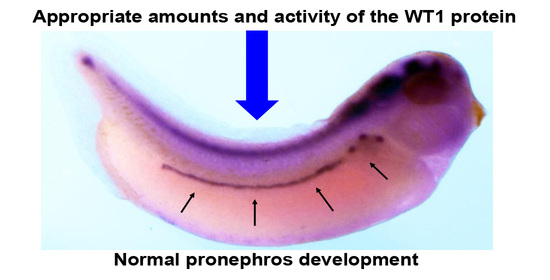
Appropriate Amounts and Activity of the Wilms’ Tumor Suppressor Gene, wt1, Are Required for Normal Pronephros Development of Xenopus Embryos
Abstract
1. Introduction
2. Materials and Methods
2.1. Embryos and Microinjection
2.2. Whole-Mount In Situ Hybridization (WISH)
2.3. Beta-Gal Staining
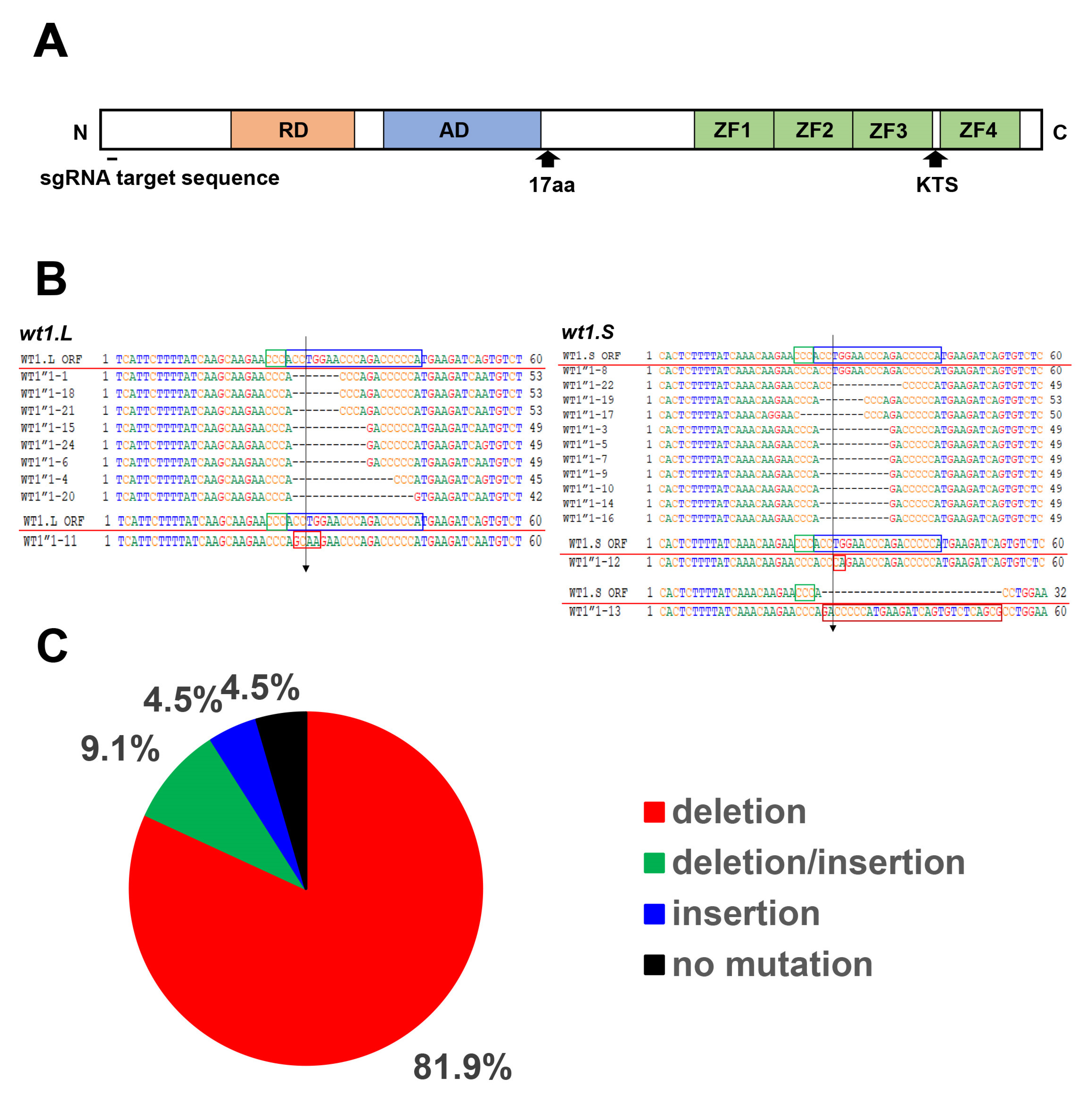
2.4. CRISPR/Cas9 Knockout of wt1 and tyrosinase
2.5. Plasmid Construction and mRNA Synthesis
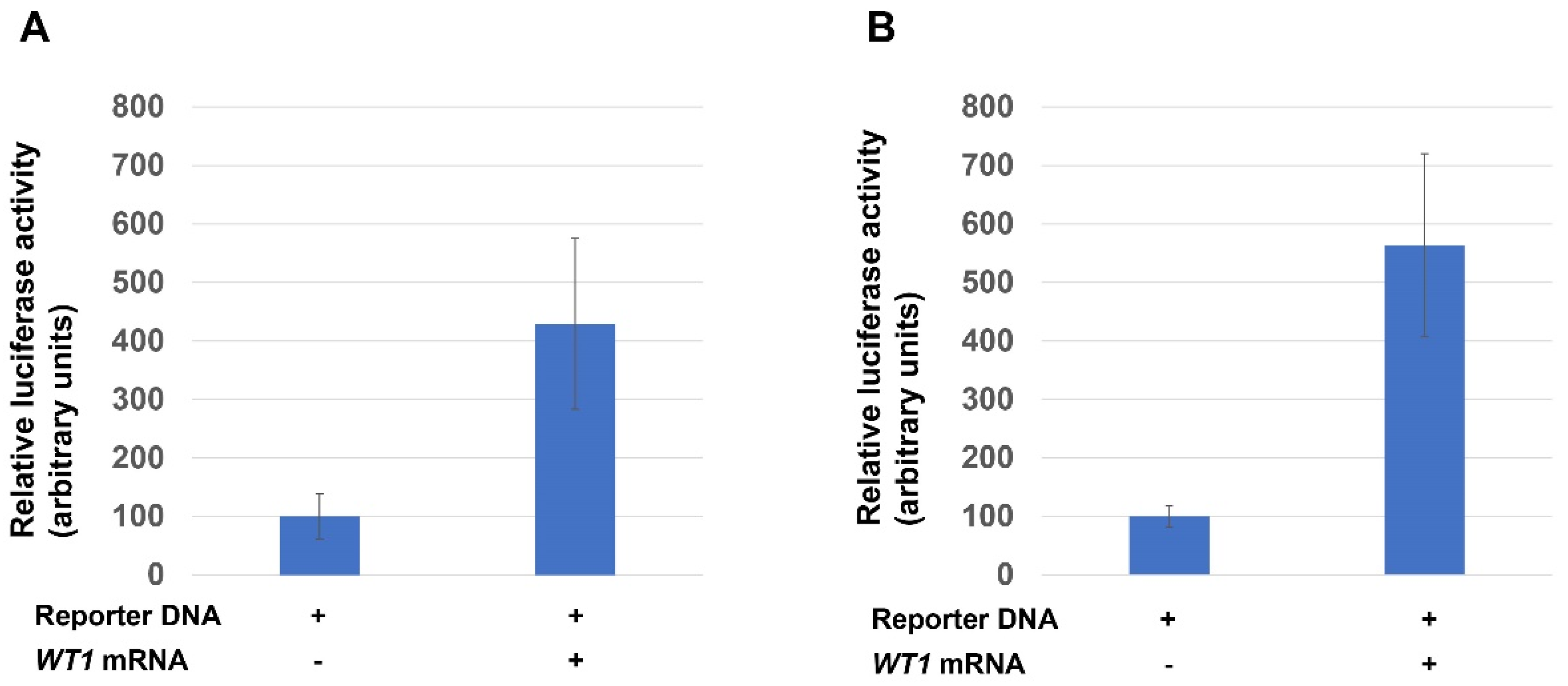
2.6. Luciferase Reporter Assay
3. Results
3.1. Reduced Expression of the Pronephros Marker Genes in the wt1 Knockout Embryos by CRISPR/Cas9 Method
3.2. Transcriptional Activity of the WT1 in Xenopus Embryo
3.3. Pronephric Phenotypes of the wt1 mRNA Injected Embryos
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bonetta, L.; Kuehn, S.E.; Huang, A.; Law, D.J.; Kalikin, L.M.; Koi, M.; Reeve, A.E.; Brownstein, B.H.; Yeger, H.; Williams, B.R.G.; et al. Wilms Tumor Locus on Llp13 Defined by Multiple CpG Island-Associated Transcripts. Science 1990, 250, 994–997. [Google Scholar] [CrossRef] [PubMed]
- Call, K.M.; Glaser, T.; Ito, C.Y.; Buckler, A.J.; Pelletier, J.; Haber, D.A.; Rose, E.A.; Kral, A.; Yeger, H.; Lewis, W.H.; et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell 1990, 60, 509–520. [Google Scholar] [CrossRef]
- Gessler, M.; Poustka, A.; Cavenee, W.; Neve, R.L.; Orkin, S.H.; Bruns, G.A. Homozygous deletion in Wilms tumors of a zinc-finger gene identified by chromosome jumping. Nature 1990, 343, 774–778. [Google Scholar] [CrossRef] [PubMed]
- Little, M.; Wells, C. A Clinical Overview of WT1 Gene Mutations. Hum. Mutat. 1997, 9, 209–225. [Google Scholar] [CrossRef]
- Rivera, M.N.; Haber, D.A. Wilms’ tumor: Connecting tumorigenesis and organ development in the kidney. Nat. Rev. Cancer 2005, 5, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Han, Y.; Saiz, F.S.; Minden, M.D. A tumor suppressor and oncogene: The WT1 story. Leukemia 2007, 21, 868–876. [Google Scholar] [CrossRef]
