Mechanisms of Specificity for Hox Factor Activity
Abstract
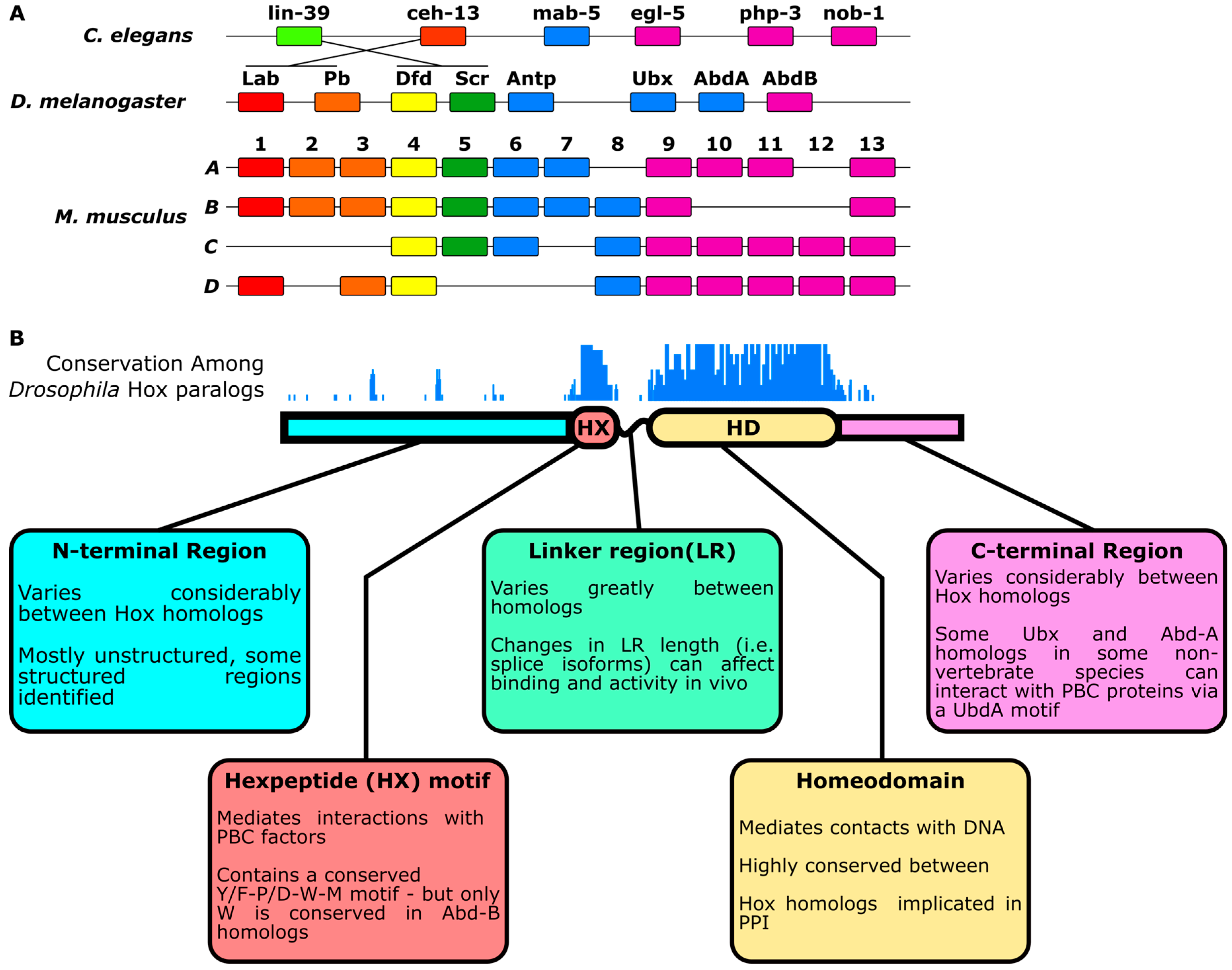
:1. Introduction
2. Differentiation of Hox Paralog Activities
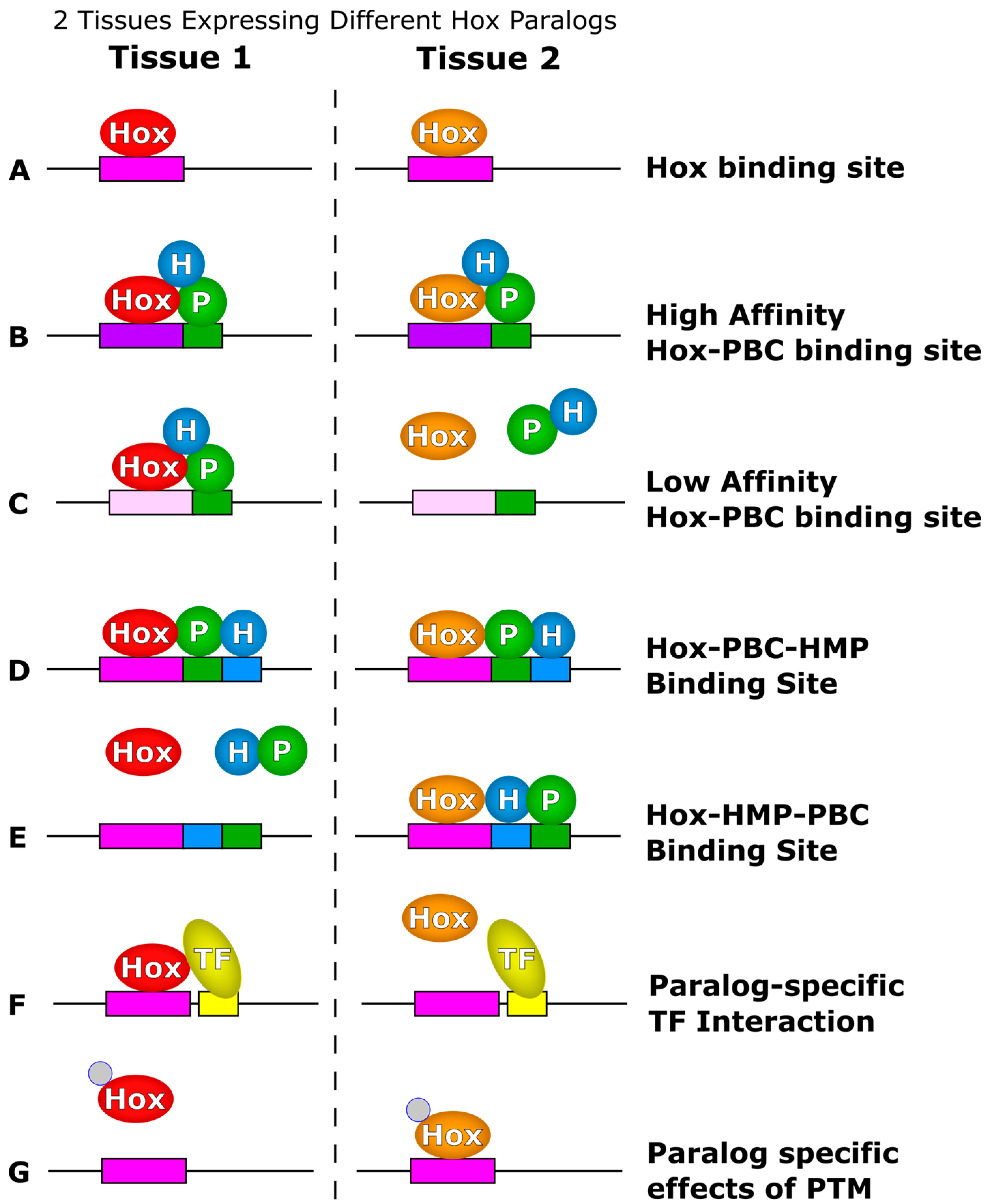
2.1. Interactions with PBC Factors Reveal Hox Latent Specificity
2.2. Low-Affinity Binding Sites Distinguish Hox-PBC Dimers
2.3. Additional PBC Interaction Surfaces Differentiate Hox Paralogs
2.4. Interactions with HMP Proteins Differentiate Hox Paralogs
2.5. Hox Paralogs Have Different Partner Preferences
2.6. Post-Translational Modifications Differentially Affect Hox Paralogs
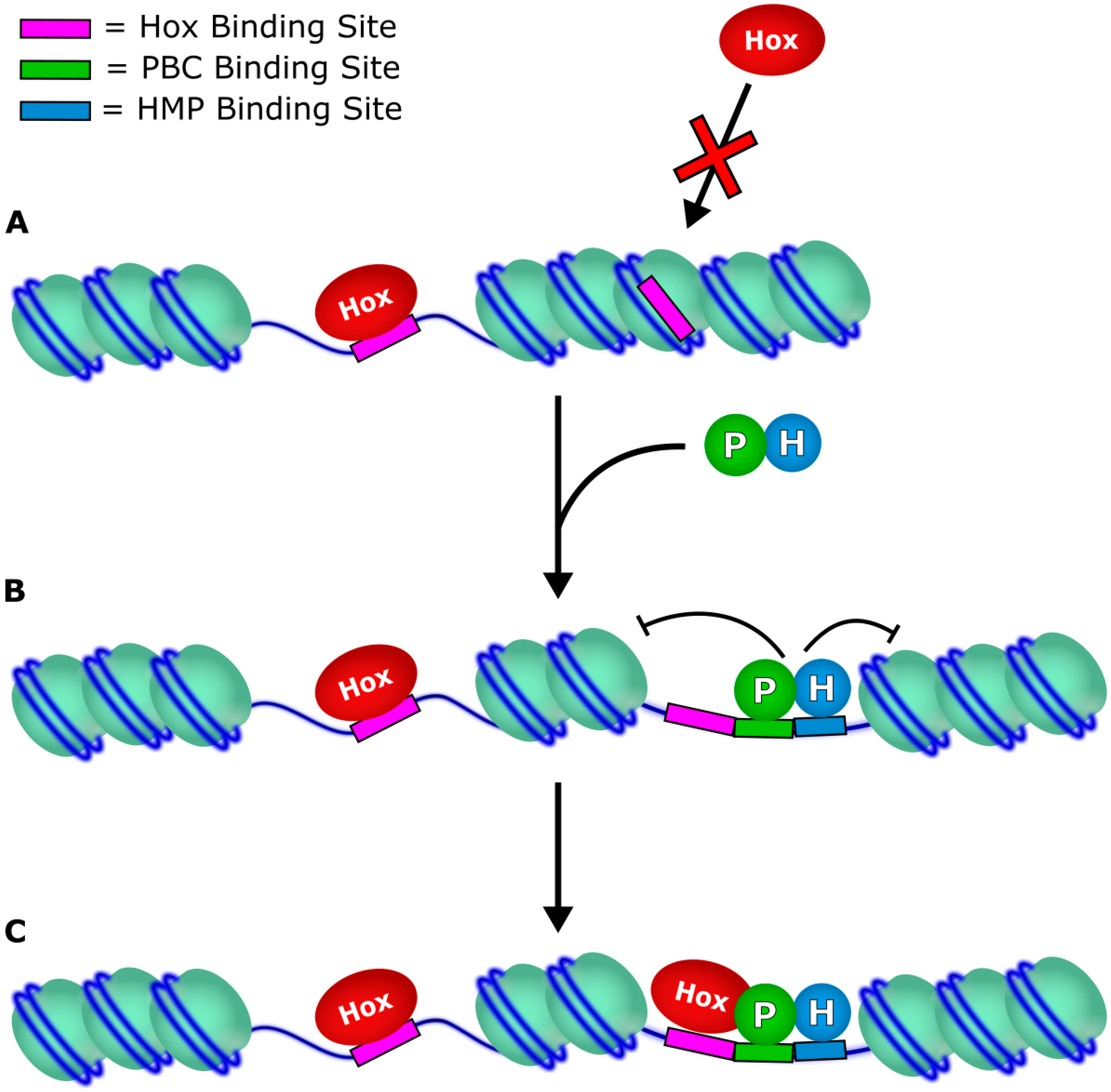
3. Target Accessibility
4. Effect on Transcription
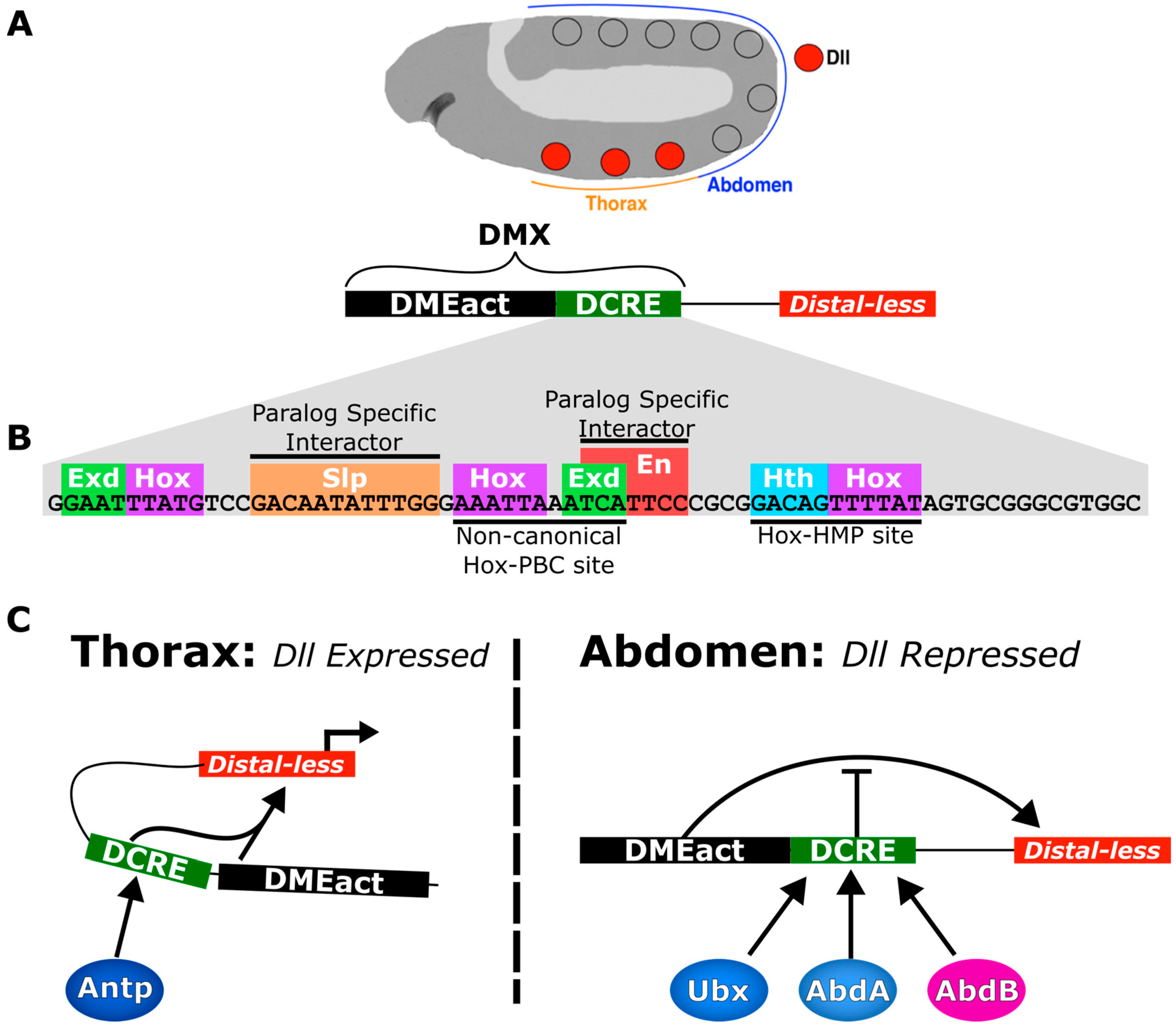
5. Case Study: The Distal-less Conserved Regulatory Element
6. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| CRM | cis-regulatory module |
| Lab | Labial |
| Scr | Sex-combs Reduced |
| Antp | Antennapedia |
| Ubx | Ultrabithorax |
| Abd-A | Abdominal-A |
| Abd-B | Abdominal-B |
| TALE | Three Amino-acid Loop Extension |
| Exd | Extradenticle |
| Hth | Homothorax |
References
- McGinnis, W.; Krumlauf, R. Homeobox genes and axial patterning. Cell 1992, 68, 283–302. [Google Scholar] [CrossRef]
- Cerdá-Esteban, N.; Spagnoli, F.M. Glimpse into Hox and tale regulation of cell differentiation and reprogramming. Dev. Dyn. 2014, 243, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Schneuwly, S.; Klemenz, R.; Gehring, W.J. Redesigning the body plan of Drosophila by ectopic expression of the homoeotic gene Antennapedia. Nature 1987, 325, 816–818. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Helms, J.A.; Chang, H.Y. Regeneration, repair and remembering identity: The three Rs of Hox gene expression. Trends Cell Biol. 2009, 19, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, J.; Sorge, S.; Bujupi, F.; Eichenlaub, M.P.; Schulz, N.G.; Wittbrodt, J.; Lohmann, I. Hox Function is Required for the Development and Maintenance of the Drosophila Feeding Motor Unit. Cell Rep. 2016, 14, 850–860. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Anaya, J.; D’Aniello, S.; Kuratani, S.; Garcia-Fernàndez, J. Evolution of Hoxgene clusters in deuterostomes. BMC Dev. Biol. 2013, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, A.S.; Ferrier, D.E.K. Hox genes are not always Colinear. Int. J. Biol. Sci. 2006, 2, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Wellik, D.M. Hox Genes and Vertebrate Axial Pattern. Curr. Top. Dev. Biol. 2009, 88, 257–278. [Google Scholar] [PubMed]
- Swalla, B.J. Building divergent body plans with similar genetic pathways. Heredity 2006, 97, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Bondos, S.E.; Swint-Kruse, L.; Matthews, K.S. Flexibility and Disorder in Gene Regulation: LacI/GalR and Hox Proteins. J. Biol. Chem. 2015, 290, 24669–24677. [Google Scholar] [CrossRef] [PubMed]
- Passner, J.M.; Ryoo, H.D.; Shen, L.; Mann, R.S.; Aggarwal, A.K. Structure of a DNA-bound Ultrabithorax–Extradenticle homeodomain complex. Nature 1999, 397, 714–719. [Google Scholar] [CrossRef] [PubMed]
- Piper, D.E.; Batchelor, A.H.; Chang, C.-P.; Cleary, M.L.; Wolberger, C. Structure of a HoxB1–Pbx1 Heterodimer Bound to DNA: Role of the Hexapeptide and a Fourth Homeodomain Helix in Complex Formation. Cell 1999, 96, 587–597. [Google Scholar] [CrossRef]
- Papadopoulos, D.K.; Reséndez-Pérez, D.; Cárdenas-Chávez, D.L.; Villanueva-Segura, K.; Canales-del-Castillo, R.; Felix, D.A.; Fünfschilling, R.; Gehring, W.J. Functional synthetic Antennapedia genes and the dual roles of YPWM motif and linker size in transcriptional activation and repression. Proc. Natl. Acad. Sci. USA 2011, 108, 11959–11964. [Google Scholar] [CrossRef] [PubMed]
- Saadaoui, M.; Merabet, S.; Litim-Mecheri, I.; Arbeille, E.; Sambrani, N.; Damen, W.; Brena, C.; Pradel, J.; Graba, Y. Selection of distinct Hox-Extradenticle interaction modes fine-tunes Hox protein activity. Proc. Natl. Acad. Sci. USA 2011, 108, 2276–2281. [Google Scholar] [CrossRef] [PubMed]
- Prince, F.; Katsuyama, T.; Oshima, Y.; Plaza, S.; Resendez-Perez, D.; Berry, M.; Kurata, S.; Gehring, W.J. The YPWM motif links Antennapedia to the basal transcriptional machinery. Development 2008, 135, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Boube, M.; Hudry, B.; Immarigeon, C.; Carrier, Y.; Bernat-Fabre, S.; Merabet, S.; Graba, Y.; Bourbon, H.-M.; Cribbs, D.L. Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of mediator subunit Med19. PLoS Genet. 2014, 10, e1004303. [Google Scholar] [CrossRef] [PubMed]
