Involvement of a Basic Helix-Loop-Helix Gene BHLHE40 in Specification of Chicken Retinal Pigment Epithelium
Abstract
1. Introduction
2. Materials and Methods
2.1. Chicken Embryos
2.2. Isolation of Chicken cDNAs and Construction of Expression Vectors
2.3. Cryosectioning
2.4. In Situ Hybridization (ISH)
2.5. Western Blot Analysis
2.6. In Ovo Electroporation
2.7. Immunofluorescence
2.8. Image Capture and Processing
3. Results
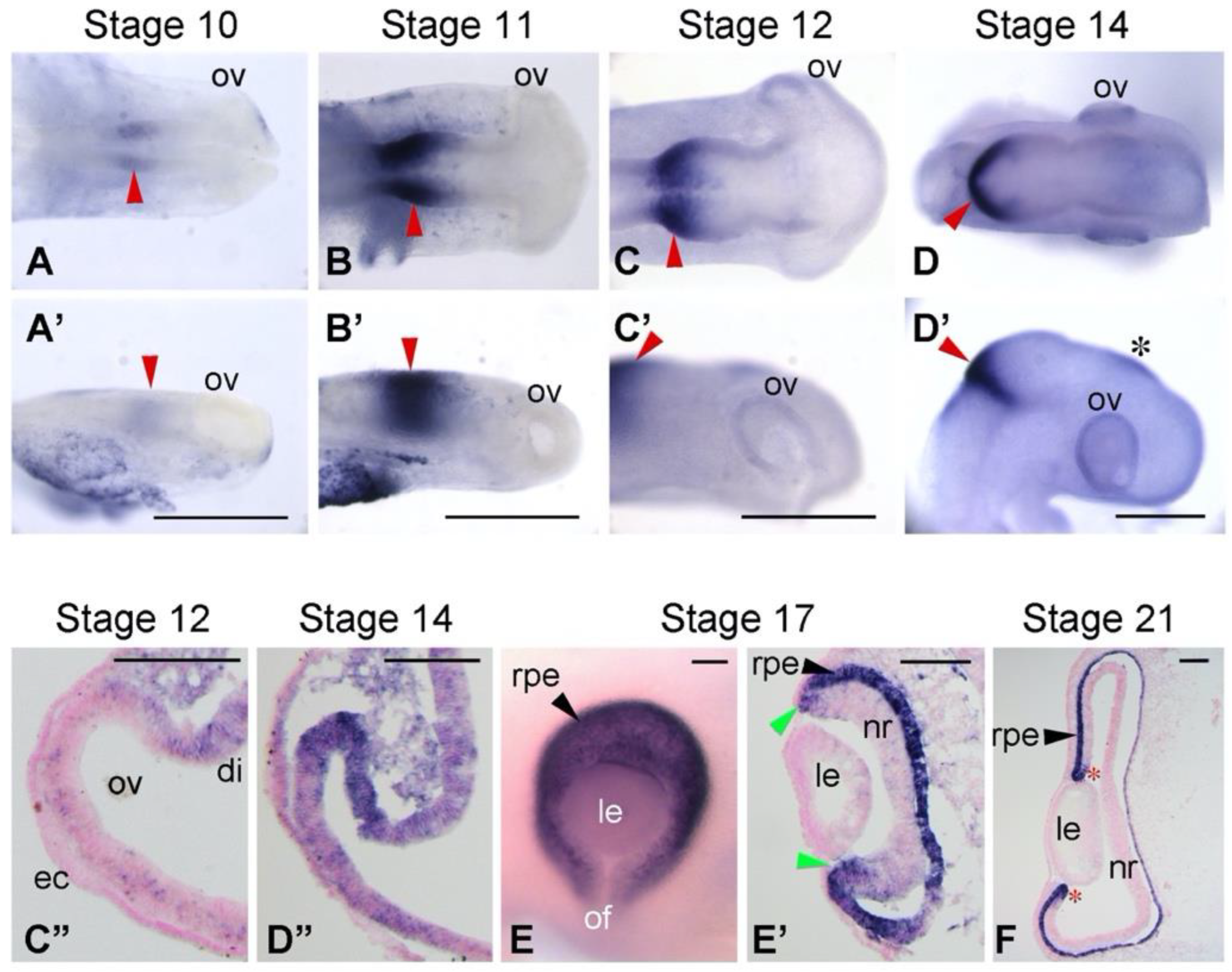
3.1. Expression Patterns of BHLHE40 in Chickens’ Early Eye Development
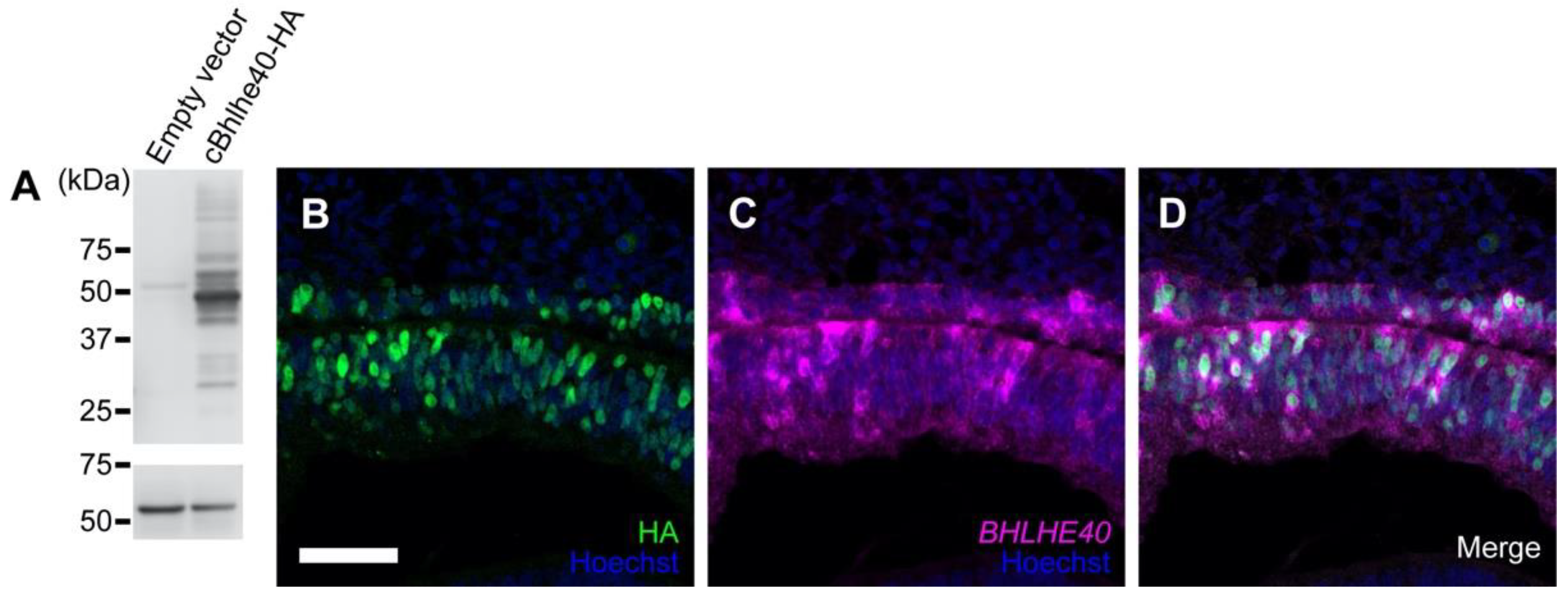
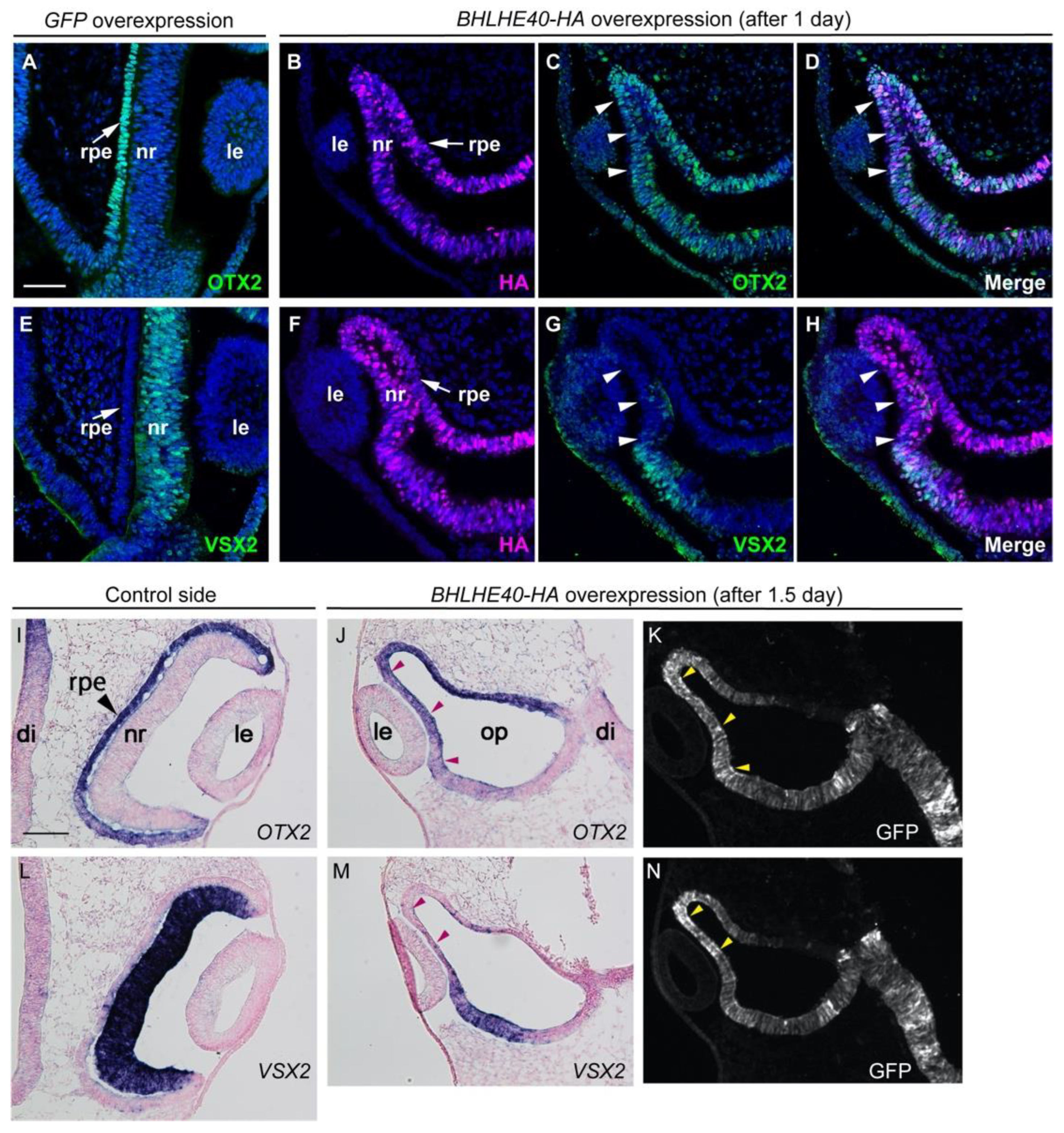
3.2. Overexpression of BHLHE40 in the OV Drives Early RPE Development
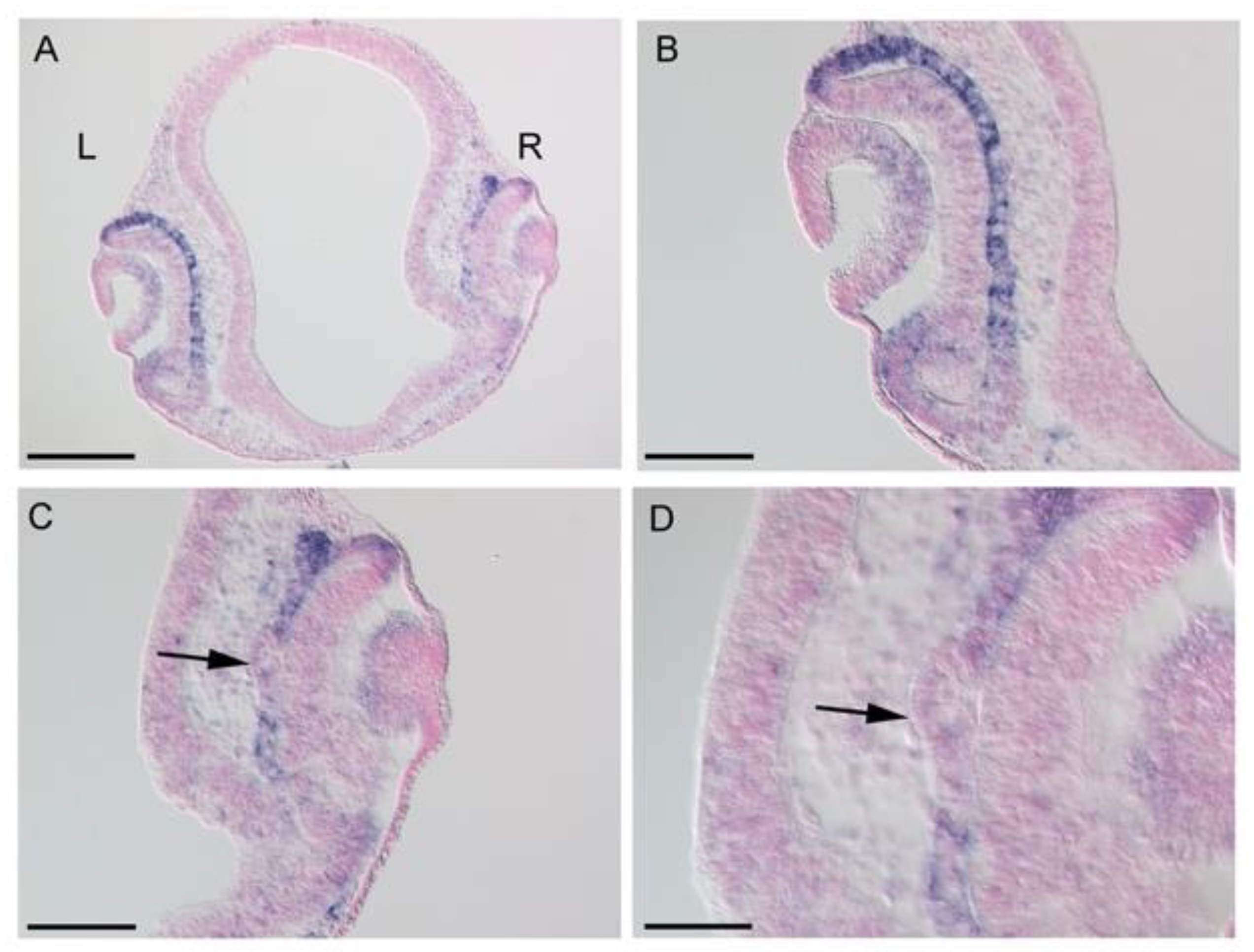
3.3. Effects of LHX1 Overexpression on BHLHE40 Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyer, J.; Mima, T.; Mikawa, T. FGF1 patterns the optic vesicle by directing the placement of the neural retina domain. Development 1998, 125, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Pandit, T.; Jidigam, V.K.; Patthey, C.; Gunhaga, L. Neural retina identity is specified by lens-derived BMP signals. Development 2015, 142, 1850–1859. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, J.; Steinfeld, I.; Bausch, A.; Coronato, N.; Hampel, M.-L.; Depner, H.; Layer, P.G.; Vogel-Höpker, A. BMP-induced reprograming of the retina into RPE requires WNT signalling in the developing chick optic cup. Biol. Open 2017, 6, 979–992. [Google Scholar] [CrossRef] [PubMed]
