Correlating Lipid Membrane Permeabilities of Imidazolium Ionic Liquids with Their Cytotoxicities on Yeast, Bacterial, and Mammalian Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Model DOPC:DOPG Lipid Vesicles Entrapped with Ru(bpy)32+
2.2. Fluorescence Lifetime-Based Leakage Assay of Ru(bpy)32+-Entrapped LUVs in the Presence of ILs
2.3. Minimum-Inhibitory Concentration (MIC) Assays of ILs on Bacterial and Yeast Cells
2.4. Bacterial Membrane Permeabilization
2.5. Fungal Membrane Permeabilization
2.6. Red Blood Cell Membrane Permeabilization
3. Results
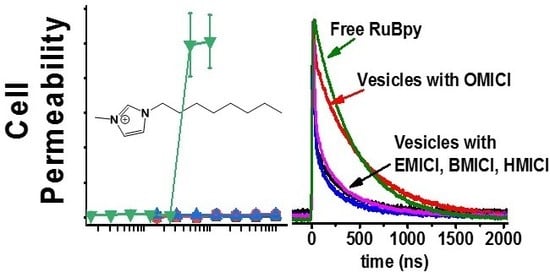
3.1. Lifetime-Based Leakage Assay Results
3.2. MIC Assay Results for ILs on Yeast and Bacteria
3.3. Direct Permeabilization Assays of ILs on E.coli
3.4. Direct Permeabilization Assays of ILs on S. cerevisae
3.5. Direct Permeabilization Assays of ILs on Red Blood Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Elgharbawy, A.A.; Alam, M.Z.; Moniruzzaman, M.; Goto, M. Ionic liquid pretreatment as emerging approaches for enhanced enzymatic hydrolysis of lignocellulosic biomass. Biochem. Eng. J. 2016, 109, 252–267. [Google Scholar] [CrossRef]
- Mikkola, S.-K.; Robciuc, A.; Lokajová, J.; Holding, A.J.; Lämmerhofer, M.; Kilpeläinen, I.; Holopainen, J.M.; King, A.W.T.; Wiedmer, S.K. Impact of Amphiphilic Biomass-Dissolving Ionic Liquids on Biological Cells and Liposomes. Environ. Sci. Technol. 2015, 49, 1870–1878. [Google Scholar] [CrossRef] [PubMed]
- Stanton, J.; Xue, Y.; Pandher, P.; Malek, L.; Brown, T.; Hu, X.; Salas-de la Cruz, D. Impact of ionic liquid type on the structure, morphology and properties of silk-cellulose biocomposite materials. Int. J. Biol. Macromol. 2018, 108, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Pinkert, A.; Marsh, K.N.; Pang, S.; Staiger, M.P. Ionic Liquids and Their Interaction with Cellulose. Chem. Rev. 2009, 109, 6712–6728. [Google Scholar] [CrossRef] [PubMed]
