and elucidation of connectivity patterns. Future functional connectome studies will require a strong push for rapid whole-brain imaging techniques that maintain a resolution which allows for detailed analysis of individual neurons. Currently, these techniques are not optimized in many organisms. In the case of C. elegans, using a technique to image a single neuron requires the animal to be moved into a separate testing environment. By this methodology, the worm is often reduced to a fixed space [44]. This idea of imaging in vivo yet removed from the natural environment rings to the same tune as MRI or other imaging techniques that require large pieces of equipment. On the other end of this spectrum, exists techniques that do not even require a microscope. These lens-free methods include an optofluidic microscope: C. elegans are still able to move around freely in solution while their behavior is monitored [130]. This technique goes further than a behavioral assay, as it obtains real time results from internal structures.
Whole-brain imaging studies suggest that a population coding mechanism allows for the smooth transitioning of network activity [38,59,60]. This allows the worm to switch between different programs (forward to backward or vice versa) during locomotor behaviors. Novel computational methods will need to be developed to verify in a quantitative manner that population-level features indeed encode behaviors. Moreover, whole-brain imaging in freely moving worms should reveal whether other possible population-level features have indeed behavioral correlates. Optogenetics is especially well suited to uncovering compartment-specific processes. Developing the optogenetic toolkit further to localize photosensitive proteins to specific subcellular locations with precise activation is an area of future research. These technologies will help decipher subcellular dynamics of sensory and interneurons.
In silico approaches that utilize the C. elegans connectome to model known behaviors and also predict novel outcomes to a known stimulus are currently being developed. One such open-source platform is the OpenWorm, with the aim of building a complete digital organism to simulate all features of C. elegans’ behavior [131,132]. Computational modeling using novel algorithms and superimposition of these models on the experimental data can provide insights into how network/s function after stimulus exposure [73,74,75]. However, developing models that can predict network function based on simulations is still an area that requires further study. Additionally, dynamics of multitudes of neurons during a certain behavior, allow for new approaches in modeling incorporating both the structural connectome data and layering it with the neurophysiological responses and interactions [77]. The ‘Dynome’ model depicts the dynamical systems overlaying the structural connectivity [77]. These models are more akin to the realistic nervous system and have amazing potential for revealing novel neural pathways and functionalities of the network [7,10,78].
There has also been a substantial amount of developments in non-invasive techniques for probing neural mechanisms. One technique utilizes ultrasound waves to stimulate neural circuits in worms and other excitatory cells [133]. This field of sonogenetics delivers ultrasound to manipulate the neural circuit through a variety of mediums. One method uses repeated exposure of low-pressure ultrasound with microbubbles, while C. elegans remain on agar plates [134]. Others turn to microfluidic chip devices to deliver a single, short pulse of ultrasound [135]. As of 2016, the use of sonogenetics has been approved by the FDA to treat essential tremors in humans. This high-intensity focused ultrasound uses the mechanisms of MRI to map structures, before ablating damaged structures exacerbating tremors, typically localized in the thalamus [136].
In conclusion, connectomics (both structural and functional) are likely to expand significantly in coming years. Several large-scale national and international projects and consortia directed at brain science are underway, including the Human Connectome Project and the BRAIN initiative in the U.S. as well as the Human Brain Project in the E.U. [137,138,139]. Given the rate of data generation, an interesting avenue is development of frameworks that can span different scales and neural systems, helping make sense of “big brain data” [140].
Author Contributions
E.M.D. wrote the entire manuscript with inputs from C.D.C. and J.S. Figures were made by E.M.D. with inputs from J.S.; S.B. provided inputs on the section pertaining to multi-sensory integration.
Funding
This work was supported in by grants from the NIH to J.S. (R01DC016058). We thank the Caenorhabditis Genetics Center (CGC), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440).
