Evaluation of the Unintended Effects of fad2-1-Gene-Edited Soybean Line AE15 Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. PCR-Based Detection of Gene-Edited Soybean Line AE15
2.3. Protein Preparation and Trypsin Digestion
2.4. Mass Spectrometry (MS) Analysis
2.5. Data Analysis
2.6. qRT-PCR
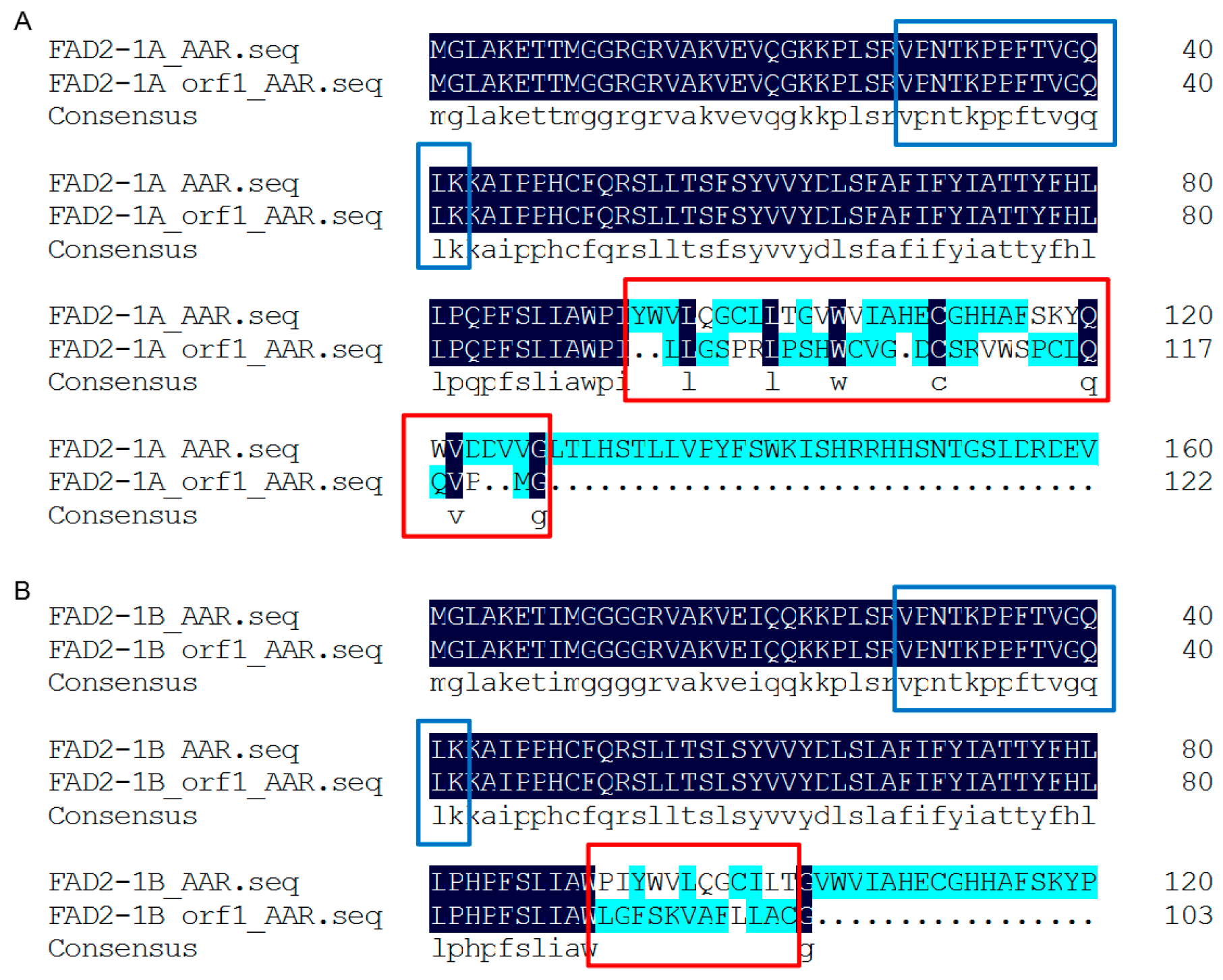
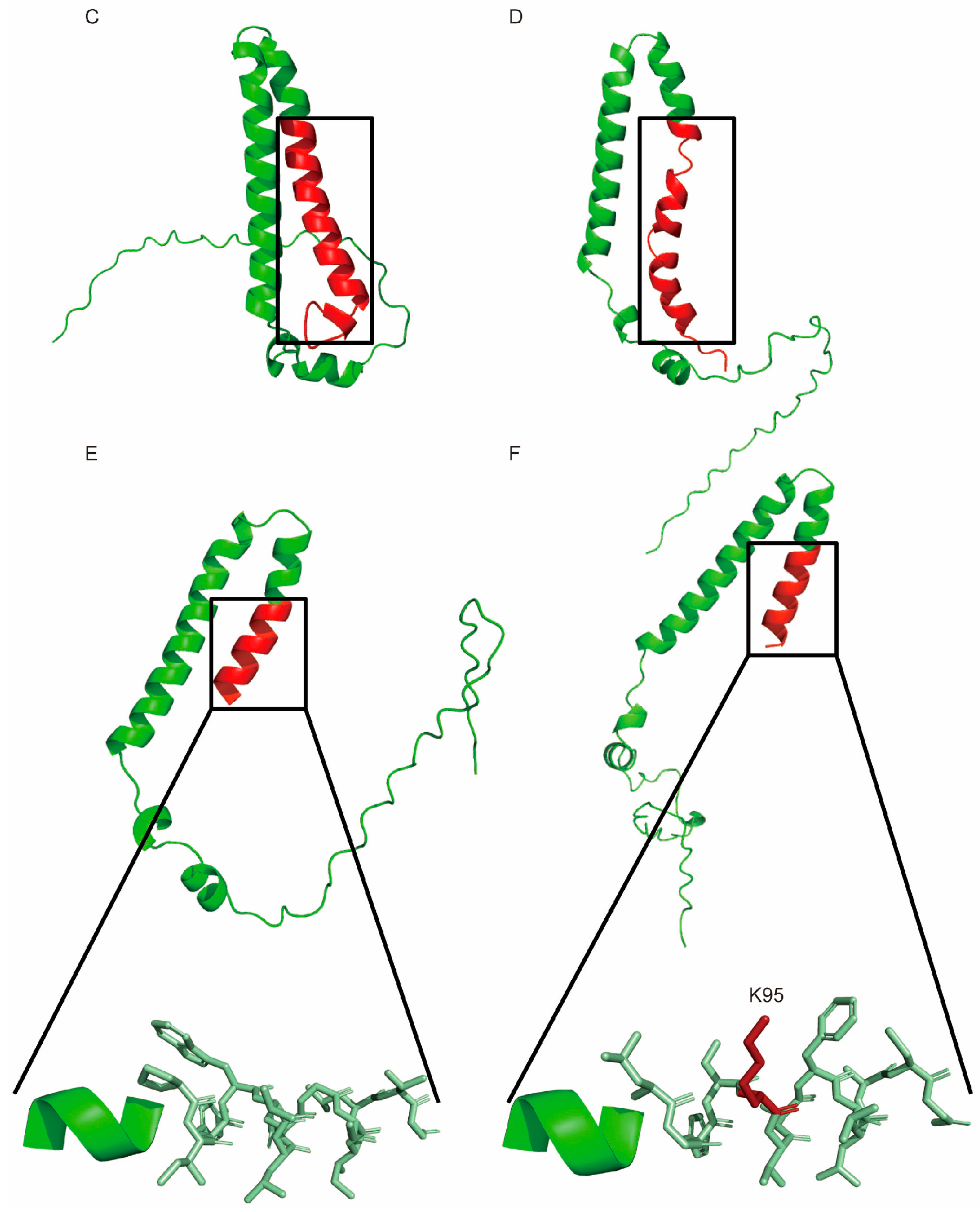
2.7. New ORF Prediction, Sequence Alignment, and Protein Structure Prediction
3. Results
3.1. Soybean Line Confirmation
3.2. Protein Profiling of Soybean Seeds
3.3. DEP Detection in Soybean Seeds
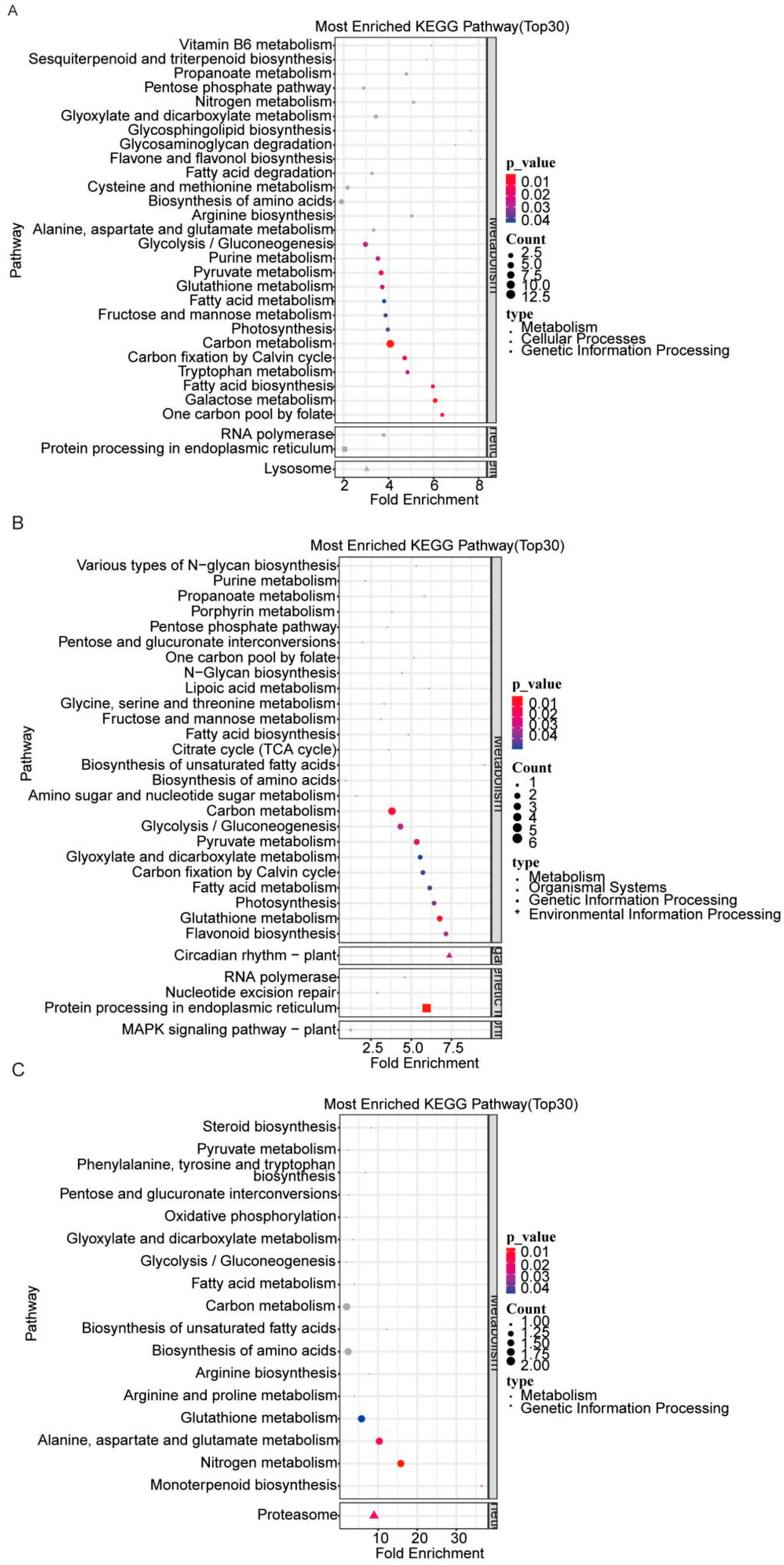
3.4. KEGG Pathway Enrichment Analysis of the Identified DEPs in AE15 Soybean Seeds
3.5. Identifying Co-DEPs and FAD2-1 in Soybean Seeds
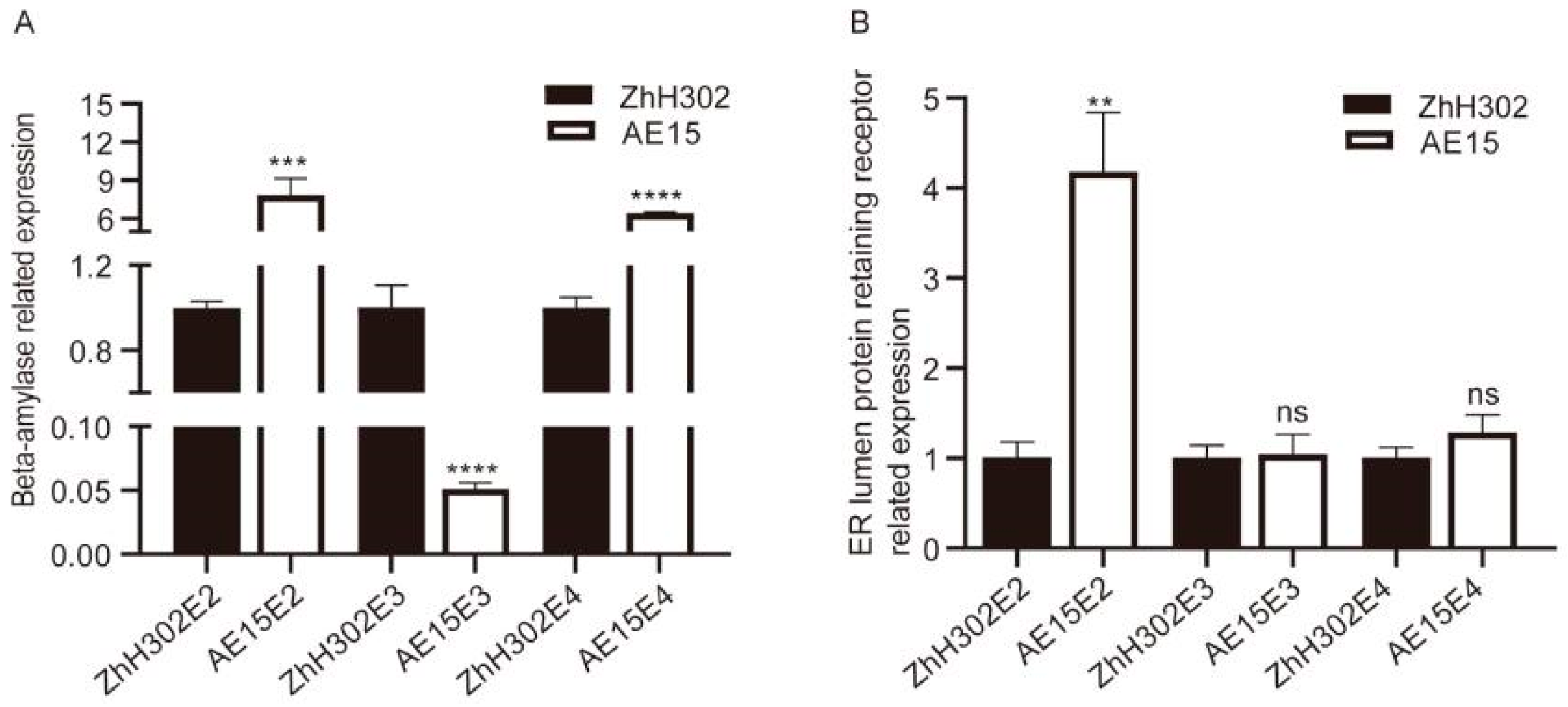
3.6. Selected Co-DEPs Further Analyzed by qRT-PCR
3.7. FAD2-1 in Studied Soybean Seeds
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FAD2-1 | Fatty acid desaturase 2-1 |
| DIA | Data-independent acquisition |
| FC | Fold change |
| DEPs | Differentially expressed proteins |
| co-DEPs | Commonly differentially expressed proteins |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| qRT-PCR | Quantitative real-time PCR |
| MS | Mass spectrometry |
| ZhH | Zhonghuang |
| AE15 | fad2-1-gene-edited soybean line AE15 |
References
