A Forest of Forests: A Spatially Weighted and Computationally Efficient Formulation of Geographical Random Forests
Abstract
:1. Introduction
2. Materials and Methods
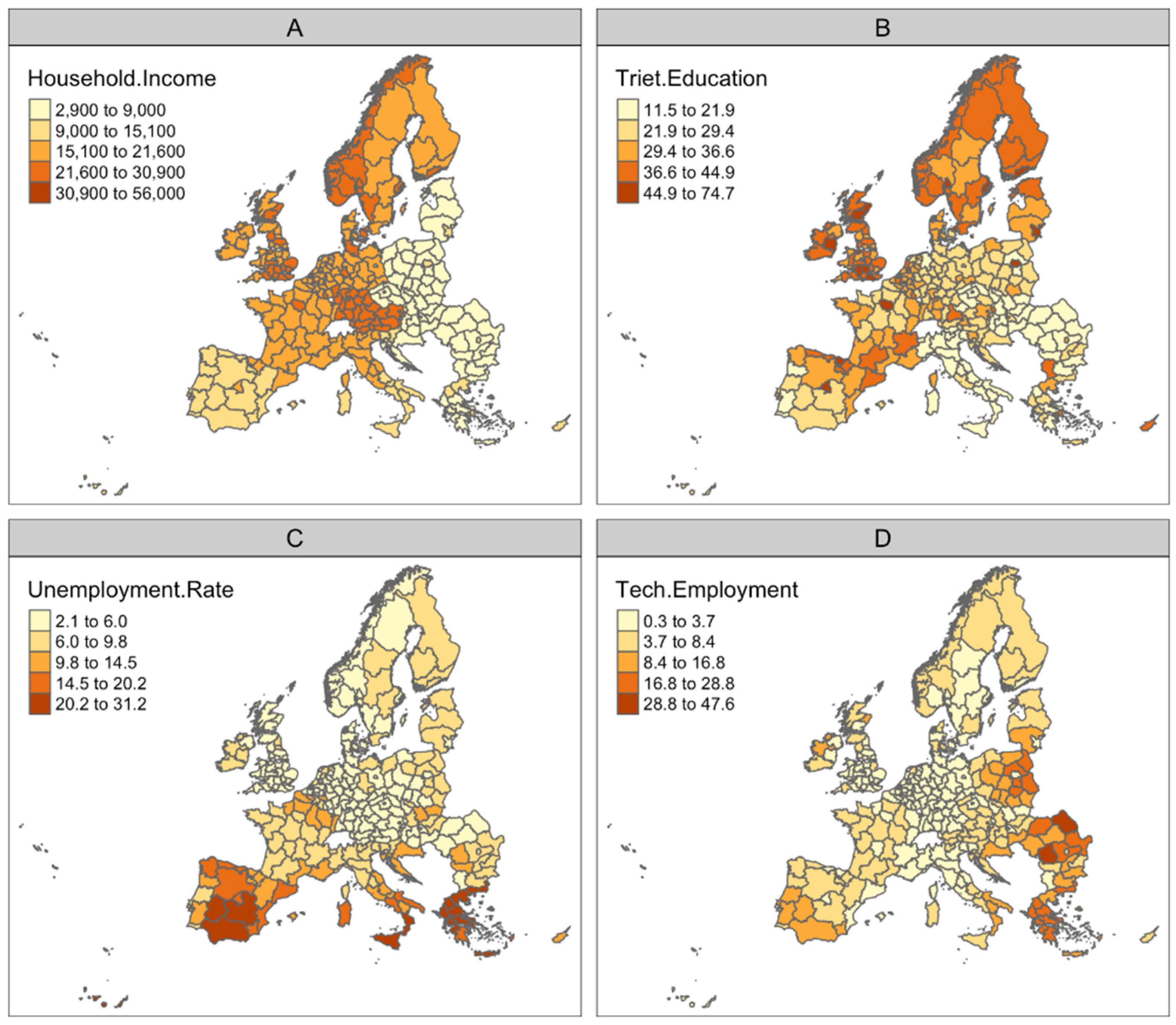
2.1. Dataset
2.2. Models
2.2.1. Geographical Random Forest
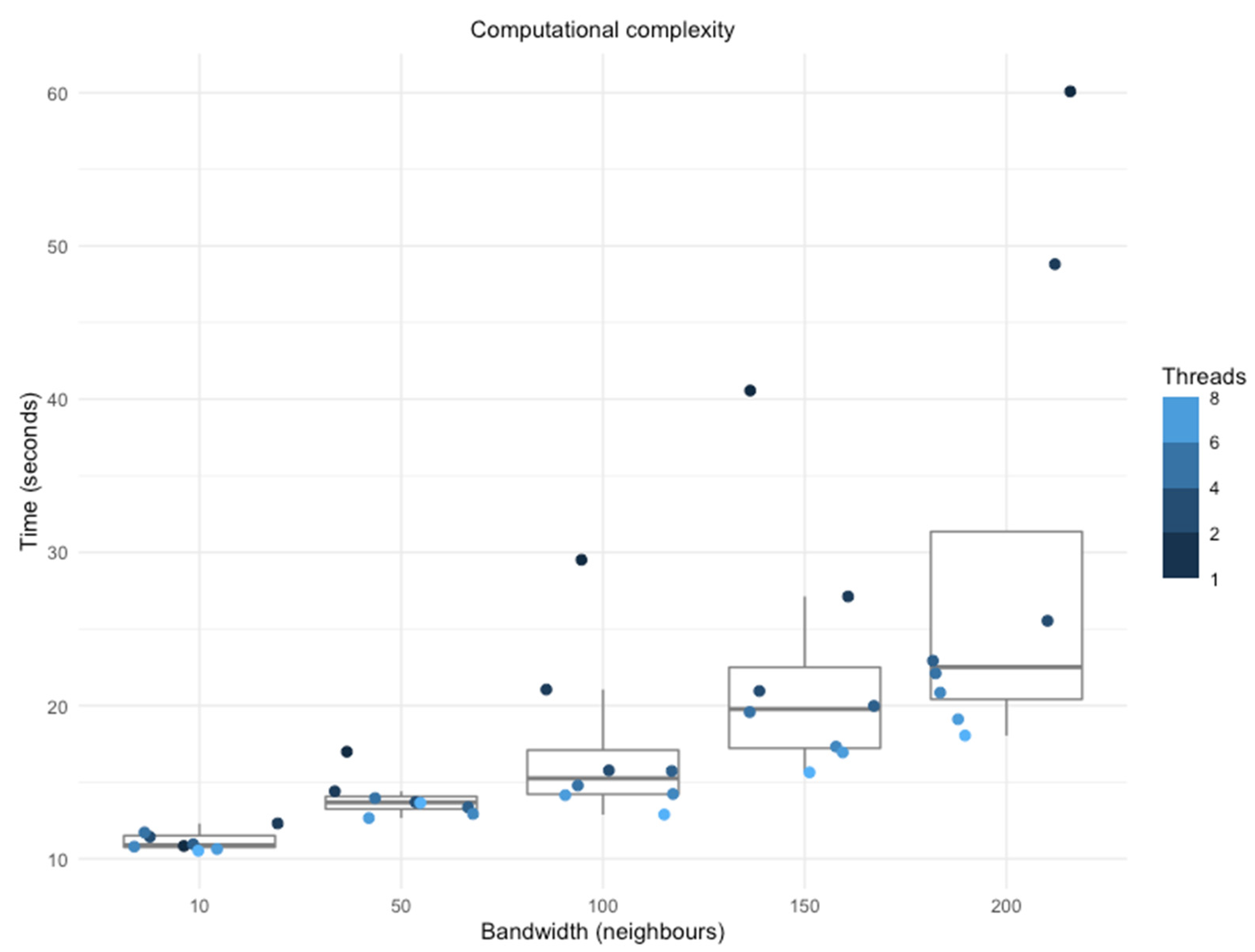
GRF Computational Efficiency
GRF Bandwidth Optimization
Spatial Weighting
2.2.2. Benchmark Models and Validation Metrics
3. Results
3.1. GRF and GWR Bandwidth Optimization
3.2. Predictive Performance
3.3. Computational Improvements
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hengl, T.; Nussbaum, M.; Wright, M.N.; Heuvelink, G.B.M.; Gräler, B. Random Forest as a Generic Framework for Predictive Modeling of Spatial and Spatio-Temporal Variables. PeerJ 2018, 6, e5518. [Google Scholar] [CrossRef] [PubMed]
- Georganos, S.; Grippa, T.; Gadiaga, A.N.; Linard, C.; Lennert, M.; Vanhuysse, S.; Mboga, N.O.; Wolff, E.; Kalogirou, S. Geographical Random Forests: A Spatial Extension of the Random Forest Algorithm to Address Spatial Heterogeneity in Remote Sensing and Population Modelling. Geocarto Int. 2019, 36, 121–136. [Google Scholar] [CrossRef]
- Mariano, C.; Mónica, B. A Random Forest-Based Algorithm for Data-Intensive Spatial Interpolation in Crop Yield Mapping. Comput. Electron. Agric. 2021, 184, 106094. [Google Scholar] [CrossRef]
- Sekulić, A.; Kilibarda, M.; Heuvelink, G.; Nikolić, M.; Bajat, B. Random Forest Spatial Interpolation. Remote Sens. 2020, 12, 1687. [Google Scholar] [CrossRef]
- Ahn, S.; Ryu, D.-W.; Lee, S. A Machine Learning-Based Approach for Spatial Estimation Using the Spatial Features of Coordinate Information. ISPRS Int. J. Geo Inf. 2020, 9, 587. [Google Scholar] [CrossRef]
- Xia, Z.; Stewart, K.; Fan, J. Incorporating Space and Time into Random Forest Models for Analyzing Geospatial Patterns of Drug-Related Crime Incidents in a Major Us Metropolitan Area. Comput. Environ. Urban Syst. 2021, 87, 101599. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Basu, S.; Datta, A. Random Forests for Spatially Dependent Data. J. Am. Stat. Assoc. 2021, 1–19. [Google Scholar] [CrossRef]