Acknowledgments
We thank Douglas K. Reilly and the Srinivasan lab for critical comments on the manuscript and the Zhen lab for the EM picture of the worm connectome.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Seung, H.S. Neuroscience: Towards functional connectomics. Nature 2011, 471, 170–172. [Google Scholar] [CrossRef] [PubMed]
- Berger, D.R.; Seung, H.S.; Lichtman, J.W. VAST (Volume Annotation and Segmentation Tool): Efficient Manual and Semi-Automatic Labeling of Large 3D Image Stacks. Front. Neural Circuits 2018, 12, 88. [Google Scholar] [CrossRef]
- Albertson, D.G.; Thomson, J.N. The pharynx of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1976, 275, 299–325. [Google Scholar] [CrossRef] [PubMed]
- White, J.G.; Southgate, E.; Thomson, J.N.; Brenner, S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986, 314, 1–340. [Google Scholar] [CrossRef] [PubMed]
- Brenner, S. The Genetics of Caenorhabsitis elegans. Genetics 1974, 77, 71–94. [Google Scholar]
- Jarrell, T.A.; Wang, Y.; Bloniarz, A.E.; Brittin, C.A.; Xu, M.; Thomson, J.N.; Albertson, D.G.; Hall, D.H.; Emmons, S.W. The connectome of a decision-making neural network. Science 2012, 337, 437–444. [Google Scholar] [CrossRef]
- Schafer, W.R. The Worm Connectome: Back to the Future. Trends Neurosci. 2018, 41, 763–765. [Google Scholar] [CrossRef]
- Colosimo, C.; Pontieri, F.E. Yawning in Parkinson’s disease. Neurology 1999, 52, 428. [Google Scholar] [CrossRef]
- Bargmann, C.; Marder, E. From the connectome to brain function. Nat. Methods 2013, 10, 483–490. [Google Scholar] [CrossRef]
- Bargmann, C.I. Beyond the connectome: How neuromodulators shape neural circuits. BioEssays News Rev. Mol. Cell. Dev. Biol. 2012, 34, 458–465. [Google Scholar] [CrossRef]
- Hall, D.H. Gap junctions in C. elegans: Their roles in behavior and development. Dev. Neurobiol. 2017, 77, 587–596. [Google Scholar] [CrossRef]
- Hall, D.H. The role of gap junctions in the C. elegans connectome. Neurosci. Lett. 2017. [Google Scholar] [CrossRef]
- Bentley, B.; Branicky, R.; Barnes, C.L.; Chew, Y.L.; Yemini, E.; Bullmore, E.T.; Vertes, P.E.; Schafer, W.R. The Multilayer Connectome of Caenorhabditis elegans. PLoS Comput. Biol. 2016, 12, e1005283. [Google Scholar] [CrossRef]
- Jékely, G.; Melzer, S.; Beets, I.; Kadow, I.C.G.; Koene, J.; Haddad, S.; Holden-Dye, L. The long and the short of it—A perspective on peptidergic regulation of circuits and behaviour. J. Exp. Biol. 2018, 221. [Google Scholar] [CrossRef]
- Gray, J.M.; Hill, J.J.; Bargmann, C.I. A circuit for navigation in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2005, 102, 3184–3191. [Google Scholar] [CrossRef]
- Chalfie, M.; Sulston, J.; White, J.; Southgate, E.; Thomson, J.; Brenner, S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J. Neurosci. 1985, 5, 956–964. [Google Scholar] [CrossRef]
- Fang-Yen, C.; Gabel, C.V.; Samuel, A.D.T.; Bargmann, C.I.; Avery, L. Laser Microsurgery in Caenorhabditis elegans. Methods Cell Biol. 2012, 107, 177–206. [Google Scholar]
- Zheng, Z.; Lauritzen, J.S.; Perlman, E.; Robinson, C.G.; Nichols, M.; Milkie, D.; Torrens, O.; Price, J.; Fisher, C.B.; Sharifi, N.; et al. A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell 2018, 174, 730–743.e22. [Google Scholar] [CrossRef]
- Piggott, B.J.; Liu, J.; Feng, Z.; Wescott, S.A.; Xu, X.Z. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell 2011, 147, 922–933. [Google Scholar] [CrossRef]
- Rakowski, F.; Karbowski, J. Optimal synaptic signaling connectome for locomotory behavior in Caenorhabditis elegans: Design minimizing energy cost. PLoS Comput. Biol. 2017, 13, e1005834. [Google Scholar] [CrossRef]
- Rakowski, F.; Srinivasan, J.; Sternberg, P.W.; Karbowski, J. Synaptic polarity of the interneuron circuit controlling C. elegans locomotion. Front. Comput. Neurosci. 2013, 7, 128. [Google Scholar] [CrossRef]
- Chew, Y.L.; Schafer, W.R. A network for swimming. eLife 2017, 6. [Google Scholar] [CrossRef]