- Schroder, C. Proteins in Ionic Liquids: Current Status of Experiments and Simulations. Top. Curr. Chem. 2017, 375, 25. [Google Scholar] [CrossRef] [PubMed]
- Van Rantwijk, F.; Sheldon, R.A. Biocatalysis in Ionic Liquids. Chem. Rev. 2007, 107, 2757–2785. [Google Scholar] [CrossRef] [PubMed]
- Liu, G. Applications of Ionic Liquids in Biomedicine. Biophys. Rev. Lett. 2012, 7, 121–134. [Google Scholar] [CrossRef]
- Jens, S. Aqueous ionic liquids and their effects on protein structures: An overview of recent theoretical and experimental results. J. Phys. Condens. Matter 2017, 29, 233001. [Google Scholar]
- Kohn, E.M.; Lee, J.Y.; Calabro, A.; Vaden, T.D.; Caputo, G.A. Heme Dissociation from Myoglobin in the Presence of the Zwitterionic Detergent N,N-Dimethyl-N-dodecylglycine betaine (Empigen BB®): Effects of Ionic Liquids. Biomolecules 2018, 8, 126. [Google Scholar] [CrossRef]
- Egorova, K.S.; Gordeev, E.G.; Ananikov, V.P. Biological Activity of Ionic Liquids and Their Application in Pharmaceutics and Medicine. Chem. Rev. 2017, 117, 7132–7189. [Google Scholar] [CrossRef]
- Hanna, S.L.; Huang, J.L.; Swinton, A.J.; Caputo, G.A.; Vaden, T.D. Synergistic effects of polymyxin and ionic liquids on lipid vesicle membrane stability and aggregation. Biophys. Chem. 2017. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gilmore, B.F. The antimicrobial potential of ionic liquids: A source of chemical diversity for infection and biofilm control. Int. J. Antimicrob. Agents 2015, 46, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Singh, U.K.; Dohare, N.; Mishra, P.; Singh, P.; Bohidar, H.B.; Patel, R. Effect of pyrrolidinium-based ionic liquid on the channel form of gramicidin in lipid vesicles. J. Photochem. Photobio. B Biol. 2015, 149, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Singh, G.; Kang, T.S. Effect of alkyl chain functionalization of ionic liquid surfactants on the complexation and self-assembling behavior of polyampholyte gelatin in aqueous medium. Phys. Chem. Chem. Phys. 2016, 18, 25993–26009. [Google Scholar] [CrossRef] [PubMed]
- Singh, G.; Kang, T.S. Ionic Liquid Surfactant Mediated Structural Transitions and Self-Assembly of Bovine Serum Albumin in Aqueous Media: Effect of Functionalization of Ionic Liquid Surfactants. J. Phys. Chem. B 2015, 119, 10573–10585. [Google Scholar] [CrossRef]
- Alves, F.; Oliveira, F.S.; Schröder, B.; Matos, C.; Marrucho, I.M. Synthesis, characterization, and liposome partition of a novel tetracycline derivative using the ionic liquids framework. J. Pharm. Sci. 2013, 102, 1504–1512. [Google Scholar] [CrossRef]
- Weaver, K.D.; Kim, H.J.; Sun, J.; MacFarlane, D.R.; Elliott, G.D. Cyto-toxicity and biocompatibility of a family of choline phosphate ionic liquids designed for pharmaceutical applications. Green Chem. 2010, 12, 507–513. [Google Scholar] [CrossRef]
- Riduan, S.N.; Zhang, Y. Imidazolium salts and their polymeric materials for biological applications. Chem. Soc. Rev. 2013, 42, 9055–9070. [Google Scholar] [CrossRef]
- Zakrewski, M.; Lovejoy, K.S.; Kern, T.L.; Miller, T.E.; Le, V.; Nagy, A.; Goumas, A.M.; Iyer, R.S.; Del Sesto, R.E.; Koppisch, A.T.; et al. Ionic liquids as a class of materials for transdermal delivery and pathogen neutralization. Proc. Natl. Acad. Sci. USA 2014, 111, 13313–13318. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Lan, J.; You, J.; Chen, L. Synthesis and biological applications of imidazolium-based polymerized ionic liquid as a gene delivery vector. Chem. Biol. Drug Des. 2009, 74, 282–288. [Google Scholar] [CrossRef]