- Dar, A.A.; Choudhury, A.R.; Kancharla, P.K.; Arumugam, N. The FAD2 Gene in Plants: Occurrence, Regulation, and Role. Front. Plant Sci. 2017, 8, 1789. [Google Scholar] [CrossRef]
- Lakhssassi, N.; Zhou, Z.; Liu, S.; Colantonio, V.; AbuGhazaleh, A.; Meksem, K. Characterization of the FAD2 Gene Family in Soybean Reveals the Limitations of Gel-Based TILLING in Genes with High Copy Number. Front. Plant Sci. 2017, 8, 324. [Google Scholar] [CrossRef] [PubMed]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. A novel FAD2-1 A allele in a soybean plant introduction offers an alternate means to produce soybean seed oil with 85% oleic acid content. Theor. Appl. Genet. 2011, 123, 793–802. [Google Scholar] [CrossRef]
- Pham, A.T.; Lee, J.D.; Shannon, J.G.; Bilyeu, K.D. Mutant alleles of FAD2-1A and FAD2-1B combine to produce soybeans with the high oleic acid seed oil trait. BMC Plant Biol. 2010, 10, 195. [Google Scholar] [CrossRef]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 2014, 12, 934–940. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Xu, Y.; Zhang, H.; Liu, X.; Cui, X.; Chen, X.; Chen, H. Creation of high oleic acid soybean lines by CRISPR/Cas9. J. Jiangsu Agric. 2023, 23, 7. [Google Scholar] [CrossRef]