- Wragg, R.T.; Hapiak, V.; Miller, S.B.; Harris, G.P.; Gray, J.; Komuniecki, P.R.; Komuniecki, R.W. Tyramine and octopamine independently inhibit serotonin-stimulated aversive behaviors in Caenorhabditis elegans through two novel amine receptors. J. Neurosci. Off. J. Soc. Neurosci. 2007, 27, 13402–13412. [Google Scholar] [CrossRef]
- De Bono, M.; Tobin, D.M.; Davis, M.W.; Avery, L.; Bargmann, C.I. Social feeding in Caenorhabditis elegans is induced by neurons that detect aversive stimuli. Nature 2002, 419, 899–903. [Google Scholar] [CrossRef]
- Hilliard, M.A.; Bergamasco, C.; Arbucci, S.; Plasterk, R.H.; Bazzicalupo, P. Worms taste bitter: ASH neurons, QUI-1, GPA-3 and ODR-3 mediate quinine avoidance in Caenorhabditis elegans. EMBO J. 2004, 23, 1101–1111. [Google Scholar] [CrossRef]
- Campbell, J.C.; Chin-Sang, I.D.; Bendena, W.G. Mechanosensation circuitry in Caenorhabditis elegans: A focus on gentle touch. Peptides 2015, 68, 164–174. [Google Scholar] [CrossRef]
- Kaplan, J.M.; Horvitz, H.R. A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 1993, 90, 2227–2231. [Google Scholar] [CrossRef]
- Hart, A.C.; Kass, J.; Shapiro, J.E.; Kaplan, J.M. Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J. Neurosci. Off. J. Soc. Neurosci. 1999, 19, 1952–1958. [Google Scholar] [CrossRef]
- Hart, A.C.; Sims, S.; Kaplan, J.M. Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 1995, 378, 82–85. [Google Scholar] [CrossRef]
- Bastiani, C.; Mendel, J. Heterotrimeric G proteins in C. elegans. WormBook 2006, 13, 1–25. [Google Scholar] [CrossRef]
- Richmond, J.E.; Jorgensen, E.M. One GABA and two acetylcholine receptors function at the C. elegans neuromuscular junction. Nat. Neurosci. 1999, 2, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Lockery, S.R.; Goodman, M.B. Tight-seal whole-cell patch clamping of Caenorhabditis elegans neurons. Methods Enzymol. 1998, 293, 201–217. [Google Scholar] [PubMed]
- Goodman, M.B.; Lindsay, T.H.; Lockery, S.R.; Richmond, J.E. Electrophysiological methods for Caenorhabditis elegans neurobiology. Methods Cell Biol. 2012, 107, 409–436. [Google Scholar] [PubMed]
- Goodman, M.B.; Hall, D.H.; Avery, L.; Lockery, S.R. Active currents regulate sensitivity and dynamic range in C. elegans neurons. Neuron 1998, 20, 763–772. [Google Scholar] [CrossRef]
- Segev, A.; Garcia-Oscos, F.; Kourrich, S. Whole-cell Patch-clamp Recordings in Brain Slices. J. Vis. Exp. 2016, 112. [Google Scholar] [CrossRef]
- Kerr, R.A. Imaging the activity of neurons and muscles. WormBook 2006, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kerr, R.A.; Schafer, W.R. Intracellular Ca2+ imaging in C. elegans. Methods Mol. Biol. 2006, 351, 253–264. [Google Scholar]
- Kato, S.; Kaplan, H.S.; Schrodel, T.; Skora, S.; Lindsay, T.H.; Yemini, E.; Lockery, S.; Zimmer, M. Global brain dynamics embed the motor command sequence of Caenorhabditis elegans. Cell 2015, 163, 656–669. [Google Scholar] [CrossRef]
- Wang, W.; Xu, Z.J.; Wu, Y.Q.; Qin, L.W.; Li, Z.Y.; Wu, Z.X. Off-response in ASH neurons evoked by CuSO4 requires the TRP channel OSM-9 in Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2015, 461, 463–468. [Google Scholar] [CrossRef]
- Hendricks, M.; Ha, H.; Maffey, N.; Zhang, Y. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature 2012, 487, 99–103. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, N.; He, Y.; Liu, Y.; Ge, L.; Zou, L.; Song, S.; Xiong, W.; Liu, X. Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP. Nat. Commun. 2018, 9, 1504. [Google Scholar] [CrossRef]
- Chen, T.W.; Wardill, T.J.; Sun, Y.; Pulver, S.R.; Renninger, S.L.; Baohan, A.; Schreiter, E.R.; Kerr, R.A.; Orger, M.B.; Jayaraman, V.; et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013, 499, 295–300. [Google Scholar] [CrossRef]
- Dana, H.; Mohar, B.; Sun, Y.; Narayan, S.; Gordus, A.; Hasseman, J.P.; Tsegaye, G.; Holt, G.T.; Hu, A.; Walpita, D.; et al. Sensitive red protein calcium indicators for imaging neural activity. eLife 2016, 5. [Google Scholar] [CrossRef]
- Chronis, N.; Zimmer, M.; Bargmann, C.I. Microfluidics for in vivo imaging of neuronal and behavioral activity in Caenorhabditis elegans. Nat. Methods 2007, 4, 727–731. [Google Scholar] [CrossRef]
