Evolutionary Rate Heterogeneity and Functional Divergence of Orthologous Genes in Pyrus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Source and Orthologous Estimation
2.2. Gene Structure Analysis
2.3. Expression Profile Analysis
2.4. Statistical Tests and Functional Divergence Analysis
3. Results
3.1. Identification of Orthologous Gene Pairs
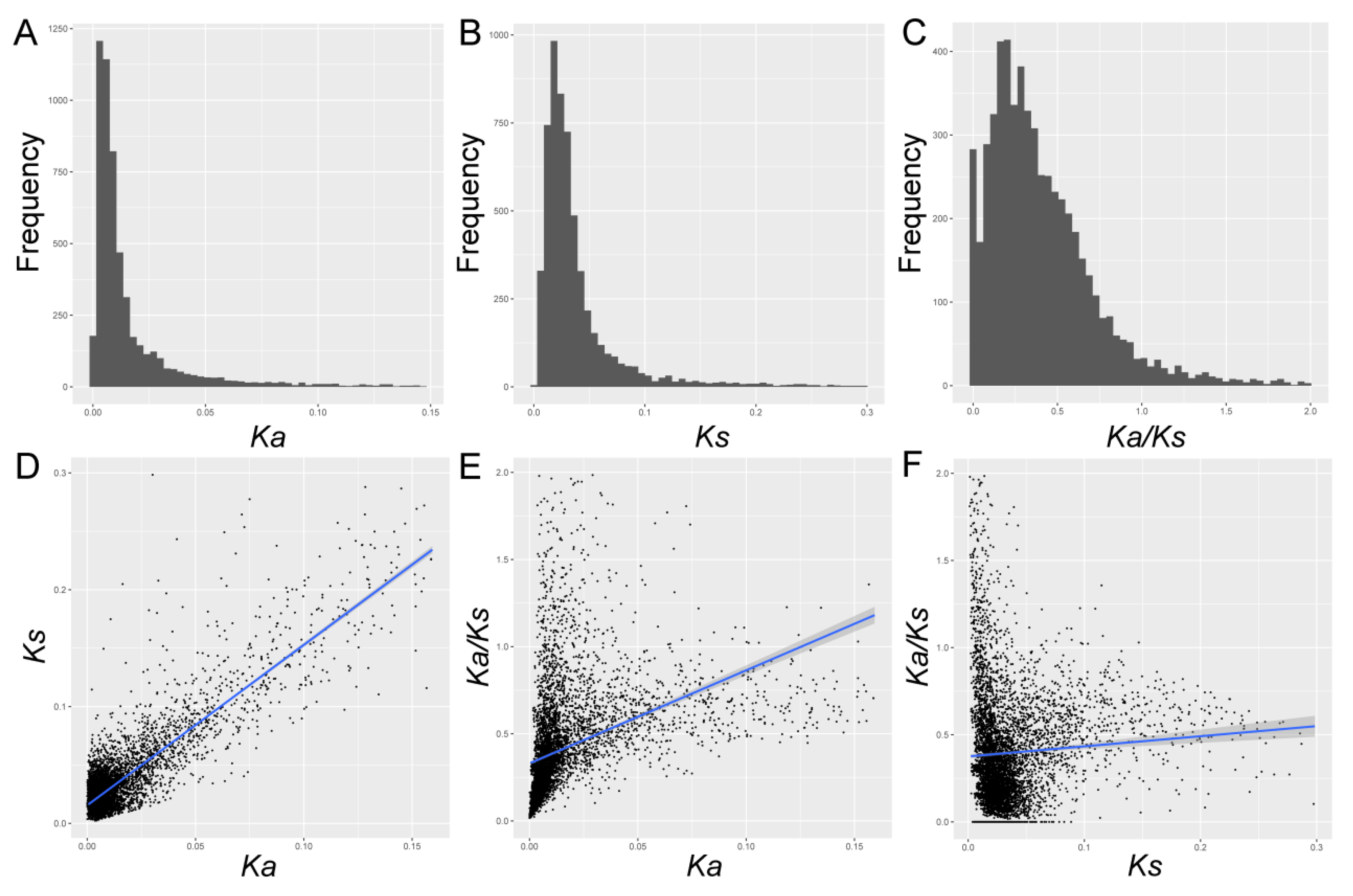
3.2. Distribution of Ka/Ks, Ks, and Ka, and Their Correlations in Pyrus
3.3. Ka, Ks, and Exon Characteristics Between PSGs and NSGs
3.4. Lower Expression Level for PSGs Than NSGs
3.5. Codon Bias Analysis of PSGs and NSGs
3.6. Gene Expression Patterns in Pyrus Fruit Revealed Subfunctionalization and Functional Redundancy for the Related to Fruit Quality Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Yang, Z. The power of phylogenetic comparison in revealing protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 3179–3180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Vision, T.J.; Gaut, B.S. Patterns of nucleotide substitution among simultaneously duplicated gene pairs in Arabidopsis thaliana. Mol. Biol. Evol. 2002, 19, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, J.; Zhang, J.; Liu, S.; Du, J. Selective modes determine evolutionary rates, gene compactness and expression patterns in brassica. Plant J. 2017, 91, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Tian, Z.; Sui, Y.; Zhao, M.; Song, Q.; Cannon, S.B.; Cregan, P.; Ma, J. Pericentromeric effects shape the patterns of divergence, retention, and expression of duplicated genes in the paleopolyploid soybean. Plant Cell 2012, 24, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Molecular evolutionary genetics; Columbia university press: New York, NY, USA, 1987. [Google Scholar]
- Wolfe, K.H.; Sharp, P.M.; Li, W.-H. Mutation rates differ among regions of the mammalian genome. Nature 1989, 337, 283–285. [Google Scholar] [CrossRef]
- Ticher, A.; Graur, D. Nucleic acid composition, codon usage, and the rate of synonymous substitution in protein-coding genes. J. Mol. Evol. 1989, 28, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Matassi, G.; Sharp, P.M.; Gautier, C. Chromosomal location effects on gene sequence evolution in mammals. Curr. Biol. 1999, 9, 786–791. [Google Scholar] [CrossRef] [Green Version]
- Civetta, A.; Singh, R.S. High divergence of reproductive tract proteins and their association with postzygotic reproductive isolation in drosophila melanogaster and drosophila virilis group species. J. Mol. Evol. 1995, 41, 1085–1095. [Google Scholar] [CrossRef]
- Coulthart, M.; Singh, R.S. High level of divergence of male-reproductive-tract proteins, between drosophila melanogaster and its sibling species, d. Simulans. Mol. Biol. Evol. 1988, 5, 182–191. [Google Scholar]
- Swanson, W.J.; Yang, Z.; Wolfner, M.F.; Aquadro, C.F. Positive darwinian selection drives the evolution of several female reproductive proteins in mammals. Proc. Natl. Acad. Sci. 2001, 98, 2509–2514. [Google Scholar] [CrossRef]