- Entine, J.; Felipe, M.S.S.; Groenewald, J.H.; Kershen, D.L.; Lema, M.; McHughen, A.; Nepomuceno, A.L.; Ohsawa, R.; Ordonio, R.L.; Parrott, W.A.; et al. Regulatory approaches for genome edited agricultural plants in select countries and jurisdictions around the world. Transgenic Res. 2021, 30, 551–584. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A.K. Global Regulation of Genetically Modified Crops Amid the Gene Edited Crop Boom—A Review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef]
- Hou, J.; Wang, J.; Yang, F.; Xu, T. DIA-MS2pep: A library-free framework for comprehensive peptide identification from data-independent acquisition data. Biophys. Rep. 2022, 8, 253–268. [Google Scholar] [CrossRef]
- Demichev, V.; Messner, C.B.; Vernardis, S.I.; Lilley, K.S.; Ralser, M. DIA-NN: Neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods 2020, 17, 41–44. [Google Scholar] [CrossRef]
- Lou, R.; Shui, W. Acquisition and Analysis of DIA-Based Proteomic Data: A Comprehensive Survey in 2023. Mol. Cell. Proteom. 2024, 23, 100712. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Rappsilber, J.; Mann, M.; Ishihama, Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007, 2, 1896–1906. [Google Scholar] [CrossRef]
- Lou, R.; Cao, Y.; Li, S.; Lang, X.; Li, Y.; Zhang, Y.; Shui, W. Benchmarking commonly used software suites and analysis workflows for DIA proteomics and phosphoproteomics. Nat. Commun. 2023, 14, 94. [Google Scholar] [CrossRef]
- Kimes, P.K.; Liu, Y.; Neil Hayes, D.; Marron, J.S. Statistical significance for hierarchical clustering. Biometrics 2017, 73, 811–821. [Google Scholar] [CrossRef]