- Huff, V. Wilms’ Tumours: About Tumour Suppressor Genes, an Oncogene and a Chameleon Gene. Nat. Rev. Cancer 2011, 11, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Pritchard-Jones, K.; Fleming, S.; Davidson, D.; Bickmore, W.; Porteous, D.; Gosden, C.; Bard, J.; Buckler, A.; Pelletier, J.; Housman, D.; et al. The candidate Wilms’ tumor gene is involved in genitourinary development. Nature 1990, 346, 194–197. [Google Scholar] [CrossRef]
- Pelletier, J.; Schalling, M.; Buckler, A.J.; Rogers, A.; Haber, D.A.; Housman, D. Expression of the Wilms’ tumor gene WT1 in the murine urogenital system. Genes Dev. 1991, 5, 1345–1356. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.F.; Pritchard-Jones, K.; Bickmore, W.A.; Hastie, N.D.; Bard, J.B. The Expression of the Wilms’ Tumour Gene, WT1, in the Developing Mammalian Embryo. Mech. Dev. 1992, 40, 85–97. [Google Scholar] [CrossRef]
- Kreidberg, J.A.; Sarioia, H.; Loring, J.M.; Maeda, M.; Pelletier, J.; Housman, D.; Jaenisch, R. WT-1 Is Required for Early Kidney Development. Cell 1993, 74, 679–691. [Google Scholar] [CrossRef]
- Hartwig, S.; Ho, J.; Pandey, P.; Macisaac, K.; Taglienti, M.; Xiang, M.; Alterovitz, G.; Ramoni, M.; Fraenkel, E.; Kreidberg, J.A. Genomic Characterization of Wilms’ Tumor Suppressor 1 Targets in Nephron Progenitor Cells during Kidney Development. Development 2010, 137, 1189–1203. [Google Scholar] [CrossRef] [PubMed]
- Rauscher, F.J.; Morris, J.F.; Tournay, O.E.; Cook, D.M.; Curran, T. Binding of the Wilms’ tumor locus zinc finger protein to the EGR-1 consensus sequence. Science 1990, 250, 1259–1262. [Google Scholar] [CrossRef]
- Morris, J.F.; Madden, S.L.; Tournay, O.E.; Cook, D.M.; Sukhatme, V.P.; Rauscher, F.J. Characterization of the zinc finger protein encoded by the WT1 Wilms’ tumor locus. Oncogene 1991, 6, 2339–2348. [Google Scholar] [PubMed]
- Wangs, Z.Y.; Qius, Q.Q.; Deuel, T.F. Communication the Wilms’ Tumor Gene Product WT1 Activates or Suppresses Transcription through Separate Functional Domains. J. Biol. Chem. 1993, 268, 9172–9175. [Google Scholar] [CrossRef] [PubMed]
