Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era
Abstract
1. Introduction
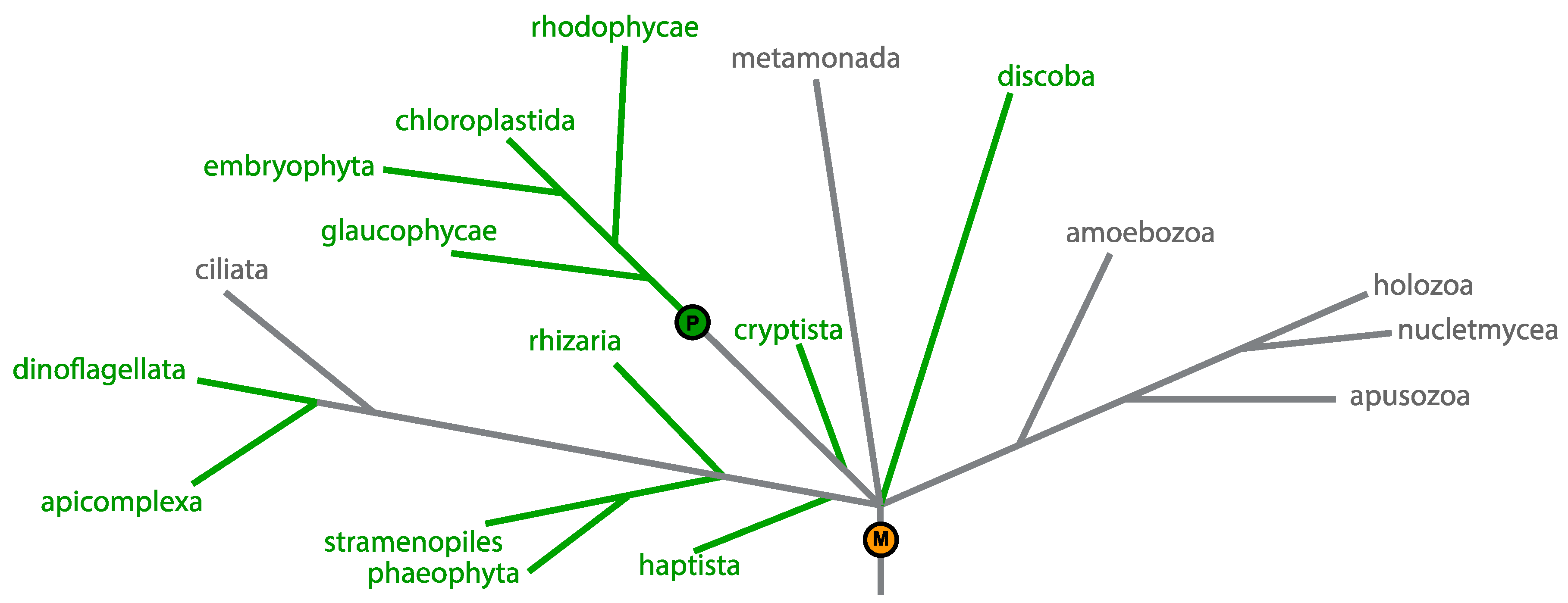
1.1. Plastid Genomes
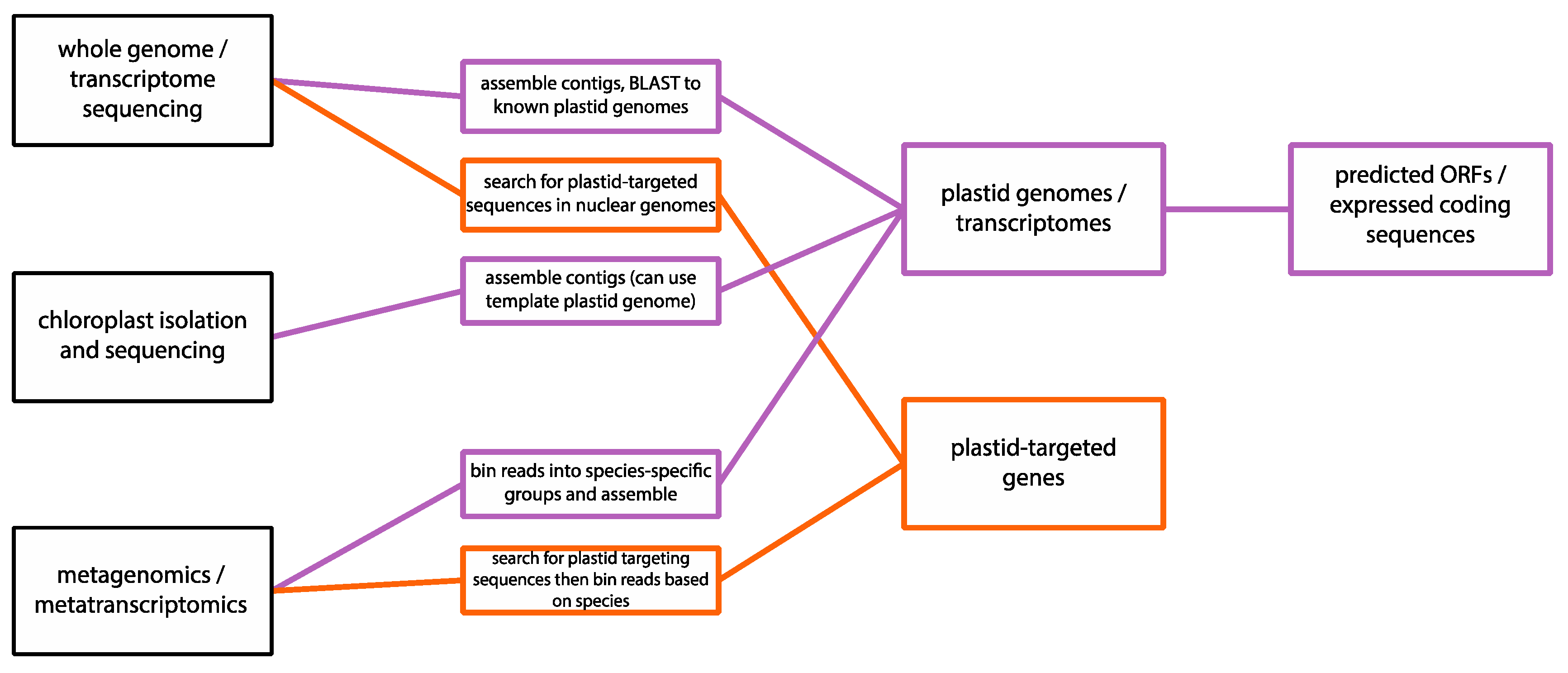
1.2. Next-Generation Sequencing
2. Case Studies
2.1. Case Study 1: Codon Usage
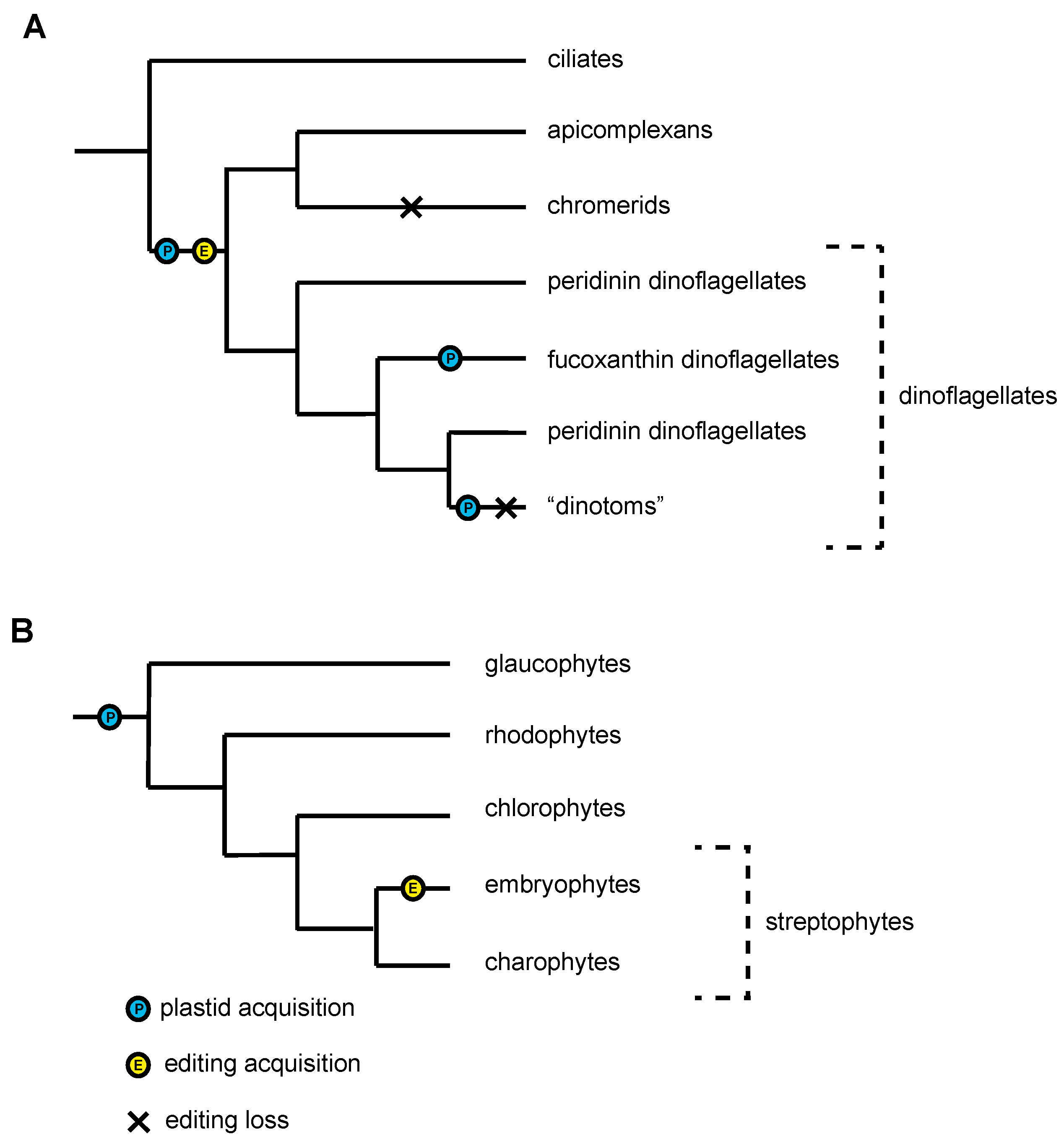
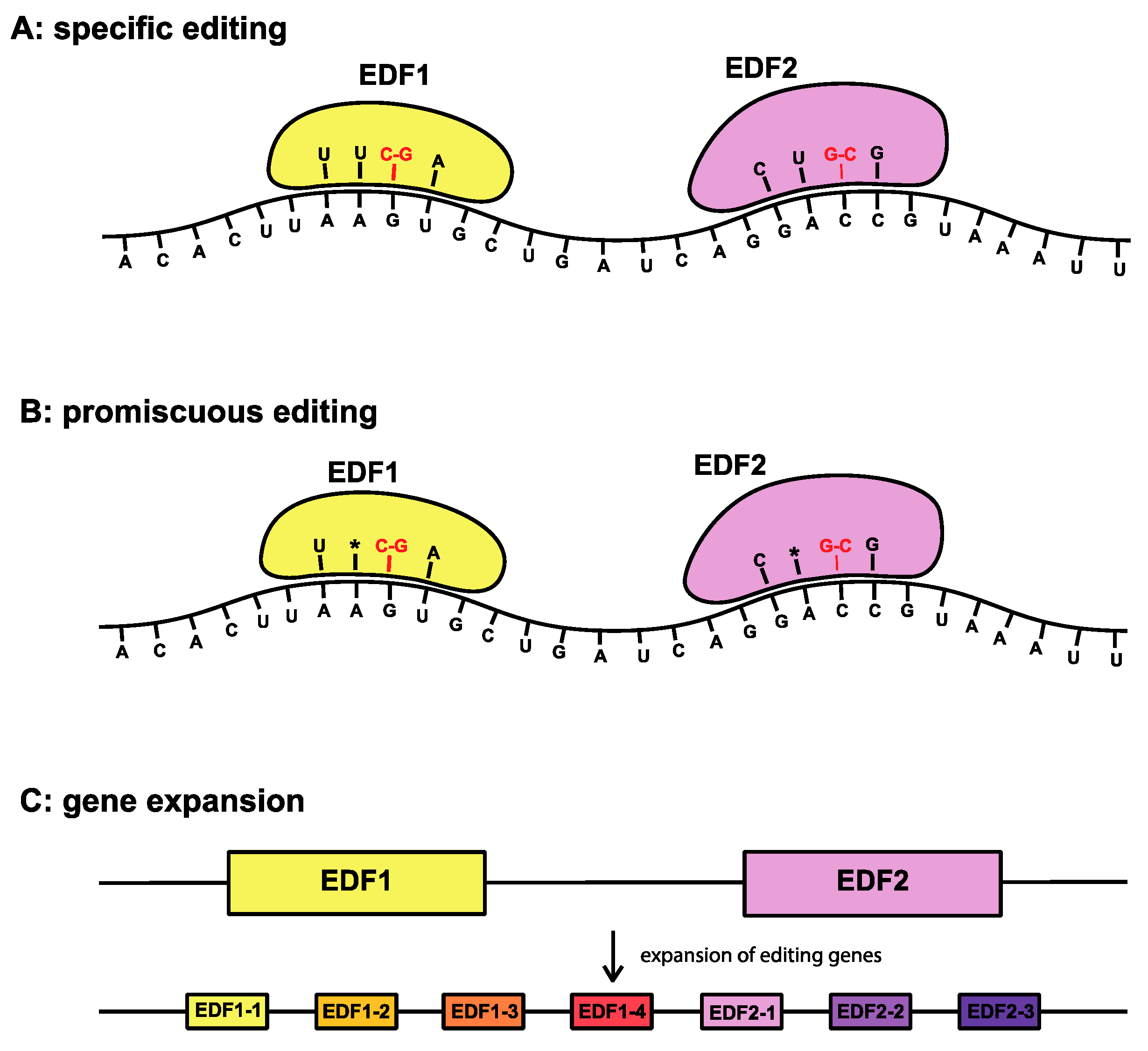
2.2. Case Study 2: RNA Editing
3. Future Possibilities
3.1. Evolutionary Biology
3.2. Ecological Research
3.3. Incorporation of New Computational Techniques and Biotechnology
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sagan, L. On the origin of mitosing cells. J. Theor. Biol. 1967, 14, 225–274. [Google Scholar] [CrossRef]
- Wallace, D.C. Structure and evolution of organelle genomes. Microbiol. Rev. 1982, 46, 208–240. [Google Scholar] [PubMed]
- Wilson, R.J.; Williamson, D.H.; Preiser, P. Malaria and other Apicomplexans: The “plant” connection. Infect. Agents Dis. 1994, 3, 29–37. [Google Scholar] [PubMed]
- Chase, M.W.; Fay, M.F. Ancient flowering plants: DNA sequences and angiosperm classification. Genome Biol. 2001, 2, reviews1012.1. [Google Scholar] [CrossRef] [PubMed]
- Ruhfel, B.R.; Gitzendanner, M.A.; Soltis, P.S.; Soltis, D.E.; Burleigh, J.G. From algae to angiosperms–inferring the phylogeny of green plants (Viridiplantae) from 360 plastid genomes. BMC Evol. Biol. 2014, 14, 23. [Google Scholar] [CrossRef] [PubMed]
- Lung, S.-C.; Smith, M.D.; Chuong, S.D. Isolation of Chloroplasts from Plant Protoplasts. Cold Spring Harb. Protoc. 2015, 2015, 074559. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef]
- Cheng, S.; Melkonian, M.; Smith, S.A.; Brockington, S.; Archibald, J.M.; Delaux, P.-M.; Li, F.-W.; Melkonian, B.; Mavrodiev, E.V.; Sun, W.; et al. 10KP: A phylodiverse genome sequencing plan. GigaScience 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Stephens, Z.D.; Lee, S.Y.; Faghri, F.; Campbell, R.H.; Zhai, C.; Efron, M.J.; Iyer, R.; Schatz, M.C.; Sinha, S.; Robinson, G.E. Big Data: Astronomical or Genomical? PLoS Biol. 2015, 13, e1002195. [Google Scholar] [CrossRef]