- Reilly, D.K.; Lawler, D.E.; Albrecht, D.R.; Srinivasan, J. Using an Adapted Microfluidic Olfactory Chip for the Imaging of Neuronal Activity in Response to Pheromones in Male C. elegans Head Neurons. J. Vis. Exp. 2017. [Google Scholar] [CrossRef]
- Larsch, J.; Flavell, S.W.; Liu, Q.; Gordus, A.; Albrecht, D.R.; Bargmann, C.I. A Circuit for Gradient Climbing in C. elegans Chemotaxis. Cell Rep. 2015, 12, 1748–1760. [Google Scholar] [CrossRef]
- Cho, Y.; Zhao, C.L.; Lu, H. Trends in high-throughput and functional neuroimaging in Caenorhabditis elegans. Wiley Interdiscip. Rev. Syst. Biol. Med. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Ben-Yakar, A.; Chronis, N.; Lu, H. Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Curr. Opin. Neurobiol. 2009, 19, 561–567. [Google Scholar] [CrossRef]
- Richmond, J.E. Electrophysiological recordings from the neuromuscular junction of C. elegans. WormBook 2006, 1–8. [Google Scholar] [CrossRef]
- Cunningham, J.P.; Yu, B.M. Dimensionality reduction for large-scale neural recordings. Nat. Neurosci. 2014, 17, 1500–1509. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Kerr, R.; Bianchi, L.; Frokjaer-Jensen, C.; Slone, D.; Xue, J.; Gerstbrein, B.; Driscoll, M.; Schafer, W.R. In vivo imaging of C. elegans mechanosensory neurons demonstrates a specific role for the MEC-4 channel in the process of gentle touch sensation. Neuron 2003, 39, 1005–1017. [Google Scholar] [CrossRef]
- Luo, L.; Wen, Q.; Ren, J.; Hendricks, M.; Gershow, M.; Qin, Y.; Greenwood, J.; Soucy, E.R.; Klein, M.; Smith-Parker, H.K.; et al. Dynamic encoding of perception, memory, and movement in a C. elegans chemotaxis circuit. Neuron 2014, 82, 1115–1128. [Google Scholar] [CrossRef]
- Chalasani, S.H.; Chronis, N.; Tsunozaki, M.; Gray, J.M.; Ramot, D.; Goodman, M.B.; Bargmann, C.I. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature 2007, 450, 63–70. [Google Scholar] [CrossRef]
- Ahrens, M.B.; Li, J.M.; Orger, M.B.; Robson, D.N.; Schier, A.F.; Engert, F.; Portugues, R. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature 2012, 485, 471–477. [Google Scholar] [CrossRef]
- Panier, T.; Romano, S.A.; Olive, R.; Pietri, T.; Sumbre, G.; Candelier, R.; Debregeas, G. Fast functional imaging of multiple brain regions in intact zebrafish larvae using selective plane illumination microscopy. Front. Neural Circuits 2013, 7, 65. [Google Scholar] [CrossRef]
- Marder, E.; Bucher, D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu. Rev. Physiol. 2007, 69, 291–316. [Google Scholar] [CrossRef] [PubMed]
- Bruno, A.M.; Frost, W.N.; Humphries, M.D. Modular deconstruction reveals the dynamical and physical building blocks of a locomotion motor program. Neuron 2015, 86, 304–318. [Google Scholar] [CrossRef]
- Arshavsky, Y.I.; Deliagina, T.G.; Orlovsky, G.N.; Panchin, Y.V.; Popova, L.B.; Sadreyev, R.I. Analysis of the central pattern generator for swimming in the mollusk Clione. Ann. N. Y. Acad. Sci. 1998, 860, 51–69. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, J.P.; Shipley, F.B.; Linder, A.N.; Plummer, G.S.; Liu, M.; Setru, S.U.; Shaevitz, J.W.; Leifer, A.M. Whole-brain calcium imaging with cellular resolution in freely behaving Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016, 113, E1074–E1081. [Google Scholar] [CrossRef]
- Venkatachalam, V.; Ji, N.; Wang, X.; Clark, C.; Mitchell, J.K.; Klein, M.; Tabone, C.J.; Florman, J.; Ji, H.; Greenwood, J.; et al. Pan-neuronal imaging in roaming Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016, 113, E1082–E1088. [Google Scholar] [CrossRef]
- Izquierdo, E.J.; Beer, R.D. The whole worm: Brain-body-environment models of C. elegans. Curr. Opin. Neurobiol. 2016, 40, 23–30. [Google Scholar] [CrossRef]
- Deisseroth, K. Optogenetics: Controlling the Brain with Light [Extended Version]. Sci. Am. 2010, 49–55. [Google Scholar]
- Akerboom, J.; Carreras Calderon, N.; Tian, L.; Wabnig, S.; Prigge, M.; Tolo, J.; Gordus, A.; Orger, M.B.; Severi, K.E.; Macklin, J.J.; et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front. Mol. Neurosci. 2013, 6, 2. [Google Scholar] [CrossRef]