- Foos, N.; Maurel-Zaffran, C.; Maté, M.J.; Vincentelli, R.; Hainaut, M.; Berenger, H.; Pradel, J.; Saurin, A.J.; Ortiz-Lombardía, M.; Graba, Y. A Flexible Extension of the Drosophila Ultrabithorax Homeodomain Defines a Novel Hox/PBC Interaction Mode. Structure 2015, 23, 270–279. [Google Scholar] [CrossRef] [PubMed]
- Livingstone, C.D.; Barton, G.J. Protein sequence alignments: A strategy for the hierarchical analysis of residue conservation. Comput. Appl. Biosci. 1993, 9, 745–756. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Noyes, M.B.; Christensen, R.G.; Wakabayashi, A.; Stormo, G.D.; Brodsky, M.H.; Wolfe, S.A. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell 2008, 133, 1277–1289. [Google Scholar] [CrossRef] [PubMed]
- Berger, M.F.; Badis, G.; Gehrke, A.R.; Talukder, S.; Philippakis, A.A.; Peña-Castillo, L.; Alleyne, T.M.; Mnaimneh, S.; Botvinnik, O.B.; Chan, E.T.; et al. Variation in Homeodomain DNA Binding Revealed by High-Resolution Analysis of Sequence Preferences. Cell 2008, 133, 1266–1276. [Google Scholar] [CrossRef] [PubMed]
- Slattery, M.; Riley, T.; Liu, P.; Abe, N.; Gomez-Alcala, P.; Dror, I.; Zhou, T.; Rohs, R.; Honig, B.; Bussemaker, H.J.; et al. Cofactor Binding Evokes Latent Differences in DNA Binding Specificity between Hox Proteins. Cell 2011, 147, 1270–1282. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Lazzarini, R.A.; Pick, L. Functional dissection of the mouse Hox-a5 gene. EMBO J. 1996, 15, 1313–1322. [Google Scholar] [PubMed]
- Tan, X.-X.; Bondos, S.; Li, L.; Matthews, K.S. Transcription Activation by Ultrabithorax Ib Protein Requires a Predicted α-Helical Region. Biochemistry 2002, 41, 2774–2785. [Google Scholar] [CrossRef] [PubMed]
- Williams, T.M.; Williams, M.E.; Innis, J.W. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev. Biol. 2005, 277, 457–471. [Google Scholar] [CrossRef] [PubMed]
- Hsiao, H.-C.; Gonzalez, K.L.; Catanese, D.J., Jr.; Jordy, K.E.; Matthews, K.S.; Bondos, S.E. The Intrinsically Disordered Regions of the Drosophila melanogaster Hox Protein Ultrabithorax Select Interacting Proteins Based on Partner Topology. PLoS ONE 2014, 9, e108217. [Google Scholar] [CrossRef] [PubMed]
- Regulski, M.; Harding, K.; Kostriken, R.; Karch, F.; Levine, M.; McGinnis, W. Homeo box genes of the Antennapedia and Bithorax Complexes of Drosophila. Cell 1985, 43, 71–80. [Google Scholar] [CrossRef]
- Rezsohazy, R. Non-transcriptional interactions of Hox proteins: inventory, facts, and future directions. Dev. Dyn. 2014, 243, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.; Serano, J.M.; Jarvis, E.; Bruce, H.S.; Wang, J.; Ray, S.; Barker, C.A.; O’Connell, L.C.; Patel, N.H. CRISPR/Cas9 Mutagenesis Reveals Versatile Roles of Hox Genes in Crustacean Limb Specification and Evolution. Curr. Biol. 2016, 26, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Lu, Z.J.; Zhong, M.; Sarov, M.; Murray, J.I.; Brdlik, C.M.; Janette, J.; Chen, C.; Alves, P.; Preston, E.; et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 2011, 21, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Bach, C.; Buhl, S.; Mueller, D.; García-Cuéllar, M.-P.; Maethner, E.; Slany, R.K. Leukemogenic transformation by HOXA cluster genes. Blood 2010, 115, 2910–2918. [Google Scholar] [CrossRef] [PubMed]
- Misra, M.; Sours, E.; Lance-Jones, C. Hox transcription factors influence motoneuron identity through the integrated actions of both homeodomain and non-homeodomain regions. Dev. Dyn. 2012, 241, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Knoch, L.; Witte, H.; Sommer, R.J. Functional specificity of the nematode Hox gene mab-5. Development 2003, 130, 983–993. [Google Scholar] [CrossRef] [PubMed]
- Hueber, S.D.; Bezdan, D.; Henz, S.R.; Blank, M.; Wu, H.; Lohmann, I. Comparative analysis of Hox downstream genes in Drosophila. Development 2007, 134, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Pearson, J.C.; Lemons, D.; McGinnis, W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005, 6, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.S.; Lelli, K.M.; Joshi, R. Hox Specificity : Unique Roles for Cofactors and Collaborators. Curr. Top. Dev. Biol. 2009, 88, 63–101. [Google Scholar] [PubMed]