- Talebi, H.; Peeters, L.J.M.; Otto, A.; Tolosana-Delgado, R. A Truly Spatial Random Forests Algorithm for Geoscience Data Analysis and Modelling. Math. Geosci. 2021, 54, 1–22. [Google Scholar] [CrossRef]
- Ancell, E.; Bean, B. Autocart--Spatially-Aware Regression Trees for Ecological and Spatial Modeling. arXiv 2021, arXiv:2101.08258. [Google Scholar]
- Meyer, H.; Reudenbach, C.; Wöllauer, S.; Nauss, T. Importance of Spatial Predictor Variable Selection in Machine Learning Applications--Moving from Data Reproduction to Spatial Prediction. Ecol. Modell. 2019, 411, 108815. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Crespo, R.; Yao, J. Geographical and Temporal Weighted Regression (GTWR). Geogr. Anal. 2015, 47, 431–452. [Google Scholar] [CrossRef] [Green Version]
- Aguirre-Gutiérrez, J.; Rifai, S.; Shenkin, A.; Oliveras, I.; Bentley, L.P.; Svátek, M.; Girardin, C.A.J.; Both, S.; Riutta, T.; Berenguer, E.; et al. Pantropical Modelling of Canopy Functional Traits Using Sentinel-2 Remote Sensing Data. Remote Sens. Environ. 2021, 252, 112122. [Google Scholar] [CrossRef]
- Urbański, J.A.; Litwicka, D. Accelerated Decline of Svalbard Coasts Fast Ice as a Result of Climate Change. Cryosph. Discuss. 2021, 1–15. [Google Scholar] [CrossRef]
- Wang, H.; Seaborn, T.; Wang, Z.; Caudill, C.C.; Link, T.E. Modeling Tree Canopy Height Using Machine Learning over Mixed Vegetation Landscapes. Int. J. Appl. Earth Obs. Geoinf. 2021, 101, 102353. [Google Scholar] [CrossRef]
- Hokstad, V.; Tiganj, D. Spatial Modelling of Unconventional Wells in the Niobrara Shale Play: A Descriptive, and a Predictive Approach. Master’s Thesis, Norwegian School of Economics, Bergen, Norway, 2020. [Google Scholar]
- Bicák, D. Geographical Random Forest Model Evaluation in Agricultural Drought Assessment. Diploma Thesis, Charles University, Prague, Czech Republic, 2021. [Google Scholar]
- Quevedo, R.P.; Maciel, D.A.; Uehara, T.D.T.; Vojtek, M.; Rennó, C.D.; Pradhan, B.; Vojteková, J.; Pham, Q.B. Consideration of Spatial Heterogeneity in Landslide Susceptibility Mapping Using Geographical Random Forest Model. Geocarto Int. 2021, 1–20. [Google Scholar] [CrossRef]