- Spang, A.; Saw, J.H.; Jørgensen, S.L.; Zaremba-Niedzwiedzka, K.; Martijn, J.; Lind, A.E.; Van Eijk, R.; Schleper, C.; Guy, L.; Ettema, T.J.G. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 2015, 521, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Zaremba-Niedzwiedzka, K.; Caceres, E.F.; Saw, J.H.; Bäckström, D.; Juzokaite, L.; Vancaester, E.; Seitz, K.W.; Anantharaman, K.; Starnawski, P.; Kjeldsen, K.U.; et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 2017, 541, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Salcher, M.M.; Andrei, A.-Ş.; Bulzu, P.-A.; Keresztes, Z.G.; Banciu, H.L.; Ghai, R. Visualization of Loki- and Heimdallarchaeia (Asgardarchaeota) by fluorescence in situ hybridization and catalyzed reporter deposition (CARD-FISH). bioRxiv 2019. [Google Scholar] [CrossRef]
- Saliba, A.-E.; Westermann, A.J.; Gorski, S.A.; Vogel, J. Single-cell RNA-seq: Advances and future challenges. Nucleic Acids Res. 2014, 42, 8845–8860. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. RNA-Seq data: A goldmine for organelle research. Brief. Funct. Genom. 2013, 12, 454–456. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Pop, M. The Theory and Practice of Genome Sequence Assembly. Annu. Rev. Genom. Hum. Genet. 2015, 16, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Sohn, J.-I.; Nam, J.-W. The present and future of de novo whole-genome assembly. Brief. Bioinform. 2018, 19, 23–40. [Google Scholar]
- Ladoukakis, E.; Kolisis, F.N.; Chatziioannou, A.A. Integrative workflows for metagenomic analysis. Front. Cell Dev. Biol. 2014, 2, 70. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2011, 12, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Lee, W.; Hottes, A.K.; Shapiro, L.; McAdams, H.H. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. USA 2004, 101, 3480–3485. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Morton, B.R. Codon Adaptation of Plastid Genes. PLoS ONE 2016, 11, e0154306. [Google Scholar] [CrossRef] [PubMed]