- Eiraku, M.; Takata, N.; Ishibashi, H.; Kawada, M.; Sakakura, E.; Okuda, S.; Sekiguchi, K.; Adachi, T.; Sasai, Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011, 472, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-Formation of Optic Cups and Storable Stratified Neural Retina from Human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef]
- Buono, L.; Corbacho, J.; Naranjo, S.; Almuedo-Castillo, M.; Moreno-Marmol, T.; de la Cerda, B.; Sanabria-Reinoso, E.; Polvillo, R.; Díaz-Corrales, F.-J.; Bogdanovic, O.; et al. Analysis of gene network bifurcation during optic cup morphogenesis in zebrafish. Nat. Commun. 2021, 12, 3866. [Google Scholar] [CrossRef]
- Moreno-Mármol, T.; Ledesma-Terrón, M.; Tabanera, N.; Martin-Bermejo, M.J.; Cardozo, M.J.; Cavodeassi, F.; Bovolenta, P. Stretching of the retinal pigment epithelium contributes to zebrafish optic cup morphogenesis. eLife 2021, 10, e63396. [Google Scholar] [CrossRef]
- Yao, J.; Wang, L.; Chen, L.; Zhang, S.; Zhao, Q.; Jia, W.; Xue, J. Cloning and developmental expression of the DEC1 ortholog gene in zebrafish. Gene Expr. Patterns 2006, 6, 919–927. [Google Scholar] [CrossRef]
- Cechmanek, P.B.; McFarlane, S. Retinal pigment epithelium expansion around the neural retina occurs in two separate phases with distinct mechanisms. Dev. Dyn. 2017, 246, 598–609. [Google Scholar] [CrossRef]