- Zeng, W.Y.; Sun, Z.D.; Cai, Z.Y.; Chen, H.Z.; Lai, Z.G.; Yang, S.Z.; Tang, X.M. Proteomic analysis by iTRAQ-MRM of soybean resistance to Lamprosema Indicate. Bmc Genomics 2017, 18, 444. [Google Scholar] [CrossRef]
- Wang, L.M.; Wang, X.C.; Jin, X.; Jia, R.Z.; Huang, Q.X.; Tan, Y.H.; Guo, A.P. Comparative proteomics of Bt-transgenic and non-transgenic cotton leaves. Proteome Sci. 2015, 13, 15. [Google Scholar] [CrossRef]
- Liu, Y.B.; Zhang, Y.X.; Song, S.Q.; Li, J.S.; Stewart, C.N.; Wei, W.; Zhao, Y.J.; Wang, W.Q. A proteomic analysis of seeds from Bt-transgenic Brassica napus and hybrids with wild B. juncea. Sci. Rep. 2015, 5, 15480. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 493–500. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, H.; Wang, W.; Shen, Y.; Ping, Z. Rapid generation of high-quality structure figures for publication with PyMOL-PUB. Bioinformatics 2024, 40, btae139. [Google Scholar] [CrossRef]
- Rigsby, R.E.; Parker, A.B. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem. Mol. Biol. Educ. 2016, 44, 433–437. [Google Scholar] [CrossRef]
- Balsamo, G.M.; Cangahuala-Inocente, G.C.; Bertoldo, J.B.; Terenzi, H.; Arisi, A.C.M. Proteomic Analysis of Four Brazilian MON810 Maize Varieties and Their Four Non-Genetically-Modified Isogenic Varieties. J. Agr. Food Chem. 2011, 59, 11553–11559. [Google Scholar] [CrossRef]