- Bergs, A.; Schultheis, C.; Fischer, E.; Tsunoda, S.P.; Erbguth, K.; Husson, S.J.; Govorunova, E.; Spudich, J.L.; Nagel, G.; Gottschalk, A.; et al. Rhodopsin optogenetic toolbox v2.0 for light-sensitive excitation and inhibition in Caenorhabditis elegans. PLoS ONE 2018, 13, e0191802. [Google Scholar] [CrossRef]
- Brown, J.; Behnam, R.; Coddington, L.; Tervo, D.G.R.; Martin, K.; Proskurin, M.; Kuleshova, E.; Park, J.; Phillips, J.; Bergs, A.C.F.; et al. Expanding the Optogenetics Toolkit by Topological Inversion of Rhodopsins. Cell 2018, 175, 1131–1140.e1111. [Google Scholar] [CrossRef]
- Husson, S.J.; Costa, W.S.; Wabnig, S.; Stirman, J.N.; Watson, J.D.; Spencer, W.C.; Akerboom, J.; Looger, L.L.; Treinin, M.; Miller, D.M., 3rd; et al. Optogenetic analysis of a nociceptor neuron and network reveals ion channels acting downstream of primary sensors. Curr. Biol. 2012, 22, 743–752. [Google Scholar] [CrossRef]
- Husson, S.J.; Gottschalk, A.; Leifer, A.M. Optogenetic manipulation of neural activity in C. elegans: From synapse to circuits and behaviour. Biol. Cell 2013, 105, 235–250. [Google Scholar] [CrossRef]
- Liewald, J.F.; Brauner, M.; Stephens, G.J.; Bouhours, M.; Schultheis, C.; Zhen, M.; Gottschalk, A. Optogenetic analysis of synaptic function. Nat. Methods 2008, 5, 895–902. [Google Scholar] [CrossRef]
- Nagel, G.; Brauner, M.; Liewald, J.F.; Adeishvili, N.; Bamberg, E.; Gottschalk, A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr. Biol. 2005, 15, 2279–2284. [Google Scholar] [CrossRef]
- Wen, Q.; Po, M.D.; Hulme, E.; Chen, S.; Liu, X.; Kwok, S.W.; Gershow, M.; Leifer, A.M.; Butler, V.; Fang-Yen, C.; et al. Proprioceptive coupling within motor neurons drives C. elegans forward locomotion. Neuron 2012, 76, 750–761. [Google Scholar] [CrossRef]
- Husson, S.J.; Liewald, J.F.; Schultheis, C.; Stirman, J.N.; Lu, H.; Gottschalk, A. Microbial light-activatable proton pumps as neuronal inhibitors to functionally dissect neuronal networks in C. elegans. PLoS ONE 2012, 7, e40937. [Google Scholar] [CrossRef] [PubMed]
- Stirman, J.N.; Crane, M.M.; Husson, S.J.; Wabnig, S.; Schultheis, C.; Gottschalk, A.; Lu, H. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat. Methods 2011, 8, 153–158. [Google Scholar] [CrossRef] [PubMed]
- Kunert-Graf, J.M.; Shlizerman, E.; Walker, A.; Kutz, J.N. Multistability and Long-Timescale Transients Encoded by Network Structure in a Model of C. elegans Connectome Dynamics. Front. Comput. Neurosci. 2017, 11, 53. [Google Scholar] [CrossRef]
- Kunert, J.; Shlizerman, E.; Kutz, J.N. Low-dimensional functionality of complex network dynamics: Neurosensory integration in the Caenorhabditis Elegans connectome. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2014, 89, 052805. [Google Scholar] [CrossRef]
- Sporns, O. The non-random brain: Efficiency, economy, and complex dynamics. Front. Comput. Neurosci. 2011, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Kim, J.; Shlizerman, E. Functional connectomics from neural dynamics: Probabilistic graphical models for neuronal network of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2018, 373. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Leahy, W.; Shlizerman, E. Neural Interactome: Interactive Simulation of a Neuronal System. BioRxiv 2018. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O. Connectome Networks: From Cells to Systems; Kennedy, H., Essen, D.V., Christen, Y., Eds.; Springer: Berlin, Germany, 2016. [Google Scholar]
- Ohyama, T.; Schneider-Mizell, C.M.; Fetter, R.D.; Aleman, J.V.; Franconville, R.; Rivera-Alba, M.; Mensh, B.D.; Branson, K.M.; Simpson, J.H.; Truman, J.W.; et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature 2015, 520, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.D.; Nitabach, M.N.; Zhang, Y.; Harris, G. Multisensory integration in C. elegans. Curr. Opin. Neurobiol. 2017, 43, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, D.D.; Sanders, T.; Hong, S.; McCurdy, L.Y.; Chase, D.L.; Cohen, N.; Koelle, M.R.; Nitabach, M.N. Neural Architecture of Hunger-Dependent Multisensory Decision Making in C. elegans. Neuron 2016, 92, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Hilliard, M.A.; Bargmann, C.I.; Bazzicalupo, P. C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr. Biol. 2002, 12, 730–734. [Google Scholar] [CrossRef]