- Bovolenta, P.; Mallamaci, A.; Briata, P.; Corte, G.; Boncinelli, E. Implication of OTX2 in Pigment Epithelium Determination and Neural Retina Differentiation. J. Neurosci. 1997, 17, 4243–4252. [Google Scholar] [CrossRef]
- Shen, M.; Kawamoto, T.; Yan, W.; Nakamasu, K.; Tamagami, M.; Koyano, Y.; Noshiro, M.; Kato, Y. Molecular Characterization of the Novel Basic Helix–Loop–Helix Protein DEC1 Expressed in Differentiated Human Embryo Chondrocytes. Biochem. Biophys. Res. Commun. 1997, 236, 294–298. [Google Scholar] [CrossRef] [PubMed]
- Boudjelal, M.; Taneja, R.; Matsubara, S.; Bouillet, P.; Dollé, P.; Chambon, P. Overexpression of Stra13, a novel retinoic acid-inducible gene of the basic helix–loop–helix family, inhibits mesodermal and promotes neuronal differentiation of P19 cells. Genes Dev. 1997, 11, 2052–2065. [Google Scholar] [CrossRef] [PubMed]
- Rossner, M.J.; Dörra, J.; Gassb, P.; Schwab, M.H.; Navea, K.A. SHARPs: MammalianEnhancer-of-Split- andHairy-Related Proteins Coupled to Neuronal Stimulation. Mol. Cell. Neurosci. 1997, 9, 460–475. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Kawamoto, T.; Fujimoto, K.; Noshiro, M.; Honda, K.K.; Honma, S.; Honma, K.-I.; Kato, Y. Functional analysis of the basic helix-loop-helix transcription factor DEC1 in circadian regulation. Interaction with BMAL1. JBIC J. Biol. Inorg. Chem. 2004, 271, 4409–4419. [Google Scholar] [CrossRef]