- Fu, W.; Wang, C.G.; Xu, W.J.; Zhu, P.Y.; Lu, Y.; Wei, S.; Wu, X.Y.; Wu, Y.P.; Zhao, Y.Q.; Zhu, S.F. Unintended effects of transgenic rice revealed by transcriptome and metabolism. Gm. Crops Food-Biotechnol. Agric. Food Chain 2019, 10, 20–34. [Google Scholar] [CrossRef]
- Gong, C.Y.; Li, Q.; Yu, H.T.; Wang, Z.Z.; Wang, T. Proteomics Insight into the Biological Safety of Transgenic Modification of Rice As Compared with Conventional Genetic Breeding and Spontaneous Genotypic Variation. J. Proteome Res. 2012, 11, 3019–3029. [Google Scholar] [CrossRef]
- Liu, W.X.; Xu, W.T.; Li, L.; Dong, M.; Wan, Y.S.; He, X.Y.; Huang, K.L.; Jin, W.J. iTRAQ-based quantitative tissue proteomic analysis of differentially expressed proteins (DEPs) in non-transgenic and transgenic soybean seeds. Sci. Rep. 2018, 8, 17681. [Google Scholar] [CrossRef]
- Liu, X.-J.; Xing, B.; Wang, M.-Y.; Li, X.-M.; Wang, X.-J.; Wang, Z.-X. Transcriptional and proteomic analysis. GM Crops Food 2023, 14, 1–16. [Google Scholar] [CrossRef]
- Pedrazzini, E.; Vitale, A. Protein Biosynthesis and Maturation in the ER. Plant Endoplasmic Reticulum 2018, 1691, 179–189. [Google Scholar] [CrossRef]
- Vitale, A.; Boston, R.S. Endoplasmic reticulum quality control and the unfolded protein response: Insights from plants. Traffic 2008, 9, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.X.; Howell, S.H. Managing the protein folding demands in the endoplasmic reticulum of plants. New Phytol. 2016, 211, 418–428. [Google Scholar] [CrossRef] [PubMed]