- Metaxakis, A.; Petratou, D.; Tavernarakis, N. Multimodal sensory processing in Caenorhabditis elegans. Open Biol. 2018, 8. [Google Scholar] [CrossRef]
- Hart, A.C.; Chao, M.Y. From Odors to Behaviors in Caenorhabditis elegans. In The Neurobiology of Olfaction; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2010. [Google Scholar]
- Walker, D.S.; Vazquez-Manrique, R.P.; Gower, N.J.; Gregory, E.; Schafer, W.R.; Baylis, H.A. Inositol 1,4,5-trisphosphate signalling regulates the avoidance response to nose touch in Caenorhabditis elegans. PLoS Genet. 2009, 5, e1000636. [Google Scholar] [CrossRef]
- Maricq, A.V.; Peckol, E.; Driscoll, M.; Bargmann, C.I. Mechanosensory signalling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 1995, 378, 78–81. [Google Scholar] [CrossRef]
- Mellem, J.E.; Brockie, P.J.; Zheng, Y.; Madsen, D.M.; Maricq, A.V. Decoding of polymodal sensory stimuli by postsynaptic glutamate receptors in C. elegans. Neuron 2002, 36, 933–944. [Google Scholar] [CrossRef]
- Edison, A.S. Caenorhabditis elegans pheromones regulate multiple complex behaviors. Curr. Opin. Neurobiol. 2009, 19, 378–388. [Google Scholar] [CrossRef]
- Srinivasan, J.; Kaplan, F.; Ajredini, R.; Zachariah, C.; Alborn, H.T.; Teal, P.E.; Malik, R.U.; Edison, A.S.; Sternberg, P.W.; Schroeder, F.C. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature 2008, 454, 1115–1118. [Google Scholar] [CrossRef]
- Von Reuss, S.H.; Bose, N.; Srinivasan, J.; Yim, J.J.; Judkins, J.C.; Sternberg, P.W.; Schroeder, F.C. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small-molecule signals in C. elegans. J. Am. Chem. Soc. 2012, 134, 1817–1824. [Google Scholar] [CrossRef] [PubMed]
- Von Reuss, S.H.; Schroeder, F.C. Combinatorial chemistry in nematodes: Modular assembly of primary metabolism-derived building blocks. Natural Prod. Rep. 2015, 32, 994–1006. [Google Scholar] [CrossRef]
- Jang, H.; Kim, K.; Neal, S.J.; Macosko, E.; Kim, D.; Butcher, R.A.; Zeiger, D.M.; Bargmann, C.I.; Sengupta, P. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron 2012, 75, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Macosko, E.Z.; Pokala, N.; Feinberg, E.H.; Chalasani, S.H.; Butcher, R.A.; Clardy, J.; Bargmann, C.I. A hub-and-spoke circuit drives pheromone attraction and social behaviour in C. elegans. Nature 2009, 458, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Sulston, J.E.; Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977, 56, 110–156. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Varshney, L.R.; Chen, B.L.; Paniagua, E.; Hall, D.H.; Chklovskii, D.B. Structural properties of the Caenorhabditis elegans neuronal network. PLoS Comput. Biol. 2011, 7, e1001066. [Google Scholar] [CrossRef] [PubMed]
- Alicea, B. The emergent connectome in Caenorhabditis elegans embryogenesis. Biosystems 2018, 173, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Wasserstrom, A.; Adar, R.; Shefer, G.; Frumkin, D.; Itzkovitz, S.; Stern, T.; Shur, I.; Zangi, L.; Kaplan, S.; Harmelin, A.; et al. Reconstruction of cell lineage trees in mice. PLoS ONE 2008, 3, e1939. [Google Scholar] [CrossRef] [PubMed]
- Azulay, A.; Itskovits, E.; Zaslaver, A. The C. elegans connectome consists of homogenous circuits with defined functional roles. PLoS Comput. Biol. 2016, 12, e1005021. [Google Scholar] [CrossRef] [PubMed]
- Muschiol, D.; Schroeder, F.; Traunspurger, W. Life cycle and population growth rate of Caenorhabditis elegans studied by a new method. BMC Ecol. 2009, 9, 14. [Google Scholar] [CrossRef]
- White, J.G.; Albertson, D.G.; Anness, M.A. Connectivity changes in a class of motoneurone during the development of a nematode. Nature 1978, 271, 764–766. [Google Scholar] [CrossRef]
- Von Stetina, S.E.; Treinin, M.; Miller, D.M., 3rd. The motor circuit. Int. Rev. Neurobiol. 2006, 69, 125–167. [Google Scholar] [PubMed]