- Quiñones, S.; Goyal, A.; Ahmed, Z.U. Geographically Weighted Machine Learning Model for Untangling Spatial Heterogeneity of Type 2 Diabetes Mellitus (T2D) Prevalence in the USA. Sci. Rep. 2021, 11, 6955. [Google Scholar] [CrossRef]
- Córdoba, M.; Carranza, J.P.; Piumetto, M.; Monzani, F.; Balzarini, M. A Spatially Based Quantile Regression Forest Model for Mapping Rural Land Values. J. Environ. Manag. 2021, 289, 112509. [Google Scholar] [CrossRef]
- Maxwell, K.; Rajabi, M.; Esterle, J. Spatial Interpolation of Coal Properties Using Geographic Quantile Regression Forest. Int. J. Coal Geol. 2021, 248, 103869. [Google Scholar] [CrossRef]
- Deng, L.; Adjouadi, M.; Rishe, N. Inverse Distance Weighted Random Forests: Modeling Unevenly Distributed Non-Stationary Geographic Data. In Proceedings of the 2020 International Conference on Advanced Computer Science and Information Systems (ICACSIS), Depok, Indonesia, 17–18 October 2020; pp. 41–46. [Google Scholar]
- Deng, L.; Adjouadi, M.; Rishe, N. Geographic Boosting Tree: Modeling Non-Stationary Spatial Data. In Proceedings of the 2020 19th IEEE International Conference on Machine Learning and Applications (ICMLA), Miami, FL, USA, 14–17 December 2020; pp. 1205–1210. [Google Scholar]
- Masrur, A.; Yu, M.; Mitra, P.; Peuquet, D.; Taylor, A. Interpretable Machine Learning for Analysing Heterogeneous Drivers of Geographic Events in Space-Time. Int. J. Geogr. Inf. Sci. 2021, 36, 692–719. [Google Scholar] [CrossRef]
- Santos, F.; Graw, V.; Bonilla, S. A Geographically Weighted Random Forest Approach for Evaluate Forest Change Drivers in the Northern Ecuadorian Amazon. PLoS ONE 2019, 14, e0226224. [Google Scholar] [CrossRef]
- Kalogirou, S.; Hatzichristos, T. A Spatial Modelling Framework for Income Estimation. Spat. Econ. Anal. 2007, 2, 297–316. [Google Scholar] [CrossRef] [Green Version]
- Wright, M.N.; Ziegler, A. ranger: A fast implementation of random forests for high dimensional data in C++ and R. arXiv 2015, arXiv:1508.04409. [Google Scholar] [CrossRef]