- Gebelein, B.; Culi, J.; Ryoo, H.D.; Zhang, W.; Mann, R.S. Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev. Cell 2002, 3, 487–498. [Google Scholar] [CrossRef]
- Li-Kroeger, D.; Witt, L.; Grimes, H.; Cook, T.A.; Gebelein, B. Hox and Senseless Antagonism Functions as a Molecular Switch to Regulate EGF Secretion in the Drosophila PNS. Dev. Cell 2008, 15, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Crocker, J.; Abe, N.; Rinaldi, L.; McGregor, A.P.; Frankel, N.; Wang, S.; Alsawadi, A.; Valenti, P.; Plaza, S.; Payre, F.; et al. Low affinity binding site clusters confer hox specificity and regulatory robustness. Cell 2015, 160, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Hirth, F.; Loop, T.; Egger, B.; Miller, D.F.B.; Kaufman, T.C.; Reichert, H. Functional equivalence of Hox gene products in the specification of the tritocerebrum during embryonic brain development of Drosophila. Development 2001, 128, 4781–4788. [Google Scholar] [PubMed]
- Raines, A.M.; Magella, B.; Adam, M.; Potter, S.S. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev. Biol. 2015, 15, 28. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.F.; Montgomery, J.C.; Rozenfeld, S.; Moskow, J.J.; Lawrence, H.J.; Buchberg, A.M.; Largman, C. AbdB-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol. Cell. Biol. 1997, 17, 6448–6458. [Google Scholar] [CrossRef] [PubMed]
- Jolma, A.; Yin, Y.; Nitta, K.R.; Dave, K.; Popov, A.; Taipale, M.; Enge, M.; Kivioja, T.; Morgunova, E.; Taipale, J. DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 2015, 527, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Gebelein, B.; McKay, D.J.; Mann, R.S. Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 2004, 431, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Li-Kroeger, D.; Cook, T.A.; Gebelein, B. Integration of an abdominal Hox complex with Pax2 yields cell-specific EGF secretion from Drosophila sensory precursor cells. Development 2012, 139, 1611–1619. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ellmann, S.; Rubin, E.; Gil, M.; Jin, K.; Han, L.; Chen, H.; Kwon, E.M.; Guo, J.; Ha, H.C.; et al. ADP ribosylation by PARP-1 suppresses HOXB7 transcriptional activity. PLoS ONE 2012, 7, e40644. [Google Scholar] [CrossRef] [PubMed]
- Eklund, E.A.; Jalava, A.; Kakar, R. Tyrosine Phosphorylation of HoxA10 Decreases DNA Binding and Transcriptional Repression during Interferon γ-induced Differentiation of Myeloid Leukemia Cell Lines. J. Biol. Chem. 2000, 275, 20117–20126. [Google Scholar] [PubMed]
- Vijapurkar, U.; Fischbach, N.; Shen, W.; Brandts, C.; Stokoe, D.; Lawrence, H.J.; Largman, C. Protein kinase C-mediated phosphorylation of the leukemia-associated HOXA9 protein impairs its DNA binding ability and induces myeloid differentiation. Mol. Cell. Biol. 2004, 24, 3827–3837. [Google Scholar] [CrossRef] [PubMed]
- Yaron, Y.; McAdara, J.K.; Lynch, M.; Hughes, E.; Gasson, J.C. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J. Immunol. 2001, 166, 5058–5067. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Wieschaus, E. Mutations in the Drosophila gene extradenticle affect the way specific homeo domain proteins regulate segmental identity. Genes Dev. 1990, 4, 1209–1223. [Google Scholar] [CrossRef] [PubMed]
- Nourse, J.; Mellentin, J.D.; Galili, N.; Wilkinson, J.; Stanbridge, E.; Smith, S.D.; Cleary, M.L. Chromosomal translocation t(1;19) results in synthesis of a homeobox fusion mRNA that codes for a potential chimeric transcription factor. Cell 1990, 60, 535–545. [Google Scholar] [CrossRef]
- Kamps, M.P.; Murre, C.; Sun, X.; Baltimore, D. A new homeobox gene contributes the DNA binding domain of the t(1;19) translocation protein in pre-B all. Cell 1990, 60, 547–555. [Google Scholar] [CrossRef]
- Bürglin, T.R. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997, 25, 4173–4180. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-K.; Jaffe, L.; Capovilla, M.; Botas, J.; Mann, R.S. The DNA binding specificity of ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 1994, 78, 603–615. [Google Scholar] [CrossRef]
- Chang, C.P.; Brocchieri, L.; Shen, W.F.; Largman, C.; Cleary, M.L. Pbx modulation of Hox homeodomain amino-terminal arms establishes different DNA-binding specificities across the Hox locus. Mol. Cell. Biol. 1996, 16, 1734–1745. [Google Scholar] [CrossRef] [PubMed]