- Cook, M.E.; Jarjour, N.N.; Lin, C.-C.; Edelson, B.T. Transcription Factor Bhlhe40 in Immunity and Autoimmunity. Trends Immunol. 2020, 41, 1023–1036. [Google Scholar] [CrossRef]
- Kiss, Z.; Mudryj, M.; Ghosh, P.M. Non-circadian aspects of BHLHE40 cellular function in cancer. Genes Cancer 2020, 11, 1–19. [Google Scholar] [CrossRef]
- Hamburger, V.; Hamilton, H.L. A series of normal stages in the development of the chick embryo. J. Morphol. 1951, 88, 49–92. [Google Scholar] [CrossRef]
- Niwa, H.; Yamamura, K.-I.; Miyazaki, J.-I. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991, 108, 193–199. [Google Scholar] [CrossRef]
- Kawaue, T.; Okamoto, M.; Matsuyo, A.; Inoue, J.; Ueda, Y.; Tomonari, S.; Noji, S.; Ohuchi, H. Lhx1 in the proximal region of the optic vesicle permits neural retina development in the chicken. Biol. Open 2012, 1, 1083–1093. [Google Scholar] [CrossRef]
- Sato, K.; Nwe, K.N.; Ohuchi, H. The Opsin 3/Teleost multiple tissue opsin system: mRNA localization in the retina and brain of medaka (Oryzias latipes). J. Comp. Neurol. 2021, 529, 2484–2516. [Google Scholar] [CrossRef]
- Inoue, J.; Ueda, Y.; Bando, T.; Mito, T.; Noji, S.; Ohuchi, H. The expression of LIM-homeobox genes, Lhx1 and Lhx5, in the forebrain is essential for neural retina differentiation. Dev. Growth Differ. 2013, 55, 668–675. [Google Scholar] [CrossRef] [PubMed]