- Tsaur, S.-C.; Wu, C.-I. Positive selection and the molecular evolution of a gene of male reproduction, acp26aa of drosophila. Mol. Biol. Evol. 1997, 14, 544–549. [Google Scholar] [CrossRef] [PubMed]
- Begun, D.J.; Whitley, P.; Todd, B.L.; Waldrip-Dail, H.M.; Clark, A.G. Molecular population genetics of male accessory gland proteins in drosophila. Genetics 2000, 156, 1879–1888. [Google Scholar] [PubMed]
- Wyckoff, G.J.; Wang, W.; Chung, W. Rapid evolution of male reproductive genes in the descent of man. Nature 2000, 403, 304. [Google Scholar] [CrossRef] [PubMed]
- Buck, L.; Axel, R. A novel multigene family may encode odorant receptors: A molecular basis for odor recognition. Cell 1991, 65, 175–187. [Google Scholar] [CrossRef]
- Klein, J.; Figueroa, F. Evolution of the major histocompatibility complex. Crit. Rev. Immunol. 1986, 6, 295–386. [Google Scholar] [CrossRef]
- Hughes, A.L. Adapt. Evol. Genes Genomes; Oxford University Press: Oxford, UK, 1999. [Google Scholar]
- Hughes, A.L.; Nei, M. Pattern of nucleotide substitution at major histocompatibility complex class i loci reveals overdominant selection. Nature 1988, 335, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Fay, J.C.; Wu, C.-I. The neutral theory in the genomic era. Curr. Opin. Genet. Dev. 2001, 11, 642–646. [Google Scholar] [CrossRef]
- Hurst, L.D. The ka/ks ratio: Diagnosing the form of sequence evolution. TRENDS Genet. 2002, 18, 486–487. [Google Scholar] [CrossRef]
- Kondrashov, F.A.; Rogozin, I.B.; Wolf, Y.I.; Koonin, E.V. Selection in the evolution of gene duplications. Genome Boil. 2002, 3, research0008. 0001. [Google Scholar]
- Nielsen, R.; Bustamante, C.; Clark, A.G.; Glanowski, S.; Sackton, T.B.; Hubisz, M.J.; Fledel-Alon, A.; Tanenbaum, D.M.; Civello, D.; White, T.J. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005, 3, e170. [Google Scholar] [CrossRef]
- Nei, M.; Suzuki, Y.; Nozawa, M. The neutral theory of molecular evolution in the genomic era. Ann. Rev. Genomics Human Genet. 2010, 11, 265–289. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S. Ancestral polyploidy in seed plants and angiosperms. Nature 2011, 473, 97. [Google Scholar] [CrossRef] [PubMed]
- Beilstein, M.A.; Nagalingum, N.S.; Clements, M.D.; Manchester, S.R.; Mathews, S. Dated molecular phylogenies indicate a miocene origin for arabidopsis thaliana. Proc. Natl. Acad. Sci. 2010, 107, 18724–18728. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Gaut, B.S. Factors that contribute to variation in evolutionary rate among arabidopsis genes. Mol. Biol. Evol. 2011, 28, 2359–2369. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (pyrus bretschneideri rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Chagné, D.; Crowhurst, R.N.; Pindo, M.; Thrimawithana, A.; Deng, C.; Ireland, H.; Fiers, M.; Dzierzon, H.; Cestaro, A.; Fontana, P. The draft genome sequence of european pear (pyrus communis l.‘Bartlett’). PloS ONE 2014, 9, e92644. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, Y.; Xu, J.; Korban, S.S.; Fei, Z.; Tao, S.; Ming, T.; Tai, S.; Khan, A.M.; Postman, J.D.; et al. Diversification and independent domestication of Asian and European pears. Genome Biol. 2018, 19, 77. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic analysis of the 4-coumarate:Coenzyme a ligase (4cl) related genes and expression profiling during fruit development in the chinese pear. Genes 2016, 7, 89. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Structural, evolutionary, and functional analysis of the class iii peroxidase gene family in chinese pear (pyrus bretschneideri). Front Plant Sci. 2016, 7, 1874. [Google Scholar] [CrossRef]
- Li, J.M.; San Huang, X.; Li, L.T.; Zheng, D.M.; Xue, C.; Zhang, S.L.; Wu, J. Proteome analysis of pear reveals key genes associated with fruit development and quality. Planta 2015, 241, 1363–1379. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. Kaks_calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom., Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J. From fastq data to high-confidence variant calls: The genome analysis toolkit best practices pipeline. Current protocols in bioinformatics 2013, 11.10. 11–11.10. 33. [Google Scholar]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of rna-seq experiments with tophat and cufflinks. Nat. Protoc. 2012, 7, 562. [Google Scholar] [CrossRef]
- Goff, L.A.; Trapnell, C.; Kelley, D. Cummerbund: Visualization and exploration of cufflinks high-throughput sequencing data. R Packag. Version 2012, 2. [Google Scholar]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef]
- Yim, W.C.; Lee, B.-M.; Jang, C.S. Expression diversity and evolutionary dynamics of rice duplicate genes. Mol. Genet. Genom. 2009, 281, 483–493. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Xue, C.; Xu, L.; Sun, H.; Qin, M.-F.; Zhang, S.; Wu, J. Distinct transcriptome profiles reveal gene expression patterns during fruit development and maturation in five main cultivated species of pear (pyrus l.). Sci. Rep. 2016, 6, 28130. [Google Scholar] [CrossRef]
- Chen, X.; Zhu, W.; Azam, S.; Li, H.; Zhu, F.; Li, H.; Hong, Y.; Liu, H.; Zhang, E.; Wu, H. Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant biotechnol. J. 2013, 11, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Gao, H.; Liu, J.; Tian, P.; Nan, Z. Comprehensive analysis of correlations among codon usage bias, gene expression, and substitution rate in arachis duranensis and arachis ipa?Nsis orthologs. Sci. Rep 2017, 7, 14853. [Google Scholar] [CrossRef] [PubMed]
- Hershberg, R.; Petrov, D.A. Selection on codon bias. Ann. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef]
- Larracuente, A.M.; Sackton, T.B.; Greenberg, A.J.; Wong, A.; Singh, N.D.; Sturgill, D.; Zhang, Y.; Oliver, B.; Clark, A.G. Evolution of protein-coding genes in drosophila. Trends Genet. 2008, 24, 114–123. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32. [Google Scholar] [CrossRef]
- Cai, Y.; Li, G.; Nie, J.; Lin, Y.; Nie, F.; Zhang, J.; Xu, Y. Study of the structure and biosynthetic pathway of lignin in stone cells of pear. Sci. Hortic. 2010, 125, 374–379. [Google Scholar] [CrossRef]