- Bulmer, M. The Selection-Mutation-Drift Theory of Synonymous Codon Usage. Genetics 1991, 129, 897–907. [Google Scholar]
- Sharp, P.M.; Emery, L.R.; Zeng, K. Forces that influence the evolution of codon bias. Philos. Trans. R. Soc. B Boil. Sci. 2010, 365, 1203–1212. [Google Scholar] [CrossRef]
- Morton, B.R. Chloroplast DNA codon use: Evidence for selection at the psb A locus based on tRNA availability. J. Mol. Evol. 1993, 37, 273–280. [Google Scholar] [CrossRef]
- Morton, B.R. Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 1998, 46, 449–459. [Google Scholar] [CrossRef]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef]
- Inagaki, Y.; Simpson, A.G.B.; Dacks, J.B.; Roger, A.J. Phylogenetic Artifacts Can be Caused by Leucine, Serine, and Arginine Codon Usage Heterogeneity: Dinoflagellate Plastid Origins as a Case Study. Syst. Biol. 2004, 53, 582–593. [Google Scholar] [CrossRef]
- Gray, M.W. Evolutionary Origin of RNA Editing. Biochemistry 2012, 51, 5235–5242. [Google Scholar] [CrossRef]
- Jackson, C.J.; Gornik, S.G.; Waller, R.F. A Tertiary Plastid Gains RNA Editing in Its New Host. Mol Biol Evol 2013, 30, 788–792. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Klinger, C.M.; Paoli, L.; Newby, R.J.; Wang, M.Y.-W.; Carroll, H.D.; Leblond, J.D.; Howe, C.J.; Dacks, J.B.; Bowler, C.; Cahoon, A.B.; et al. Plastid Transcript Editing across Dinoflagellate Lineages Shows Lineage-Specific Application but Conserved Trends. Genome Biol. Evol. 2018, 10, 1019–1038. [Google Scholar] [CrossRef] [PubMed]
- Zauner, S.; Greilinger, D.; Laatsch, T.; Kowallik, K.V.; Maier, U.-G. Substitutional editing of transcripts from genes of cyanobacterial origin in the dinoflagellateCeratium horridum. FEBS Lett. 2004, 577, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Morse, D. Rampant polyuridylylation of plastid gene transcripts in the dinoflagellate Lingulodinium. Nucleic Acids Res. 2006, 34, 613–619. [Google Scholar] [CrossRef] [PubMed]