- Jing, B.; Lan, N.; Qiu, J.; Zhu, Y. Interaction of Ionic Liquids with a Lipid Bilayer: A Biophysical Study of Ionic Liquid Cytotoxicity. J. Phys. Chem. B 2016, 120, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.; Shah, J.K.; Zhu, Y.; Maginn, E.J. Amphiphilic interactions of ionic liquids with lipid biomembranes: A molecular simulation study. Soft Matter 2014, 10, 8641–8651. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.; Zhu, Y.; Maginn, E.J. Molecular Mechanism of Ionic-Liquid-Induced Membrane Disruption: Morphological Changes to Bilayers, Multilayers, and Vesicles. Langmuir 2016, 32, 5403–5411. [Google Scholar] [CrossRef] [PubMed]
- Gal, N.; Malferarri, D.; Kolusheva, S.; Galletti, P.; Tagliavini, E.; Jelinek, R. Membrane interactions of ionic liquids: Possible determinants for biological activity and toxicity. Biochim. Biophys. Acta Biomembr. 2012, 1818, 2967–2974. [Google Scholar] [CrossRef] [PubMed]
- Benedetto, A.; Heinrich, F.; Gonzalez, M.A.; Fragneto, G.; Watkins, E.; Ballone, P. Structure and stability of phospholipid bilayers hydrated by a room-temperature ionic liquid/water solution: A neutron reflectometry study. J. Phys. Chem. B 2014, 118, 12192–12206. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.; Ha, S.H.; Han, S.-H.; Lim, M.-C.; Kim, S.M.; Kim, Y.-R.; Koo, Y.-M.; Therefore, J.-S.; Jeon, T.-J. Elucidation of molecular interactions between lipid membranes and ionic liquids using model cell membranes. Soft Matter 2012, 8, 5501–5506. [Google Scholar] [CrossRef]
- Gayet, F.; Marty, J.-D.; Brûlet, A.; Viguerie, N.L.-D. Vesicles in Ionic Liquids. Langmuir 2011, 27, 9706–9710. [Google Scholar] [CrossRef] [PubMed]
- Rengstl, D.; Kraus, B.; Van Vorst, M.; Elliot, G.D.; Kunz, W. Effect of choline carboxylate ionic liquids on biological membranes. Colloids Surf. B Biointerfaces 2014, 123, 575–581. [Google Scholar] [CrossRef]