- Jin, Q.; Yan, C.; Qiu, J.; Zhang, N.; Lin, Y.; Cai, Y. Structural characterization and deposition of stone cell lignin in dangshan su pear. Sci. Hortic. 2013, 155, 123–130. [Google Scholar] [CrossRef]
- Hanada, K.; Kuromori, T.; Myouga, F.; Toyoda, T.; Li, W.-H.; Shinozaki, K. Evolutionary persistence of functional compensation by duplicate genes in arabidopsis. Genome Biol. Evol. 2009, 1, 409–414. [Google Scholar] [CrossRef]
- Wright, S.I.; Yau, C.K.; Looseley, M.; Meyers, B.C. Effects of gene expression on molecular evolution in arabidopsis thaliana and arabidopsis lyrata. Mol. Biol. Evol. 2004, 21, 1719–1726. [Google Scholar] [CrossRef]
- Wang, Y.; Diehl, A.; Wu, F.; Vrebalov, J.; Giovannoni, J.; Siepel, A.; Tanksley, S.D. Sequencing and comparative analysis of a conserved syntenic segment in the solanaceae. Genetics 2008, 180, 391–408. [Google Scholar] [CrossRef]
- Duret, L.; Mouchiroud, D. Expression pattern and, surprisingly, gene length shape codon usage in caenorhabditis, drosophila, and arabidopsis. Proc. Natl. Acad. Sci. 1999, 96, 4482–4487. [Google Scholar] [CrossRef] [PubMed]
- Castillo-Davis, C.I.; Mekhedov, S.L.; Hartl, D.L.; Koonin, E.V.; Kondrashov, F.A. Selection for short introns in highly expressed genes. Nat. Genet. 2002, 31, 415. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes are compact. TRENDS Genet. 2003, 19, 362–365. [Google Scholar] [CrossRef] [Green Version]
- Urrutia, A.O.; Hurst, L.D. The signature of selection mediated by expression on human genes. Genome Res. 2003, 13, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Vinogradov, A.E. Compactness of human housekeeping genes: Selection for economy or genomic design? TRENDS Genet. 2004, 20, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-Y.; Vorst, O.; Fiers, M.W.; Stiekema, W.J.; Nap, J.-P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Kosiol, C.; Vinař, T.; da Fonseca, R.R.; Hubisz, M.J.; Bustamante, C.D.; Nielsen, R.; Siepel, A. Patterns of positive selection in six mammalian genomes. PLoS Genet. 2008, 4, e1000144. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Y.; Jiang, L.; Wang, L.; Cai, Y. Evolutionary Rate Heterogeneity and Functional Divergence of Orthologous Genes in Pyrus. Biomolecules 2019, 9, 490. https://doi.org/10.3390/biom9090490
Cao Y, Jiang L, Wang L, Cai Y. Evolutionary Rate Heterogeneity and Functional Divergence of Orthologous Genes in Pyrus. Biomolecules. 2019; 9(9):490. https://doi.org/10.3390/biom9090490
Chicago/Turabian StyleCao, Yunpeng, Lan Jiang, Lihu Wang, and Yongping Cai. 2019. "Evolutionary Rate Heterogeneity and Functional Divergence of Orthologous Genes in Pyrus" Biomolecules 9, no. 9: 490. https://doi.org/10.3390/biom9090490
APA StyleCao, Y., Jiang, L., Wang, L., & Cai, Y. (2019). Evolutionary Rate Heterogeneity and Functional Divergence of Orthologous Genes in Pyrus. Biomolecules, 9(9), 490. https://doi.org/10.3390/biom9090490