- Pöpperl, H.; Bienz, M.; Studer, M.; Chan, S.K.; Aparicio, S.; Brenner, S.; Mann, R.S.; Krumlauf, R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 1995, 81, 1031–1042. [Google Scholar] [CrossRef]
- Rauskolb, C.; Peifer, M.; Wieschaus, E. Extradenticle, a regulator of homeotic gene activity, is a homolog of the homeobox-containing human proto-oncogene pbx1. Cell 1993, 74, 1101–1112. [Google Scholar] [CrossRef]
- Rauskolb, C.; Wieschaus, E. Coordinate regulation of downstream genes by extradenticle and the homeotic selector proteins. EMBO J. 1994, 13, 3561–3569. [Google Scholar] [PubMed]
- Sun, B.; Hursh, D.A.; Jackson, D.; Beachy, P.A. Ultrabithorax protein is necessary but not sufficient for full activation of decapentaplegic expression in the visceral mesoderm. EMBO J. 1995, 14, 520–535. [Google Scholar] [PubMed]
- Izpisúa-Belmonte, J.C.; Falkenstein, H.; Dollé, P.; Renucci, A.; Duboule, D. Murine genes related to the Drosophila AbdB homeotic genes are sequentially expressed during development of the posterior part of the body. EMBO J. 1991, 10, 2279–2289. [Google Scholar] [PubMed]
- Van Dijk, M.A.; Murre, C. Extradenticle Raises the DNA binding specificity of homeotic selector gene products. Cell 1994, 78, 617–624. [Google Scholar] [CrossRef]
- Ebner, A.; Cabernard, C.; Affolter, M.; Merabet, S. Recognition of distinct target sites by a unique Labial/Extradenticle/Homothorax complex. Development 2005, 132, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Ryoo, H.D.; Marty, T.; Casares, F.; Affolter, M.; Mann, R.S. Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 1999, 126, 5137–5148. [Google Scholar] [PubMed]
- Ferretti, E.; Cambronero, F.; Tümpel, S.; Longobardi, E.; Wiedemann, L.M.; Blasi, F.; Krumlauf, R. Hoxb1 Enhancer and Control of Rhombomere 4 Expression: Complex Interplay between PREP1-PBX1-HOXB1 Binding Sites. Mol. Cell. Biol. 2005, 25, 8541–8552. [Google Scholar] [CrossRef] [PubMed]
- Ferretti, E.; Marshall, H.; Popperl, H.; Maconochie, M.; Krumlauf, R.; Blasi, F. Segmental expression of Hoxb2 in r4 requires two separate sites that integrate cooperative interactions between Prep1, Pbx and Hox proteins. Development 2000, 127, 155–166. [Google Scholar] [PubMed]
- Tümpel, S.; Cambronero, F.; Ferretti, E.; Blasi, F.; Wiedemann, L.M.; Krumlauf, R. Expression of Hoxa2 in rhombomere 4 is regulated by a conserved cross-regulatory mechanism dependent upon Hoxb1. Dev. Biol. 2007, 302, 646–660. [Google Scholar] [CrossRef] [PubMed]
- Manzanares, M.; Bel-Vialar, S.; Ariza-McNaughton, L.; Ferretti, E.; Marshall, H.; Maconochie, M.M.; Blasi, F.; Krumlauf, R. Independent regulation of initiation and maintenance phases of Hoxa3 expression in the vertebrate hindbrain involve auto- and cross-regulatory mechanisms. Development 2001, 128, 3595–3607. [Google Scholar] [PubMed]
- Uhl, J.D.; Zandvakili, A.; Gebelein, B. A Hox Transcription Factor Collective Binds a Highly Conserved Distal-less cis-Regulatory Module to Generate Robust Transcriptional Outcomes. PLoS Genet 2016, 12, e1005981. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.; Passner, J.M.; Rohs, R.; Jain, R.; Sosinsky, A.; Crickmore, M.A.; Jacob, V.; Aggarwal, A.K.; Honig, B.; Mann, R.S. Functional Specificity of a Hox Protein Mediated by the Recognition of Minor Groove Structure. Cell 2007, 131, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Broströmer, E.; Xing, D.; Jin, J.; Chong, S.; Ge, H.; Wang, S.; Gu, C.; Yang, L.; Gao, Y.Q.; et al. Probing Allostery Through DNA. Science 2013, 339, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, K.; Pillay, S.; Huang, Y.-H.; Jayabal, S.; Udayasuryan, B.; Veerapandian, V.; Kolatkar, P.; Cojocaru, V.; Pervushin, K.; Jauch, R. DNA-mediated cooperativity facilitates the co-selection of cryptic enhancer sequences by SOX2 and PAX6 transcription factors. Nucleic Acids Res. 2015, 43, 1513–1528. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.P.; Shen, W.F.; Rozenfeld, S.; Lawrence, H.J.; Largman, C.; Cleary, M.L. Pbx proteins display hexapeptide-dependent cooperative DNA binding with a subset of Hox proteins. Genes Dev. 1995, 9, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Kamps, M.P. Heterodimerization of Hox proteins with Pbx1 and oncoprotein E2a-Pbx1 generates unique DNA-binding specifities at nucleotides predicted to contact the N-terminal arm of the Hox homeodomain--demonstration of Hox-dependent targeting of E2a-Pbx1 in vivo. Oncogene 1997, 14, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Abe, N.; Dror, I.; Yang, L.; Slattery, M.; Zhou, T.; Bussemaker, H.J.; Rohs, R.; Mann, R.S. Deconvolving the recognition of DNA shape from sequence. Cell 2015, 161, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Tanay, A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006, 16, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Scardigli, R.; Bäumer, N.; Gruss, P.; Guillemot, F.; Roux, I.L. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development 2003, 130, 3269–3281. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.I.; Barolo, S. Low-affinity transcription factor binding sites shape morphogen responses and enhancer evolution. Phil. Trans. R. Soc. B 2013, 368, 20130018. [Google Scholar] [CrossRef] [PubMed]
- Evans, N.C.; Swanson, C.I.; Barolo, S. Sparkling insights into enhancer structure, function, and evolution. Curr. Top. Dev. Biol. 2012, 98, 97–120. [Google Scholar] [PubMed]
- Crocker, J.; Noon, E.P.-B.; Stern, D.L. The Soft Touch: Low-Affinity Transcription Factor Binding Sites in Development and Evolution. Curr. Top. Dev. Biol. 2016, 17, 455–469. [Google Scholar]
- Chan, S.K.; Pöpperl, H.; Krumlauf, R.; Mann, R.S. An extradenticle-induced conformational change in a HOX protein overcomes an inhibitory function of the conserved hexapeptide motif. EMBO J. 1996, 15, 2476–2487. [Google Scholar] [PubMed]
- Galant, R.; Walsh, C.M.; Carroll, S.B. Hox repression of a target gene: Extradenticle-independent, additive action through multiple monomer binding sites. Development 2002, 129, 3115–3126. [Google Scholar] [PubMed]
- Lelli, K.M.; Noro, B.; Mann, R.S. Variable motif utilization in homeotic selector (Hox)–cofactor complex formation controls specificity. Proc. Natl. Acad. Sci. USA 2011, 108, 21122–21127. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Kambris, Z.; Capovilla, M.; Bérenger, H.; Pradel, J.; Graba, Y. The Hexapeptide and Linker Regions of the AbdA Hox Protein Regulate Its Activating and Repressive Functions. Dev. Cell 2003, 4, 761–768. [Google Scholar] [CrossRef]
- Merabet, S.; Saadaoui, M.; Sambrani, N.; Hudry, B.; Pradel, J.; Affolter, M.; Graba, Y. A unique Extradenticle recruitment mode in the Drosophila Hox protein Ultrabithorax. Proc. Natl. Acad. Sci. USA 2007, 104, 16946–16951. [Google Scholar] [CrossRef] [PubMed]
- Tour, E.; Hittinger, C.T.; McGinnis, W. Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development 2005, 132, 5271–5281. [Google Scholar] [CrossRef] [PubMed]
- Hudry, B.; Remacle, S.; Delfini, M.-C.; Rezsohazy, R.; Graba, Y.; Merabet, S. Hox Proteins Display a Common and Ancestral Ability to Diversify Their Interaction Mode with the PBC Class Cofactors. PLoS Biol. 2012, 10, e1001351. [Google Scholar] [CrossRef] [PubMed]
- Jürgens, G.; Wieschaus, E.; Nüsslein-Volhard, C.; Kluding, H. Mutations affecting the pattern of the larval cuticle inDrosophila melanogaster. Wilhelm Rouxs Arch. Dev. Biol. 1984, 193, 283–295. [Google Scholar] [CrossRef]
- Moskow, J.J.; Bullrich, F.; Huebner, K.; Daar, I.O.; Buchberg, A.M. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol. Cell. Biol. 1995, 15, 5434–5443. [Google Scholar] [CrossRef] [PubMed]
- Rieckhof, G.E.; Casares, F.; Ryoo, H.D.; Abu-Shaar, M.; Mann, R.S. Nuclear Translocation of Extradenticle Requires homothorax, which Encodes an Extradenticle-Related Homeodomain Protein. Cell 1997, 91, 171–183. [Google Scholar] [CrossRef]
- Abu-Shaar, M.; Ryoo, H.D.; Mann, R.S. Control of the nuclear localization of extradenticle by competing nuclear import and export signals. Genes Dev. 1999, 13, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Berthelsen, J.; Kilstrup-Nielsen, C.; Blasi, F.; Mavilio, F.; Zappavigna, V. The subcellular localization of PBX1 and EXD proteins depends on nuclear import and export signals and is modulated by association with PREP1 and HTH. Genes Dev. 1999, 13, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Jaw, T.J.; You, L.R.; Knoepfler, P.S.; Yao, L.C.; Pai, C.Y.; Tang, C.Y.; Chang, L.P.; Berthelsen, J.; Blasi, F.; Kamps, M.P.; et al. Direct interaction of two homeoproteins, homothorax and extradenticle, is essential for EXD nuclear localization and function. Mech. Dev. 2000, 91, 279–291. [Google Scholar] [CrossRef]