- Nicosia, V.; Vértes, P.E.; Schafer, W.R.; Latora, V.; Bullmore, E.T. Phase transition in the economically modeled growth of a cellular nervous system. Proc. Natl. Acad. Sci. USA 2013, 110, 7880–7885. [Google Scholar] [CrossRef] [PubMed]
- Fagan, K.A.; Luo, J.; Lagoy, R.C.; Schroeder, F.C.; Albrecht, D.R.; Portman, D.S. A Single-Neuron Chemosensory Switch Determines the Valence of a Sexually Dimorphic Sensory Behavior. Curr. Biol. 2018, 28, 902–914.e905. [Google Scholar] [CrossRef] [PubMed]
- White, J.Q.; Nicholas, T.J.; Gritton, J.; Truong, L.; Davidson, E.R.; Jorgensen, E.M. The sensory circuitry for sexual attraction in C. elegans males. Curr. Biol. 2007, 17, 1847–1857. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, P.; Berkseth, M.; Zarkower, D.; Hobert, O. Sexually Dimorphic unc-6/Netrin Expression Controls Sex-Specific Maintenance of Synaptic Connectivity. Curr. Biol. 2018, 28, 623–629.e623. [Google Scholar] [CrossRef] [PubMed]
- Hilbert, Z.A.; Kim, D.H. PDF-1 neuropeptide signaling regulates sexually dimorphic gene expression in shared sensory neurons of C. elegans. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Barr, M.M.; Garcia, L.R.; Portman, D.S. Sexual dimorphism and sex differences in Caenorhabditis elegans neuronal development and behavior. Genetics 2018, 208, 909–935. [Google Scholar] [CrossRef]
- Barrios, A.; Ghosh, R.; Fang, C.; Emmons, S.W.; Barr, M.M. PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nat. Neurosci. 2012, 15, 1675–1682. [Google Scholar] [CrossRef]
- Lee, K.; Portman, D.S. Neural sex modifies the function of a C. elegans sensory circuit. Curr. Biol. 2007, 17, 1858–1863. [Google Scholar] [CrossRef]
- Mowrey, W.R.; Bennett, J.R.; Portman, D.S. Distributed effects of biological sex define sex-typical motor behavior in Caenorhabditis elegans. J. Neurosci. 2014, 34, 1579–1591. [Google Scholar] [CrossRef]
- Sakai, N.; Iwata, R.; Yokoi, S.; Butcher, R.A.; Clardy, J.; Tomioka, M.; Iino, Y. A sexually conditioned switch of chemosensory behavior in C. elegans. PLoS ONE 2013, 8, e68676. [Google Scholar] [CrossRef]
- Sulston, J.; Albertson, D.; Thomson, J. The Caenorhabditis elegans male: Postembryonic development of nongonadal structures. Dev. Biol. 1980, 78, 542–576. [Google Scholar] [CrossRef]
- Narayan, A.; Venkatachalam, V.; Durak, O.; Reilly, D.K.; Bose, N.; Schroeder, F.C.; Samuel, A.D.T.; Srinivasan, J.; Sternberg, P.W. Contrasting responses within a single neuron class enable sex-specific attraction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 2016, 113, E1392–E1401. [Google Scholar] [CrossRef]
- Conradt, B.; Horvitz, H.R. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell 1999, 98, 317–327. [Google Scholar] [CrossRef]
- Singhal, A.; Shaham, S. Infrared laser-induced gene expression for tracking development and function of single C. elegans embryonic neurons. Nat. Commun. 2017, 8, 14100. [Google Scholar] [CrossRef]
- Oren-Suissa, M.; Bayer, E.A.; Hobert, O. Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature 2016, 533, 206–211. [Google Scholar] [CrossRef]
- Bayer, E.A.; Hobert, O. Past experience shapes sexually dimorphic neuronal wiring through monoaminergic signalling. Nature 2018, 561, 117–121. [Google Scholar] [CrossRef]
- Ruta, V.; Datta, S.R.; Vasconcelos, M.L.; Freeland, J.; Looger, L.L.; Axel, R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature 2010, 468, 686–690. [Google Scholar] [CrossRef]
- Kohl, J.; Ostrovsky, A.D.; Frechter, S.; Jefferis, G.S.X.E. A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell 2013, 155, 1610–1623. [Google Scholar] [CrossRef]
- Datta, S.R.; Vasconcelos, M.L.; Ruta, V.; Luo, S.; Wong, A.; Demir, E.; Flores, J.; Balonze, K.; Dickson, B.J.; Axel, R. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature 2008, 452, 473–477. [Google Scholar] [CrossRef]
- Chute, C.D.; Srinivasan, J. Chemical mating cues in C. elegans. Semin. Cell Dev. Biol. 2014, 33, 18–24. [Google Scholar] [CrossRef]