- Toska, E.; Roberts, S.G.E. Mechanisms of Transcriptional Regulation by WT1 (Wilms’ Tumor 1). Biochem. J. 2014, 461, 15–32. [Google Scholar] [CrossRef] [PubMed]
- Hastie, N.D. Life, Sex, and WT1 Isoforms—Three Amino Acids Can Make All the Difference. Cell 2001, 106, 391–394. [Google Scholar] [CrossRef]
- Carroll, T.J.; Vize, P.D. Wilms’ Tumor Suppressor Gene Is Involved in the Development of Disparate Kidney Forms: Evidence from Expression in the Xenopus Pronephros. Dev. Dyn. 1996, 206, 131–138. [Google Scholar] [CrossRef]
- Semba, K.; Saito-Ueno, R.; Takayama, G.; Kondo, M. cDNA Cloning and Its Pronephros-Specific Expression of the Wilms’ Tumor. Gene 1996, 175, 167–172. [Google Scholar] [CrossRef]
- Wallingford, J.B.; Carroll, T.J.; Vize, P.D. Precocious Expression of the Wilms’ Tumor Gene XWT1 Inhibits Embryonic Kidney Development in Xenopus Laevis. Dev. Biol. 1998, 202, 103–112. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delay, B.D.; Corkins, M.E.; Hanania, H.L.; Salanga, M.; Deng, J.M.; Sudou, N.; Taira, M.; Horb, M.E.; Miller, K.M. Tissue-Specific Gene Inactivation in Xenopus Laevis: Knockout of Lhx1 in the Kidney with CRISPR/Cas9. Genetics 2018, 208, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Nieuwkoop, P.D.; Faber, J. Normal Table of Xenopus laevis (Daudin): A systematical and Chronological Survey of the Development from the Fertilized Egg till the End of Metamorphosis; Garland Publishing, Inc.: New York, NY, USA; London, UK, 1984. [Google Scholar]
- Sive, H.L.; Grainger, R.M.; Harland, R.M. Early Development of Xenopus Laevis: A Laboratory Manual; Cold Spring Harbor Lab. Press: New York, NY, USA, 2000; pp. 249–274. [Google Scholar]
- Session, A.M.; Uno, Y.; Kwon, T.; Chapman, J.A.; Toyoda, A.; Takahashi, S.; Fukui, A.; Hikosaka, A.; Suzuki, A.; Kondo, M.; et al. Genome Evolution in the Allotetraploid Frog Xenopus Laevis. Nature 2016, 538, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Fortriede, J.D.; Pells, T.J.; Chu, S.; Chaturvedi, P.; Wang, D.Z.; Fisher, M.E.; James-Zorn, C.; Wang, Y.; Nenni, M.J.; A Burns, K.; et al. Xenbase: Deep integration of GEO & SRA RNA-seq and ChIP-seq data in a model organism database. Nucleic Acids Res. 2020, 48, D776–D782. [Google Scholar] [CrossRef] [PubMed]