- Pai, C.Y.; Kuo, T.S.; Jaw, T.J.; Kurant, E.; Chen, C.T.; Bessarab, D.A.; Salzberg, A.; Sun, Y.H. The Homothorax homeoprotein activates the nuclear localization of another homeoprotein, extradenticle, and suppresses eye development in Drosophila. Genes Dev. 1998, 12, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Hudry, B.; Thomas-Chollier, M.; Volovik, Y.; Duffraisse, M.; Dard, A.; Frank, D.; Technau, U.; Merabet, S. Molecular insights into the origin of the Hox-TALE patterning system. eLife 2014, 3, e01939. [Google Scholar] [CrossRef] [PubMed]
- Berthelsen, J.; Zappavigna, V.; Ferretti, E.; Mavilio, F.; Blasi, F. The novel homeoprotein Prep1 modulates Pbx–Hox protein cooperativity. EMBO J. 1998, 17, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, Y.; Schnabel, C.A.; Cleary, M.L. Trimeric Association of Hox and TALE Homeodomain Proteins Mediates Hoxb2 Hindbrain Enhancer Activity. Mol. Cell. Biol. 1999, 19, 5134–5142. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, K.; Green, N.C.; Rambaldi, I.; Saragovi, H.U.; Featherstone, M.S. PBX and MEIS as Non-DNA-Binding Partners in Trimeric Complexes with HOX Proteins. Mol. Cell. Biol. 1999, 19, 7577–7588. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.F.; Rozenfeld, S.; Kwong, A.; Kömüves, L.G.; Lawrence, H.J.; Largman, C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol. Cell. Biol. 1999, 19, 3051–3061. [Google Scholar] [CrossRef] [PubMed]
- Uhl, J.D.; Cook, T.A.; Gebelein, B. Comparing anterior and posterior Hox complex formation reveals guidelines for predicting cis-regulatory elements. Dev. Biol. 2010, 343, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Grice, J.; Noyvert, B.; Doglio, L.; Elgar, G. A Simple Predictive Enhancer Syntax for Hindbrain Patterning Is Conserved in Vertebrate Genomes. PLoS ONE 2015, 10, e0130413. [Google Scholar] [CrossRef] [PubMed]
- Baëza, M.; Viala, S.; Heim, M.; Dard, A.; Hudry, B.; Duffraisse, M.; Rogulja-Ortmann, A.; Brun, C.; Merabet, S. Inhibitory activities of short linear motifs underlie Hox interactome specificity in vivo. eLife 2015, 4, e06034. [Google Scholar] [CrossRef] [PubMed]
- Gong, K.-Q.; Yallowitz, A.R.; Sun, H.; Dressler, G.R.; Wellik, D.M. A Hox-Eya-Pax complex regulates early kidney developmental gene expression. Mol. Cell. Biol. 2007, 27, 7661–7668. [Google Scholar] [CrossRef] [PubMed]
- Gavis, E.R.; Hogness, D.S. Phosphorylation, expression and function of the Ultrabithorax protein family in Drosophila melanogaster. Development 1991, 112, 1077–1093. [Google Scholar] [PubMed]
- Hsia, C.C.; Paré, A.C.; Hannon, M.; Ronshaugen, M.; McGinnis, W. Silencing of an abdominal Hox gene during early development is correlated with limb development in a crustacean trunk. Evol. Dev. 2010, 12, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, L.; Ryoo, H.D.; Mann, R.S. A role for phosphorylation by casein kinase II in modulating Antennapedia activity in Drosophila. Genes Dev. 1997, 11, 1327–1340. [Google Scholar] [CrossRef] [PubMed]
- Taghli-Lamallem, O.; Hsia, C.; Ronshaugen, M.; McGinnis, W. Context-dependent regulation of Hox protein functions by CK2 phosphorylation sites. Dev. Genes Evol. 2008, 218, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Eklund, E.A.; Goldenberg, I.; Lu, Y.; Andrejic, J.; Kakar, R. SHP1 protein-tyrosine phosphatase regulates HoxA10 DNA binding and transcriptional repression activity in undifferentiated myeloid cells. J. Biol. Chem. 2002, 277, 36878–36888. [Google Scholar] [CrossRef] [PubMed]
- Lindsey, S.; Huang, W.; Wang, H.; Horvath, E.; Zhu, C.; Eklund, E.A. Activation of SHP2 protein-tyrosine phosphatase increases HoxA10-induced repression of the genes encoding gp91(PHOX) and p67(PHOX). J. Biol. Chem. 2007, 282, 2237–2249. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lindsey, S.; Konieczna, I.; Bei, L.; Horvath, E.; Huang, W.; Saberwal, G.; Eklund, E.A. Constitutively active SHP2 cooperates with HoxA10 overexpression to induce acute myeloid leukemia. J. Biol. Chem. 2009, 284, 2549–2567. [Google Scholar] [CrossRef] [PubMed]