- Lim, G.S.; Jaenicke, S.; Klahn, M. How the spontaneous insertion of amphiphilic imidazolium-based cations changes biological membranes: A molecular simulation study. Phys. Chem. Chem. Phys. 2015, 17, 29171–29183. [Google Scholar] [CrossRef]
- Kontro, I.; Svedstrom, K.; Dusa, F.; Ahvenainen, P.; Ruokonen, S.K.; Witos, J.; Wiedmer, S.K. Effects of phosphonium-based ionic liquids on phospholipid membranes studied by small-angle X-ray scattering. Chem. Phys. Lipids 2016, 201, 59–66. [Google Scholar] [CrossRef]
- Cole, M.R. Chemical and Biological Evaluation of Antibiotic-Based Ionic Liquids and Gumbos Against Pathogenic Bacteria. Ph.D. Dissertation, Louisiana State University, Baton Rouge, LA, USA, 2012. Volume 487. [Google Scholar]
- Bhattacharya, G.; Giri, R.P.; Saxena, H.; Agrawal, V.V.; Gupta, A.; Mukhopadhyay, M.K.; Ghosh, S.K. X-ray Reflectivity Study of the Interaction of an Imidazolium-Based Ionic Liquid with a Soft Supported Lipid Membrane. Langmuir 2017, 33, 1295–1304. [Google Scholar] [CrossRef] [PubMed]
- Galletti, P.; Malferrari, D.; Samorì, C.; Sartor, G.; Tagliavini, E. Effects of ionic liquids on membrane fusion and lipid aggregation of egg-PC liposomes. Colloids Surf. B Biointerfaces 2015, 125, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Jungnickel, C.; Łuczak, J.; Ranke, J.; Fernández, J.F.; Müller, A.; Thöming, J. Micelle formation of imidazolium ionic liquids in aqueous solution. Colloids Surf. A Physicochem. Eng. Asp. 2008, 316, 278–284. [Google Scholar] [CrossRef]
- Evans, K.O. Room-temperature ionic liquid cations act as short-chain surfactants and disintegrate a phospholipid bilayer. Colloids Surf. A Physicochem. Eng. Asp. 2006, 274, 11–17. [Google Scholar] [CrossRef]