- Fotheringham, A.S.; Brunsdon, C.; Charlton, M. Geographically Weighted Regression: The Analysis of Spatially Varying Relationships; John Wiley & Sons: Hoboken, NJ, USA, 2003. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar] [CrossRef]
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; Team, R.C.; et al. R Package; Version 6.0–21; Caret: Classification and Regression Training; CRAN: Wien, Austria, 2014. [Google Scholar]
- Chicco, D.; Warrens, M.J.; Jurman, G. The Coefficient of Determination R-Squared Is More Informative than SMAPE, MAE, MAPE, MSE and RMSE in Regression Analysis Evaluation. PeerJ Comput. Sci. 2021, 7, e623. [Google Scholar] [CrossRef]
- Janitza, S.; Hornung, R. On the Overestimation of Random Forest’s out-of-Bag Error. PLoS ONE 2018, 13, e0201904. [Google Scholar] [CrossRef]


| OLS | RF | GRF | GRF-W | GWR | |
|---|---|---|---|---|---|
| RMSE | 5024.7 | 4104.6 | 3071.2 | 2801.5 | 3259.00 |
| MAE | 3949.6 | 2578.7 | 1763.2 | 1580.4 | 1933.6 |
| R2 | 0.45 | 0.61 | 0.79 | 0.82 | 0.77 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Georganos, S.; Kalogirou, S. A Forest of Forests: A Spatially Weighted and Computationally Efficient Formulation of Geographical Random Forests. ISPRS Int. J. Geo-Inf. 2022, 11, 471. https://doi.org/10.3390/ijgi11090471
Georganos S, Kalogirou S. A Forest of Forests: A Spatially Weighted and Computationally Efficient Formulation of Geographical Random Forests. ISPRS International Journal of Geo-Information. 2022; 11(9):471. https://doi.org/10.3390/ijgi11090471
Chicago/Turabian StyleGeorganos, Stefanos, and Stamatis Kalogirou. 2022. "A Forest of Forests: A Spatially Weighted and Computationally Efficient Formulation of Geographical Random Forests" ISPRS International Journal of Geo-Information 11, no. 9: 471. https://doi.org/10.3390/ijgi11090471
APA StyleGeorganos, S., & Kalogirou, S. (2022). A Forest of Forests: A Spatially Weighted and Computationally Efficient Formulation of Geographical Random Forests. ISPRS International Journal of Geo-Information, 11(9), 471. https://doi.org/10.3390/ijgi11090471