- Taira, M.; Otani, H.; Jamrich, M.; Dawid, I.B. Expression of the LIM Class Homeobox Gene Xlim-1 in Pronephros and CNS Cell Lineages of Xenopus Embryos Is Affected by Retinoic Acid and Exogastrulation. Development 1994, 120, 1525–1536. [Google Scholar] [CrossRef] [PubMed]
- Heller, N.; Brändli, A.W. Xenopus Pax-2 Displays Multiple Splice Forms during Embryogenesis and Pronephric Kidney Development. Mech. Dev. 1997, 69, 83–104. [Google Scholar] [CrossRef]
- Carroll, T.J.; Wallingford, J.B.; Vize, P.D. Dynamic Patterns of Gene Expression in the Developing Pronephros of Xenopus Laevis. Dev. Genet. 1999, 207, 199–207. [Google Scholar] [CrossRef]
- Nakagama, H.; Heinrich, G.; Pelletier, J.; Housman, D.E. Sequence and Structural Requirements for High-Affinity DNA Binding by the WT1 Gene Product. Mol. Cell. Biol. 1995, 15, 1489–1498. [Google Scholar] [CrossRef]
- Dale, L.; Slack, J.M.W. Fate Map for the 32-Cell Stage of Xenopus Laevis. Development 1987, 551, 527–551. [Google Scholar] [CrossRef]
- Delay, B.D.; Krneta-stankic, V.; Miller, R.K. 2016. Technique to Target Microinjection to the Developing Xenopus Kidney. J. Vis. Exp. 2016, 111, 1–9. [Google Scholar] [CrossRef]
- Watanabe, M.; Whitman, M. FAST-1 Is a Key Maternal Effector of Mesoderm Inducers in the Early Xenopus Embryo. Development 1999, 126, 5621–5634. [Google Scholar] [CrossRef]
- Li, L.; Wen, L.; Gong, Y.; Mei, G.; Liu, J.; Chen, Y.; Peng, T. Xenopus as a Model System for the Study of GOLPH2/GP73 Function: Xenopus Golph2 Is Required for Pronephros Development. PLoS ONE 2012, 7, e38939. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, T.M.; Miller, R.K. Modeling Congenital Kidney Diseases in Xenopus Laevis. Dis. Model. Mech. 2019, 12, 038604. [Google Scholar] [CrossRef] [PubMed]
- Vize, P.D.; Jones, E.A.; Pfister, R. Development of the Xenopus pronephric system. Dev. Biol. 1995, 171, 531–540. [Google Scholar] [CrossRef]