- Kumar, R.A.; Papaïconomou, N.; Lee, J.-M.; Salminen, J.; Clark, D.S.; Prausnitz, J.M. In vitro cytotoxicities of ionic liquids: Effect of cation rings, functional groups, and anions. Environ. Toxicol. Int. J. 2009, 24, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Stolte, S.; Matzke, M.; Arning, J.; Boschen, A.; Pitner, W.-R.; Welz-Biermann, U.; Jastorff, B.; Ranke, J. Effects of different head groups and functionalised side chains on the aquatic toxicity of ionic liquids. Green Chem. 2007, 9, 1170–1179. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef] [PubMed]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Yoo, B.; Jing, B.; Jones, S.E.; Lamberti, G.A.; Zhu, Y.; Shah, J.K.; Maginn, E.J. Molecular mechanisms of ionic liquid cytotoxicity probed by an integrated experimental and computational approach. Sci. Rep. 2016, 6, 19889. [Google Scholar] [CrossRef]
- Saint Jean, K.D.; Henderson, K.D.; Chrom, C.L.; Abiuso, L.E.; Renn, L.M.; Caputo, G.A. Effects of Hydrophobic Amino Acid Substitutions on Antimicrobial Peptide Behavior. Probiotics Antimicrob. Proteins 2018, 10, 408–419. [Google Scholar] [CrossRef]
- Chrom, C.L.; Renn, L.M.; Caputo, G.A. Characterization and Antimicrobial Activity of Amphiphilic Peptide AP3 and Derivative Sequences. Antibiotics 2019, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Burman, L.G.; Nordstrom, K.; Boman, H.G. Resistance of Escherichia coli to penicillins. V. Physiological comparison of two isogenic strains, one with chromosomally and one with episomally mediated ampicillin resistance. J. Bacteriol. 1968, 96, 438–446. [Google Scholar] [PubMed]
- Kohn, E.M.; Shirley, D.J.; Arotsky, L.; Picciano, A.M.; Ridgway, Z.; Urban, M.W.; Carone, B.R.; Caputo, G.A. Role of Cationic Side Chains in the Antimicrobial Activity of C18G. Molecules 2018, 23, 329. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, I.; Hilpert, K.; Hancock, R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Shirley, D.J.; Chrom, C.L.; Richards, E.A.; Carone, B.R.; Caputo, G.A. Antimicrobial activity of a porphyrin binding peptide. Pept. Sci. 2018, 110. [Google Scholar] [CrossRef]