- Rishi, V.; Vinson, C. Dominant-Negative Mutants of Helix-Loop-Helix Proteins: Transcriptional Inhibition. In Methods in Enzymology; RNA Polymerases and Associated Factors, Part C; Academic Press: Cambridge, MA, USA, 2003; Volume 370, pp. 454–466. [Google Scholar] [CrossRef]
- Venters, S.J.; Mikawa, T.; Hyer, J. Central and Peripheral Retina Arise through Distinct Developmental Paths. PLoS ONE 2013, 8, e61422. [Google Scholar] [CrossRef] [PubMed]
- Ehata, S.; Hanyu, A.; Hayashi, M.; Aburatani, H.; Kato, Y.; Fujime, M.; Saitoh, M.; Miyazawa, K.; Imamura, T.; Miyazono, K. Transforming growth factor-beta promotes survival of mammary carcinoma cells through induction of antiapoptotic transcription factor DEC1. Cancer Res. 2007, 67, 9694–9703. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.-H.; Yarmishyn, A.A.; Hwang, D.-K.; Hsu, C.-C.; Wang, M.-L.; Yang, Y.-P.; Chien, K.-H.; Chiou, S.-H.; Peng, C.-H.; Chen, S.-J. Expression profiling of cell-intrinsic regulators in the process of differentiation of human iPSCs into retinal lineages. Stem Cell Res. Ther. 2018, 9, 140. [Google Scholar] [CrossRef] [PubMed]
- Sato, F.; Bhawal, U.K.; Yoshimura, T.; Muragaki, Y. DEC1 and DEC2 Crosstalk between Circadian Rhythm and Tumor Progression. J. Cancer 2016, 7, 153–159. [Google Scholar] [CrossRef]
- Lange, C.A.K.; Luhmann, U.F.O.; Mowat, F.; Georgiadis, A.; West, E.; Abraham, S.; Sayed, H.; Powner, M.B.; Fruttiger, M.; Smith, A.J.; et al. Von Hippel-Lindau protein in the RPE is essential for normal ocular growth and vascular development. Development 2012, 139, 2340–2350. [Google Scholar] [CrossRef]