- Liu, Z.; Kariya, M.J.; Chute, C.D.; Pribadi, A.K.; Leinwand, S.G.; Tong, A.; Curran, K.P.; Bose, N.; Schroeder, F.C.; Srinivasan, J.; et al. Predator-secreted sulfolipids induce defensive responses in C. elegans. Nat. Commun. 2018, 9, 1128. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, A.; Fair, D.A.; Kelly, C.; Satterthwaite, T.D.; Castellanos, F.X.; Thomason, M.E.; Craddock, R.C.; Luna, B.; Leventhal, B.L.; Zuo, X.-N.; et al. Unraveling the miswired connectome: A developmental perspective. Neuron 2014, 83, 1335–1353. [Google Scholar] [CrossRef] [PubMed]
- Sherlekar, A.L.; Janssen, A.; Siehr, M.S.; Koo, P.K.; Caflisch, L.; Boggess, M.; Lints, R. The C. elegans male exercises directional control during mating through cholinergic regulation of sex-shared command interneurons. PLoS ONE 2013, 8, e60597. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.Y.; Komatsu, H.; Fukuto, H.S.; Dionne, H.M.; Hart, A.C. Feeding status and serotonin rapidly and reversibly modulate a Caenorhabditis elegans chemosensory circuit. Proc. Natl. Acad. Sci. USA 2004, 101, 15512–15517. [Google Scholar] [CrossRef] [PubMed]
- Harris, G.; Korchnak, A.; Summers, P.; Hapiak, V.; Law, W.J.; Stein, A.M.; Komuniecki, P.; Komuniecki, R. Dissecting the serotonergic food signal stimulating sensory-mediated aversive behavior in C. elegans. PLoS ONE 2011, 6, e21897. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Wu, T.H.; Song, Y.X.; Ge, M.H.; Su, C.M.; Niu, W.P.; Li, L.L.; Xu, Z.J.; Ge, C.L.; Al-Mhanawi, M.T.; et al. Reciprocal inhibition between sensory ASH and ASI neurons modulates nociception and avoidance in Caenorhabditis elegans. Nat. Commun. 2015, 6, 5655. [Google Scholar] [CrossRef] [PubMed]
- Flavell, S.W.; Pokala, N.; Macosko, E.Z.; Albrecht, D.R.; Larsch, J.; Bargmann, C.I. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell 2013, 154, 1023–1035. [Google Scholar] [CrossRef]
- Kim, S.B.; Bae, H.; Koo, K.-I.; Dokmeci, M.R.; Ozcan, A.; Khademhosseini, A. Lens-Free Imaging for Biological Applications. J. Lab. Autom. 2012, 17, 43–49. [Google Scholar] [CrossRef]
- Palyanov, A.; Khayrulin, S.; Larson, S.D. Application of smoothed particle hydrodynamics to modeling mechanism of biological tissue. Adv. Eng. Softw. 2016, 98, 1–11. [Google Scholar] [CrossRef]
- Gleeson, P.; Cantarelli, M.; Currie, M.; Hokanson, J.; Idili, G.; Khayrulin, S.; Palyanov, A.; Szigeti, B.; Larson, S. The OpenWorm Project: Currently available resources and future plans. BMC Neurosci. 2015, 16, P141. [Google Scholar] [CrossRef]
- Kubanek, J.; Shukla, P.; Das, A.; Baccus, S.A.; Goodman, M.B. Ultrasound Elicits Behavioral Responses through Mechanical Effects on Neurons and Ion Channels in a Simple Nervous System. J. Neurosci. Off. J. Soc. Neurosci. 2018, 38, 3081–3091. [Google Scholar] [CrossRef]
- Ibsen, S.; Tong, A.; Schutt, C.; Esener, S.; Chalasani, S.H. Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat. Commun. 2015, 6, 8264. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, J.; Wang, K.; Huang, B.; Niu, L.; Li, F.; Cai, F.; Chen, Y.; Liu, X.; Zhang, X.; et al. Ultrasound neuro-modulation chip: Activation of sensory neurons in Caenorhabditis elegans by surface acoustic waves. Lab Chip 2017, 17, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Fishman, P.S. Thalamotomy for essential tremor: FDA approval brings brain treatment with FUS to the clinic. J. Ther. Ultrasound 2017, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Yuste, R.; Bargmann, C. Toward a Global BRAIN Initiative. Cell 2017, 168, 956–959. [Google Scholar] [CrossRef]
- Amunts, K.; Ebell, C.; Muller, J.; Telefont, M.; Knoll, A.; Lippert, T. The Human Brain Project: Creating a European Research Infrastructure to Decode the Human Brain. Neuron 2016, 92, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Van Essen, D.C.; Smith, S.M.; Barch, D.M.; Behrens, T.E.J.; Yacoub, E.; Ugurbil, K.; Consortium, W.U.-M.H. The WU-Minn Human Connectome Project: An overview. NeuroImage 2013, 80, 62–79. [Google Scholar] [CrossRef] [PubMed]
- Sporns, O. Making sense of brain network data. Nat. Methods 2013, 10, 491–493. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).