- Nicoletti, I.; Migliorati, G.; Pagliacci, M.C.; Grignani, F.; Riccardi, C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J. Immunol. Methods 1991, 139, 271–279. [Google Scholar] [CrossRef]
- Botstein, D.; Chervitz, S.A.; Cherry, J.M. Yeast as a model organism. Science 1997, 277, 1259–1260. [Google Scholar] [CrossRef]
- Lipke, P.N.; Ovalle, R. Cell wall architecture in yeast: New structure and new challenges. J. Bacteriol. 1998, 180, 3735–3740. [Google Scholar]
- Riccardi, C.; Nicoletti, I. Analysis of apoptosis by propidium iodide staining and flow cytometry. Nat. Protoc. 2006, 1, 1458–1461. [Google Scholar] [CrossRef]
- Jikuya, T.; Tsutsui, T.; Shigeta, O.; Sankai, Y.; Mitsui, T. Species differences in erythrocyte mechanical fragility: Comparison of human, bovine, and ovine cells. ASAIO J. 1998, 44, M452–M455. [Google Scholar] [CrossRef]
- Schultz, G.W. Animal influence on man-biting rates at a malarious site in Palawan, Philippines. Southeast Asian J. Trop. Med. Public Health 1989, 20, 49–53. [Google Scholar] [PubMed]
- Rantamäki, A.H.; Ruokonen, S.-K.; Sklavounos, E.; Kyllönen, L.; King, A.W.T.; Wiedmer, S.K. Impact of Surface-Active Guanidinium-, Tetramethylguanidinium-, and Cholinium-Based Ionic Liquids on Vibrio Fischeri Cells and Dipalmitoylphosphatidylcholine Liposomes. Sci. Rep. 2017, 7, 46673. [Google Scholar] [CrossRef] [PubMed]
- Ruokonen, S.-K.; Sanwald, C.; Robciuc, A.; Hietala, S.; Rantamäki, A.H.; Witos, J.; King, A.W.T.; Lämmerhofer, M.; Wiedmer, S.K. Correlation between Ionic Liquid Cytotoxicity and Liposome–Ionic Liquid Interactions. Chem. Eur. J. 2018, 24, 2669–2680. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, K.; Caputo, G.A.; DeGrado, W.F. The Role of Hydrophobicity in the Antimicrobial and Hemolytic Activities of Polymethacrylate Derivatives. Chemistry 2009, 15, 1123–1133. [Google Scholar] [CrossRef] [PubMed]