- Dang, Y.; Green, B.R. Substitutional editing of Heterocapsa triquetra chloroplast transcripts and a folding model for its divergent chloroplast 16S rRNA. Gene 2009, 442, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Iida, S.; Kobiyama, A.; Ogata, T.; Murakami, A. Identification of transcribed and persistent variants of the psbA gene carried by plastid minicircles in a dinoflagellate. Curr. Genet. 2009, 55, 583–591. [Google Scholar] [CrossRef]
- Mungpakdee, S.; Shinzato, C.; Takeuchi, T.; Kawashima, T.; Koyanagi, R.; Hisata, K.; Tanaka, M.; Goto, H.; Fujie, M.; Lin, S.; et al. Massive Gene Transfer and Extensive RNA Editing of a Symbiotic Dinoflagellate Plastid Genome. Genome Biol. Evol. 2014, 6, 1408–1422. [Google Scholar] [CrossRef]
- Liew, Y.J.; Li, Y.; Baumgarten, S.; Voolstra, C.R.; Aranda, M. Condition-specific RNA editing in the coral symbiont Symbiodinium microadriaticum. PLoS Genet. 2017, 13, e1006619. [Google Scholar] [CrossRef]
- Hicks, J.; Lassadi, I.; Carpenter, E.; Eno, M.; Vardakis, A.; Waller, R.F.; Howe, C.; Nisbet, R.E.R. A Pentatricopeptide Repeat Protein in the Plasmodium apicoplast is essential and shows sequence-specific RNA binding. bioRxiv 2018. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yamaguchi, K.; Tsuji-Tsukinoki, S.; Knie, N.; Knoop, V. Chloroplast RNA editing going extreme: More than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. RNA 2014, 20, 1499–1506. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. RNA editing in plants: Machinery and flexibility of site recognition. Biochim. Biophys. Acta 2015, 1847, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Howe, C.J. Integration of plastids with their hosts: Lessons learned from dinoflagellates. Proc. Natl. Acad. Sci. USA 2015, 112, 10247–10254. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W.; Lukeš, J.; Archibald, J.M.; Keeling, P.J.; Doolittle, W.F. Irremediable Complexity? Science 2010, 330, 920–921. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.W.; Schreiber, J.; Yue, J.; Guo, H.; Ding, Q.; Huang, J. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 2014, 5, 5764. [Google Scholar] [CrossRef] [PubMed]