- Fuhrmann, S.; Levine, E.; Reh, T. Extraocular mesenchyme patterns the optic vesicle during early eye development in the embryonic chick. Development 2000, 127, 4599–4609. [Google Scholar] [CrossRef]
- Westenskow, P.D.; Mckean, J.B.; Kubo, F.; Nakagawa, S.; Fuhrmann, S. Ectopic Mitf in the Embryonic Chick Retina by Co-transfection of β-Catenin and Otx2. Investig. Opthalmology Vis. Sci. 2010, 51, 5328–5335. [Google Scholar] [CrossRef]
- Rowan, S.; Chen, C.-M.A.; Young, T.L.; Fisher, D.E.; Cepko, C.L. Transdifferentiation of the retina into pigmented cells in ocular retardation mice defines a new function of the homeodomain gene Chx10. Development 2004, 131, 5139–5152. [Google Scholar] [CrossRef]
- Zou, C.; Levine, E.M. Vsx2 Controls Eye Organogenesis and Retinal Progenitor Identity via Homeodomain and Non-Homeodomain Residues Required for High Affinity DNA Binding. PLoS Genet. 2012, 8, e1002924. [Google Scholar] [CrossRef]
- Green, E.S.; Stubbs, J.L.; Levine, E. Genetic rescue of cell number in a mouse model of microphthalmia:interactions between Chx10 and G1-phase cell cycle regulators. Development 2003, 130, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a therapeutic target in cancer. Nat. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Bi, H.; Li, S.; Qu, X.; Wang, M.; Bai, X.; Xu, Z.; Ao, X.; Jia, Z.; Jiang, X.; Yang, Y.; et al. DEC1 regulates breast cancer cell proliferation by stabilizing cyclin E protein and delays the progression of cell cycle S phase. Cell Death Dis. 2015, 6, e1891. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kinuhata, T.; Sato, K.; Bando, T.; Mito, T.; Miyaishi, S.; Nohno, T.; Ohuchi, H. Involvement of a Basic Helix-Loop-Helix Gene BHLHE40 in Specification of Chicken Retinal Pigment Epithelium. J. Dev. Biol. 2022, 10, 45. https://doi.org/10.3390/jdb10040045
Kinuhata T, Sato K, Bando T, Mito T, Miyaishi S, Nohno T, Ohuchi H. Involvement of a Basic Helix-Loop-Helix Gene BHLHE40 in Specification of Chicken Retinal Pigment Epithelium. Journal of Developmental Biology. 2022; 10(4):45. https://doi.org/10.3390/jdb10040045
Chicago/Turabian StyleKinuhata, Toshiki, Keita Sato, Tetsuya Bando, Taro Mito, Satoru Miyaishi, Tsutomu Nohno, and Hideyo Ohuchi. 2022. "Involvement of a Basic Helix-Loop-Helix Gene BHLHE40 in Specification of Chicken Retinal Pigment Epithelium" Journal of Developmental Biology 10, no. 4: 45. https://doi.org/10.3390/jdb10040045
APA StyleKinuhata, T., Sato, K., Bando, T., Mito, T., Miyaishi, S., Nohno, T., & Ohuchi, H. (2022). Involvement of a Basic Helix-Loop-Helix Gene BHLHE40 in Specification of Chicken Retinal Pigment Epithelium. Journal of Developmental Biology, 10(4), 45. https://doi.org/10.3390/jdb10040045