| Sample | Genotype/Authorized Number |
|---|---|
| ZhH302E2 | wild-type(Yudou22xAg31)/Wanshendou2018006 |
| ZhH302E3 | |
| ZhH302E4 | |
| AE15E2 | fad2-1 edited in ZhH302 |
| AE15E3 | |
| AE15E4 | |
| ZhH10 | wild-type(Wenfeng7xLudou4)/(96)jingshenjingzi2 |
| ZhH42 | wild-type(Youchu4xJindou33)/Guoshengdou2007002 |
| Comparison Group | No. of DEPs | No. Upregulated | No. Downregulated |
|---|---|---|---|
| AE15E2/ZhH302E2 | 561 | 292 | 269 |
| AE15E3/ZhH302E3 | 269 | 78 | 191 |
| AE15E4/ZhH302E4 | 227 | 101 | 126 |
| ZhH302E3/ZhH10 | 1063 | 901 | 162 |
| ZhH10/ZhH42 | 989 | 407 | 582 |
| ZhH302E3/ZhH42 | 671 | 108 | 563 |
| ZhH302E2/ZhH302E3 | 442 | 154 | 288 |
| ZhH302E3/ZhH302E4 | 242 | 151 | 91 |
| ZhH302E4/ZhH302E2 | 545 | 305 | 240 |
| AE15E2/AE15E3 | 623 | 349 | 274 |
| AE15E3/AE15E4 | 666 | 292 | 374 |
| AE15E4/AE15E2 | 214 | 95 | 119 |
| ID | Name | AE15/ZhH302 Comparison Groups | ||
|---|---|---|---|---|
| E2 | E3 | E4 | ||
| GLYMA_09G168300_A0A0R4J443 | Beta-amylase (EC 3.2.1.2) | Up | Up | Up |
| LOC100790733 GLYMA_03G084600_I1JM56 | ER lumen protein-retaining receptor | Up | Up | Up |
| GLYMA_15G256000_K7MDY4 | Cysteine-rich transmembrane domain-containing protein | Up | Up | Down |