- Sevcikova, T.; Horák, A.; Klimes, V.; Zbránková, V.; Demir-Hilton, E.; Sudek, S.; Jenkins, J.; Schmutz, J.; Přibyl, P.; Fousek, J.; et al. Updating algal evolutionary relationships through plastid genome sequencing: Did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 2015, 5, 10134. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.G.; Gile, G.; McCallum, G.; Méheust, R.; Bapteste, E.P.; Klinger, C.M.; Brillet-Guéguen, L.; Freeman, K.D.; Richter, D.J.; Bowler, C. Chimeric origins of ochrophytes and haptophytes revealed through an ancient plastid proteome. eLife 2017, 6, e23717. [Google Scholar] [CrossRef] [PubMed]
- Barbrook, A.C.; Howe, C.J.; Purton, S. Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 2006, 11, 101–108. [Google Scholar] [CrossRef]
- Smith, D.R.; Crosby, K.; Lee, R.W. Correlation between Nuclear Plastid DNA Abundance and Plastid Number Supports the Limited Transfer Window Hypothesis. Genome Biol. Evol. 2011, 3, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R. Extending the Limited Transfer Window Hypothesis to Inter-organelle DNA Migration. Genome Biol. Evol. 2011, 3, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, R.; Kuo, A.; Ibarbalz, F.; Rocha Jimenez Vieira, F.; Pierella Karlusich, J.J.; McFarlane, J.; Richardson, E.; Edgar, R.M.; Potvin, M.; Peng, Y.; et al. Arctic algae genomes libraries. Apollo 2018. [Google Scholar] [CrossRef]
- Ebenezer, T.E.; Carrington, M.; Lebert, M.; Kelly, S.; Field, M.C. Euglena gracilis Genome and Transcriptome: Organelles, Nuclear Genome Assembly Strategies and Initial Features. Adv. Exp. Med. Biol. 2017, 979, 125–140. [Google Scholar]
- Curtis, B.A.; Tanifuji, G.; Burki, F.; Gruber, A.; Irimia, M.; Maruyama, S.; Arias, M.C.; Ball, S.G.; Gile, G.H.; Hirakawa, Y.; et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 2012, 492, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, K.R. Marine microorganisms and global nutrient cycles. Nature 2005, 437, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Ashworth, M.P.; Hajrah, N.H.; Khiyami, M.A.; Sabir, M.J.; Alhebshi, A.M.; Al-Malki, A.L.; Sabir, J.S.M.; Theriot, E.C.; Jansen, R.K. Chapter Five—Evolution of the Plastid Genomes in Diatoms. In Advances in Botanical Research. Plastid Genome Evolution; Chaw, S.