- Basnet, H.; Su, X.B.; Tan, Y.; Meisenhelder, J.; Merkurjev, D.; Ohgi, K.A.; Hunter, T.; Pillus, L.; Rosenfeld, M.G. Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 2014, 516, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Kraus, W.L.; Hottiger, M.O. PARP-1 and gene regulation: Progress and puzzles. Mol. Aspects Med. 2013, 34, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Jones, K.A. CK2 Controls the Recruitment of Wnt Regulators to Target Genes in Vivo. Curr. Biol. 2006, 16, 2239–2244. [Google Scholar] [CrossRef] [PubMed]
- Biggin, M.D. Animal Transcription Networks as Highly Connected, Quantitative Continua. Dev. Cell 2011, 21, 611–626. [Google Scholar] [CrossRef] [PubMed]
- Carroll, S.B.; Grenier, J.K.; Weatherbee, S.D. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design, 2nd ed.; Blackwell Pub: Malden, MA, USA, 2005. [Google Scholar]
- Casares, F.; Calleja, M.; Sánchez-Herrero, E. Functional similarity in appendage specification by the Ultrabithorax and abdominal-A Drosophila HOX genes. EMBO J. 1996, 15, 3934–3942. [Google Scholar] [PubMed]
- McKay, D.J.; Lieb, J.D. A Common Set of DNA Regulatory Elements Shapes Drosophila Appendages. Dev. Cell 2013, 27, 306–318. [Google Scholar] [CrossRef] [PubMed]
- Beh, C.Y.; El-Sharnouby, S.; Chatzipli, A.; Russell, S.; Choo, S.W.; White, R. Roles of cofactors and chromatin accessibility in Hox protein target specificity. Epigenetics Chromatin 2016, 9. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.-K.; Ladam, F.; Sagerström, C.G. TALE factors poise promoters for activation by Hox proteins. Dev. Cell 2014, 28, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Choe, S.-K.; Sagerström, C.G. Variable Meis-dependence among paralog group-1 Hox proteins. Biochem. Biophys. Res. Commun. 2005, 331, 1384–1391. [Google Scholar] [CrossRef] [PubMed]
- Shogren-Knaak, M.; Ishii, H.; Sun, J.-M.; Pazin, M.J.; Davie, J.R.; Peterson, C.L. Histone H4-K16 Acetylation Controls Chromatin Structure and Protein Interactions. Science 2006, 311, 844–847. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Donaldson, I.J.; Zannino, D.A.; Hensman, J.; Rattray, M.; Losa, M.; Spitz, F.; Ladam, F.; Sagerström, C.; Bobola, N. Hoxa2 Selectively Enhances Meis Binding to Change a Branchial Arch Ground State. Dev. Cell 2015, 32, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Sitwala, K.; Bronstein, J.; Sanders, D.; Dandekar, M.; Collins, C.; Robertson, G.; MacDonald, J.; Cezard, T.; Bilenky, M.; et al. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood 2012, 119, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Choo, S.W.; White, R.; Russell, S. Genome-Wide Analysis of the Binding of the Hox Protein Ultrabithorax and the Hox Cofactor Homothorax in Drosophila. PLoS ONE 2011, 6, e14778. [Google Scholar] [CrossRef] [PubMed]
- Ladam, F.; Sagerström, C.G. Hox regulation of transcription: More complex(es). Dev. Dyn. 2014, 243, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Wang, J.; Miao, H.; Bronstein, J.; Nawer, H.; Xu, T.; Figueroa, M.; Muntean, A.G.; Hess, J.L. C/EBPα is an essential collaborator in Hoxa9/Meis1-mediated leukemogenesis. Proc. Natl. Acad. Sci. USA 2014, 111, 9899–9904. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.A.R.; Hassan, M.Q.; Saini, S.; Montecino, M.; van Wijnen, A.J.; Stein, G.S.; Stein, J.L.; Lian, J.B. Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol. Cell. Biol. 2010, 30, 3531–3541. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-H.; Park, S.J.; Han, J.-H.; Pathak, C.; Cheong, H.-K.; Lee, B.-J. Structural insight into the interaction between the Hox and HMGB1 and understanding of the HMGB1-enhancing effect of Hox-DNA binding. Biochim. Biophys. Acta 2015, 1854, 449–459. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Goldenberg, I.; Bei, L.; Andrejic, J.; Eklund, E.A. HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J. Biol. Chem. 2003, 278, 47792–47802. [Google Scholar] [CrossRef] [PubMed]
- Zappavigna, V.; Falciola, L.; Helmer-Citterich, M.; Mavilio, F.; Bianchi, M.E. HMG1 interacts with HOX proteins and enhances their DNA binding and transcriptional activation. EMBO J. 1996, 15, 4981–4991. [Google Scholar] [PubMed]