| GLYMA_13G132400_A0A0R0GZH8 | Inorganic diphosphatase (EC 3.6.1.1) | Up | Down | Up |
| GLYMA_06G101700_I1K9W3 | Phospho-2-dehydro-3-deoxyheptonate aldolase (EC 2.5.1.54) | Up | Down | Down |
| GLYMA_03G244600_I1JRK7 | Benzyl alcohol O-benzoyltransferase | Down | Up | Down |
| LOC100527208 GLYMA_15G218900_A0A0R0GE22 | Bet v I/Major latex protein domain-containing protein | Down | Up | Down |
| GLYMA_08G274500_A0A0R0IT88 | Uncharacterized protein | Down | Up | Down |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Guo, C.; Zhang, J.; Wang, Z.; Jin, W.; Liu, W. Evaluation of the Unintended Effects of fad2-1-Gene-Edited Soybean Line AE15 Seeds. Biomolecules 2026, 16, 8. https://doi.org/10.3390/biom16010008
Wang R, Guo C, Zhang J, Wang Z, Jin W, Liu W. Evaluation of the Unintended Effects of fad2-1-Gene-Edited Soybean Line AE15 Seeds. Biomolecules. 2026; 16(1):8. https://doi.org/10.3390/biom16010008
Chicago/Turabian StyleWang, Ruizhe, Chang Guo, Jihong Zhang, Zhanchao Wang, Wujun Jin, and Weixiao Liu. 2026. "Evaluation of the Unintended Effects of fad2-1-Gene-Edited Soybean Line AE15 Seeds" Biomolecules 16, no. 1: 8. https://doi.org/10.3390/biom16010008
APA StyleWang, R., Guo, C., Zhang, J., Wang, Z., Jin, W., & Liu, W. (2026). Evaluation of the Unintended Effects of fad2-1-Gene-Edited Soybean Line AE15 Seeds. Biomolecules, 16(1), 8. https://doi.org/10.3390/biom16010008