-M., Jansen, R.K., Eds.; Academic Press: Cambridge, MA, USA, 2018; Volume 85, pp. 129–155. [Google Scholar]
- Geisen, S. Thorough high-throughput sequencing analyses unravels huge diversities of soil parasitic protists. Environ. Microbiol. 2016, 18, 1669–1672. [Google Scholar] [CrossRef] [PubMed]
- Bik, H.M.; Porazinska, D.L.; Creer, S.; Caporaso, J.G.; Knight, R.; Thomas, W.K. Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol. Evol. 2012, 27, 233–243. [Google Scholar] [CrossRef]
- Stoeck, T.; Bass, D.; Nebel, M.; Christen, R.; Jones, M.D.M.; Breiner, H.; Richards, T.A. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Mol. Ecol. 2010, 19, 21–31. [Google Scholar] [CrossRef]
- Wylezich, C.; Herlemann, D.; Jürgens, K. Improved 18S rDNA amplification protocol for assessing protist diversity in oxygen-deficient marine systems. Aquat. Microb. Ecol. 2018, 81, 83–94. [Google Scholar] [CrossRef]
- Porter, T.M.; Shokralla, S.; Baird, D.; Golding, G.B.; Hajibabaei, M. Ribosomal DNA and Plastid Markers Used to Sample Fungal and Plant Communities from Wetland Soils Reveals Complementary Biotas. PLoS ONE 2016, 11, e0142759. [Google Scholar] [CrossRef][Green Version]
- Woo, Y.H.; Ansari, H.R.; Otto, T.D.; Klinger, C.M.; Kolisko, M.; Michálek, J.; Saxena, A.; Shanmugam, D.; Tayyrov, A.; Veluchamy, A.; et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 2015, 4, e06974. [Google Scholar] [CrossRef]
- Nowack, E.C.; Melkonian, M.; Glöckner, G. Chromatophore Genome Sequence of Paulinella Sheds Light on Acquisition of Photosynthesis by Eukaryotes. Curr. Biol. 2008, 18, 410–418. [Google Scholar] [CrossRef] [PubMed]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-Generation Machine Learning for Biological Networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [PubMed]