- Hassan, M.Q.; Tare, R.; Lee, S.H.; Mandeville, M.; Weiner, B.; Montecino, M.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. HOXA10 controls osteoblastogenesis by directly activating bone regulatory and phenotypic genes. Mol. Cell. Biol. 2007, 27, 3337–3352. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.A.R.; Hassan, M.Q.; Koss, M.; Montecino, M.; Selleri, L.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; Lian, J.B. Epigenetic regulation of early osteogenesis and mineralized tissue formation by a HOXA10-PBX1-associated complex. Cells Tissues Organs 2011, 194, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Andrioli, L.P.; Oberstein, A.L.; Corado, M.S.G.; Yu, D.; Small, S. Groucho-dependent repression by sloppy-paired 1 differentially positions anterior pair-rule stripes in the Drosophila embryo. Dev. Biol. 2004, 276, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Roth, M.; Bonev, B.; Lindsay, J.; Lea, R.; Panagiotaki, N.; Houart, C.; Papalopulu, N. FoxG1 and TLE2 act cooperatively to regulate ventral telencephalon formation. Development 2010, 137, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Verginelli, F.; Perin, A.; Dali, R.; Fung, K.H.; Lo, R.; Longatti, P.; Guiot, M.-C.; del Maestro, R.F.; Rossi, S.; di Porzio, U.; et al. Transcription factors FOXG1 and Groucho/TLE promote glioblastoma growth. Nat. Commun. 2013, 4, 2956. [Google Scholar] [CrossRef] [PubMed]
- Cohen, S.M.; Brönner, G.; Küttner, F.; Jürgens, G.; Jäckle, H. Distal-less encodes a homoeodomain protein required for limb development in Drosophila. Nature 1989, 338, 432–434. [Google Scholar] [CrossRef] [PubMed]
- Vachon, G.; Cohen, B.; Pfeifle, C.; McGuffin, M.E.; Botas, J.; Cohen, S.M. Homeotic genes of the bithorax complex repress limb development in the abdomen of the Drosophila embryo through the target gene Distal-less. Cell 1992, 71, 437–450. [Google Scholar] [CrossRef]
- Sambrani, N.; Hudry, B.; Maurel-Zaffran, C.; Zouaz, A.; Mishra, R.; Merabet, S.; Graba, Y. Distinct Molecular Strategies for Hox-Mediated Limb Suppression in Drosophila: From Cooperativity to Dispensability/Antagonism in TALE Partnership. PLoS Genet 2013, 9, e1003307. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, P.A. The present status of the parasegment. Development 1988, 104, 61–65. [Google Scholar]
- Cadigan, K.M.; Grossniklaus, U.; Gehring, W.J. Localized expression of sloppy paired protein maintains the polarity of Drosophila parasegments. Genes Dev. 1994, 8, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Grossniklaus, U.; Pearson, R.K.; Gehring, W.J. The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 1992, 6, 1030–1051. [Google Scholar] [CrossRef] [PubMed]
- Agelopoulos, M.; McKay, D.J.; Mann, R.S. Developmental regulation of chromatin conformation by Hox proteins in Drosophila. Cell Rep. 2012, 1, 350–359. [Google Scholar] [CrossRef] [PubMed]
- Merabet, S.; Hudry, B. Hox transcriptional specificity despite a single class of cofactors: are flexible interaction modes the key? Plasticity in Hox/PBC interaction modes as a common molecular strategy for shaping Hox transcriptional activities. BioEssays 2013, 35, 88–92. [Google Scholar] [CrossRef] [PubMed]
- Grenier, J.K.; Carroll, S.B. Functional evolution of the Ultrabithorax protein. Proc. Natl. Acad. Sci. USA 2000, 97, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Ronshaugen, M.; McGinnis, N.; McGinnis, W. Hox protein mutation and macroevolution of the insect body plan. Nature 2002, 415, 914–917. [Google Scholar] [CrossRef] [PubMed]
- Averof, M.; Akam, M. Hox genes and the diversification of insect and crustacean body plans. Nature 1995, 376, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Averof, M.; Patel, N.H. Crustacean appendage evolution associated with changes in Hox gene expression. Nature 1997, 388, 682–686. [Google Scholar] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zandvakili, A.; Gebelein, B. Mechanisms of Specificity for Hox Factor Activity. J. Dev. Biol. 2016, 4, 16. https://doi.org/10.3390/jdb4020016
Zandvakili A, Gebelein B. Mechanisms of Specificity for Hox Factor Activity. Journal of Developmental Biology. 2016; 4(2):16. https://doi.org/10.3390/jdb4020016
Chicago/Turabian StyleZandvakili, Arya, and Brian Gebelein. 2016. "Mechanisms of Specificity for Hox Factor Activity" Journal of Developmental Biology 4, no. 2: 16. https://doi.org/10.3390/jdb4020016
APA StyleZandvakili, A., & Gebelein, B. (2016). Mechanisms of Specificity for Hox Factor Activity. Journal of Developmental Biology, 4(2), 16. https://doi.org/10.3390/jdb4020016