- Madden, S.L.; Cook, D.M.; Morris, J.F.; Gashler, A.; Sukhatme, V.P.; Rauscher, F.J. Transcriptional Repression Mediated by the WT1 Wilms Tumor Gene Product. Science 1991, 253, 1550–1553. [Google Scholar] [CrossRef]
- Roberts, S.G.E. Transcriptional Regulation by WT1 in Development. Curr. Opin. Genet. Dev. 2005, 15, 542–547. [Google Scholar] [CrossRef] [PubMed]
- Ryan, G.; Steele-Perkins, V.; Morris, J.F.; Rauscher, F.J.; Dressler, G.R. Repression of Pax-2 by WT1 during Normal Kidney Development. Development 1995, 875, 867–875. [Google Scholar] [CrossRef]
- Reddy, J.C.; Hosono, S.; Licht, J.D. The Transcriptional Effect of WT1 Is Modulated by Choice of Expression Vector. J. Biol. Chem. 1995, 270, 29976–29982. [Google Scholar] [CrossRef] [PubMed]
- Darken, R.S.; Scola, A.M.; Rakeman, A.S.; Das, G.; Mlodzik, M.; Wilson, P.A. The Planar Polarity Gene Strabismus Regulates Convergent Extension Movements in Xenopus. EMBO J. 2002, 21, 976–985. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiraki, T.; Hayashi, T.; Ozue, J.; Watanabe, M. Appropriate Amounts and Activity of the Wilms’ Tumor Suppressor Gene, wt1, Are Required for Normal Pronephros Development of Xenopus Embryos. J. Dev. Biol. 2022, 10, 46. https://doi.org/10.3390/jdb10040046
Shiraki T, Hayashi T, Ozue J, Watanabe M. Appropriate Amounts and Activity of the Wilms’ Tumor Suppressor Gene, wt1, Are Required for Normal Pronephros Development of Xenopus Embryos. Journal of Developmental Biology. 2022; 10(4):46. https://doi.org/10.3390/jdb10040046
Chicago/Turabian StyleShiraki, Taisei, Takuma Hayashi, Jotaro Ozue, and Minoru Watanabe. 2022. "Appropriate Amounts and Activity of the Wilms’ Tumor Suppressor Gene, wt1, Are Required for Normal Pronephros Development of Xenopus Embryos" Journal of Developmental Biology 10, no. 4: 46. https://doi.org/10.3390/jdb10040046
APA StyleShiraki, T., Hayashi, T., Ozue, J., & Watanabe, M. (2022). Appropriate Amounts and Activity of the Wilms’ Tumor Suppressor Gene, wt1, Are Required for Normal Pronephros Development of Xenopus Embryos. Journal of Developmental Biology, 10(4), 46. https://doi.org/10.3390/jdb10040046