- Qu, K.; Guo, F.; Liu, X.; Lin, Y.; Zou, Q. Application of Machine Learning in Microbiology. Front. Microbiol. 2019, 10, 827. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A Combinatorial Amino Acid Code for RNA Recognition by Pentatricopeptide Repeat Proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef] [PubMed]
- Giudice, C.L.; Hernández, I.; Ceci, L.R.; Pesole, G.; Picardi, E. RNA editing in plants: A comprehensive survey of bioinformatics tools and databases. Plant Physiol. Biochem. 2019, 137, 53–61. [Google Scholar] [CrossRef]
- Higgins, P.A.T.; Harte, J. Carbon Cycle Uncertainty Increases Climate Change Risks and Mitigation Challenges. J. Clim. 2012, 25, 7660–7668. [Google Scholar] [CrossRef]
- Mapelli, F.; Michoud, G.; Aulenta, F.; Boon, N.; Borin, S.; Kalogerakis, N.; Scoma, A.; Daffonchio, D. Biotechnologies for Marine Oil Spill Cleanup: Indissoluble Ties with Microorganisms. Trends Biotechnol. 2017, 35, 860–870. [Google Scholar] [CrossRef]
- Georgianna, D.R.; Mayfield, S.P. Exploiting diversity and synthetic biology for the production of algal biofuels. Nature 2012, 488, 329–335. [Google Scholar] [CrossRef]
- Gan, Q.; Jiang, J.; Han, X.; Wang, S.; Lu, Y. Engineering the Chloroplast Genome of Oleaginous Marine Microalga Nannochloropsis oceanica. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klinger, C.M.; Richardson, E. Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era. Biomolecules 2019, 9, 299. https://doi.org/10.3390/biom9080299
Klinger CM, Richardson E. Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era. Biomolecules. 2019; 9(8):299. https://doi.org/10.3390/biom9080299
Chicago/Turabian StyleKlinger, Christen M., and Elisabeth Richardson. 2019. "Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era" Biomolecules 9, no. 8: 299. https://doi.org/10.3390/biom9080299
APA StyleKlinger, C. M., & Richardson, E. (2019). Small Genomes and Big Data: Adaptation of Plastid Genomics to the High-Throughput Era. Biomolecules, 9(8), 299. https://doi.org/10.3390/biom9080299