- Geiger, O.; Sohlenkamp, C. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2015, 40, 133–159. [Google Scholar] [CrossRef]
- Guan, X.L.; Wenk, M.R. Mass spectrometry-based profiling of phospholipids and sphingolipids in extracts from Saccharomyces cerevisiae. Yeast 2006, 23, 465–477. [Google Scholar] [CrossRef]
- Klose, C.; Surma, M.A.; Gerl, M.J.; Meyenhofer, F.; Shevchenko, A.; Simons, K. Flexibility of a Eukaryotic Lipidome – Insights from Yeast Lipidomics. PLoS ONE 2012, 7, e35063. [Google Scholar] [CrossRef]
- Bowman, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. Bioessays 2006, 28, 799–808. [Google Scholar] [CrossRef]
- Aoun, M.; Corsetto, P.A.; Nugue, G.; Montorfano, G.; Ciusani, E.; Crouzier, D.; Hogarth, P.; Gregory, A.; Hayflick, S.; Zorzi, G.; et al. Changes in Red Blood Cell membrane lipid composition: A new perspective into the pathogenesis of PKAN. Mol. Genet. Metab. 2017, 121, 180–189. [Google Scholar] [CrossRef]
- Virtanen, J.A.; Cheng, K.H.; Somerharju, P. Phospholipid composition of the mammalian red cell membrane can be rationalized by a superlattice model. Proc. Natl. Acad. Sci. USA 1998, 95, 4964–4969. [Google Scholar] [CrossRef]
- Nelson, G.J. Studies on the lipids of sheep red blood cells. I. Lipid composition in low and high potassium red cells. Lipids 1967, 2, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Avci, F.G.; Sariyar Akbulut, B.; Ozkirimli, E. Membrane Active Peptides and Their Biophysical Characterization. Biomolecules 2018, 8, 77. [Google Scholar] [CrossRef] [PubMed]
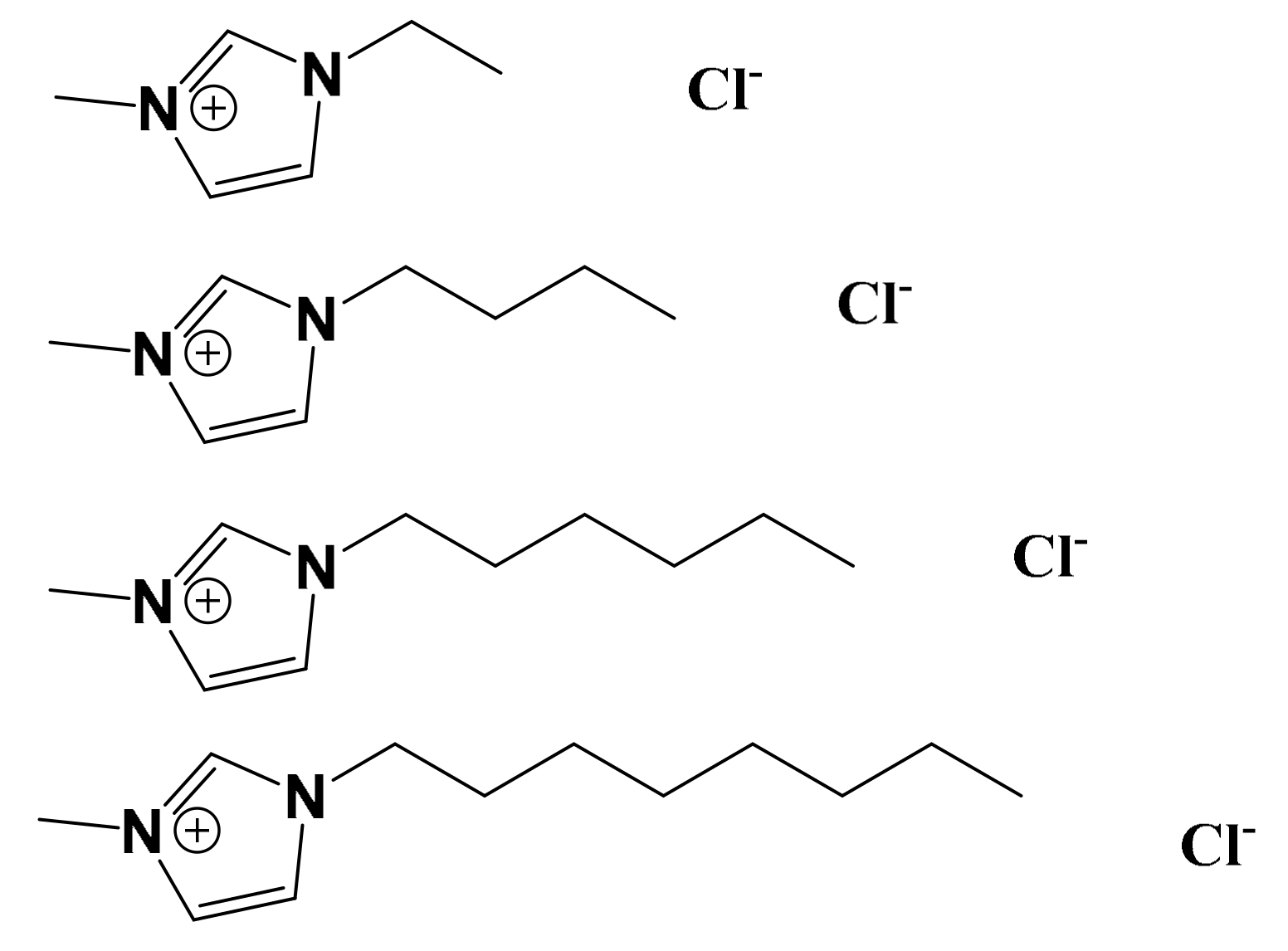
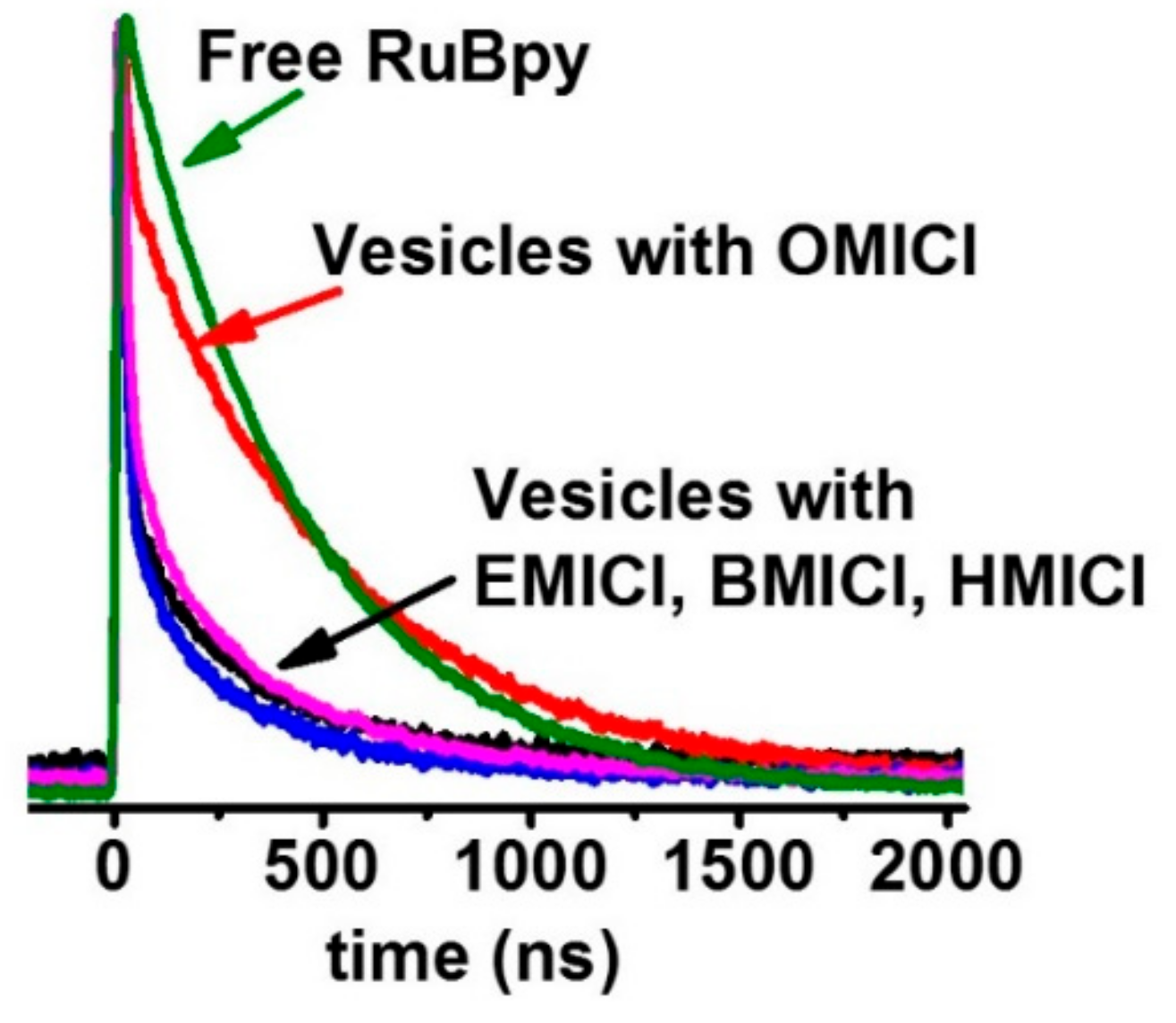
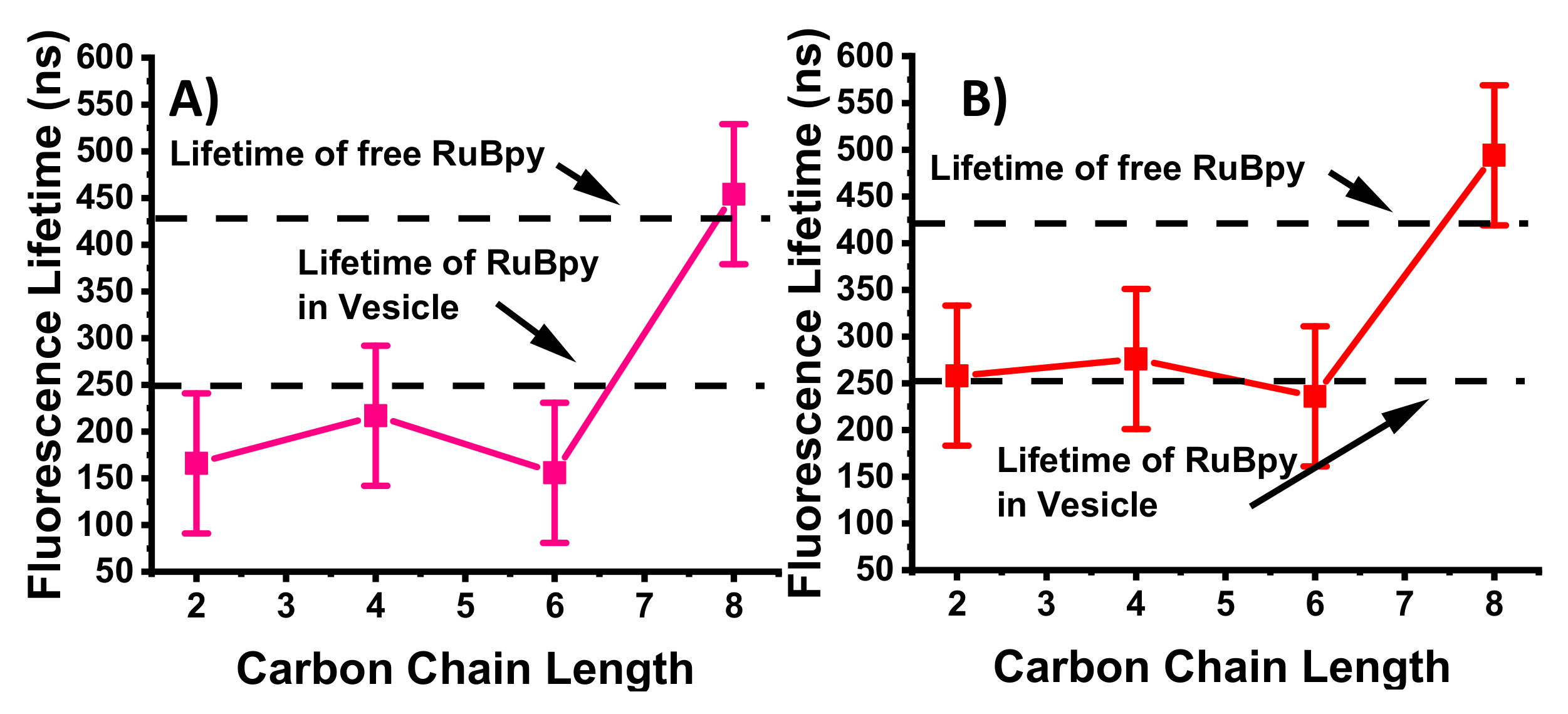
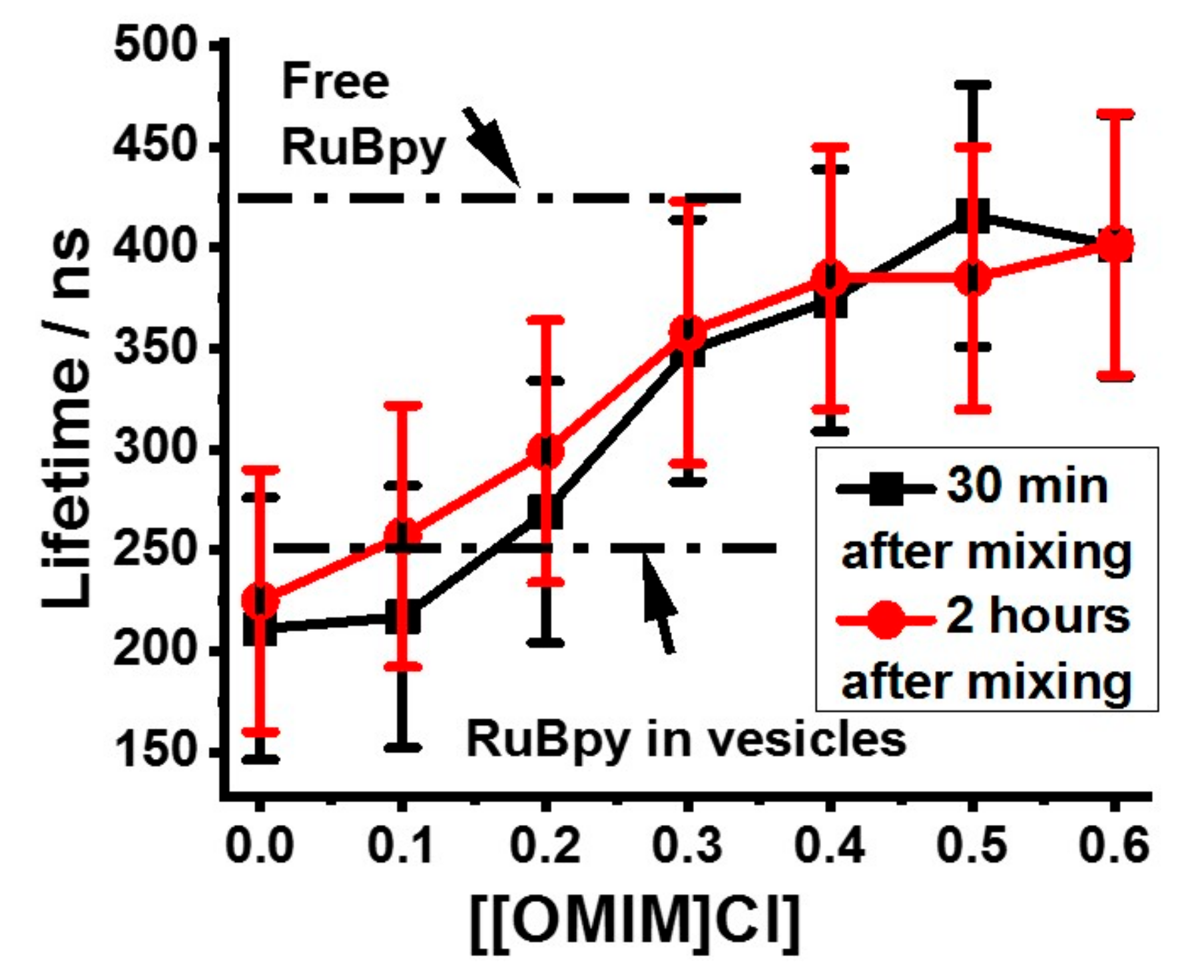
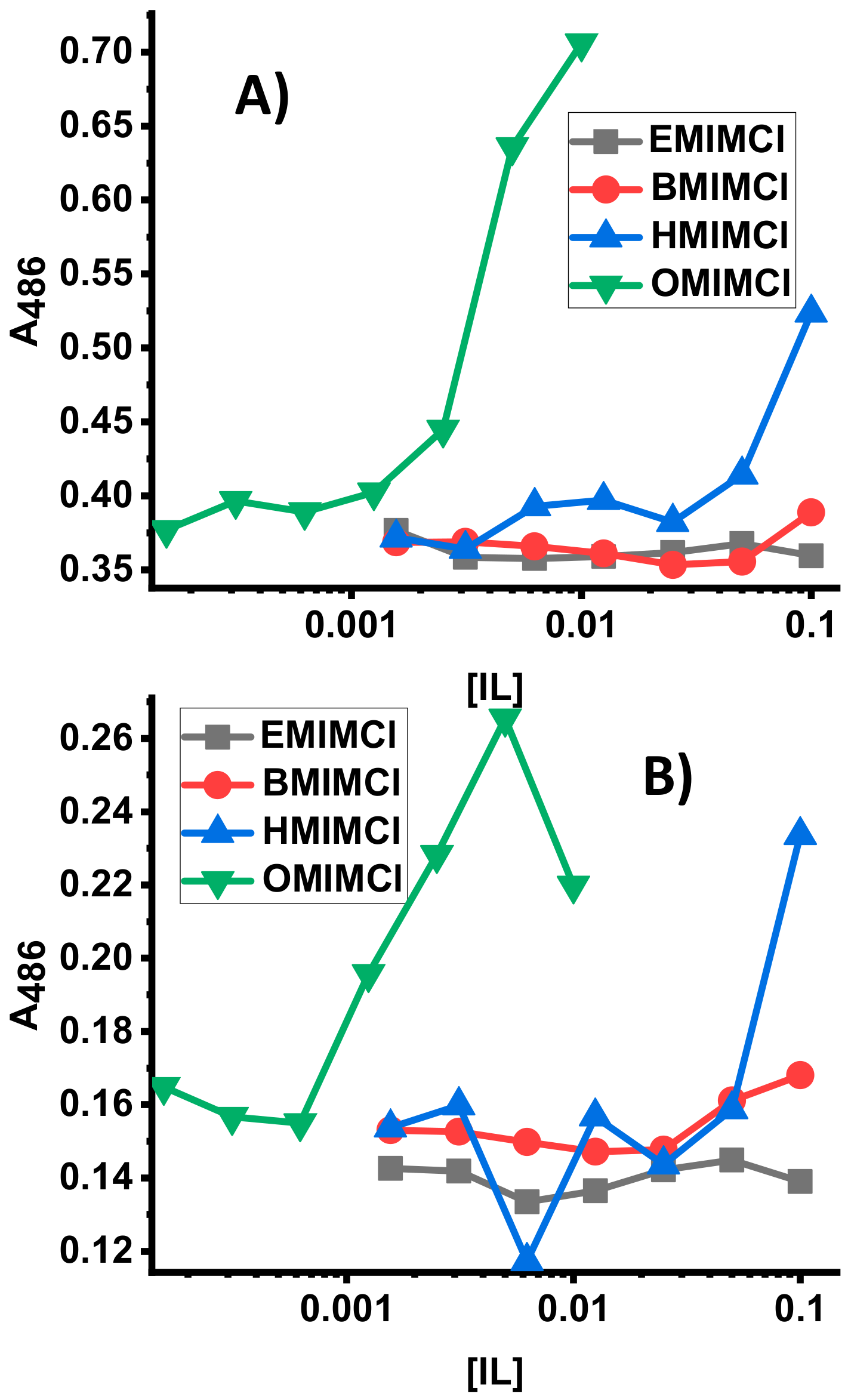
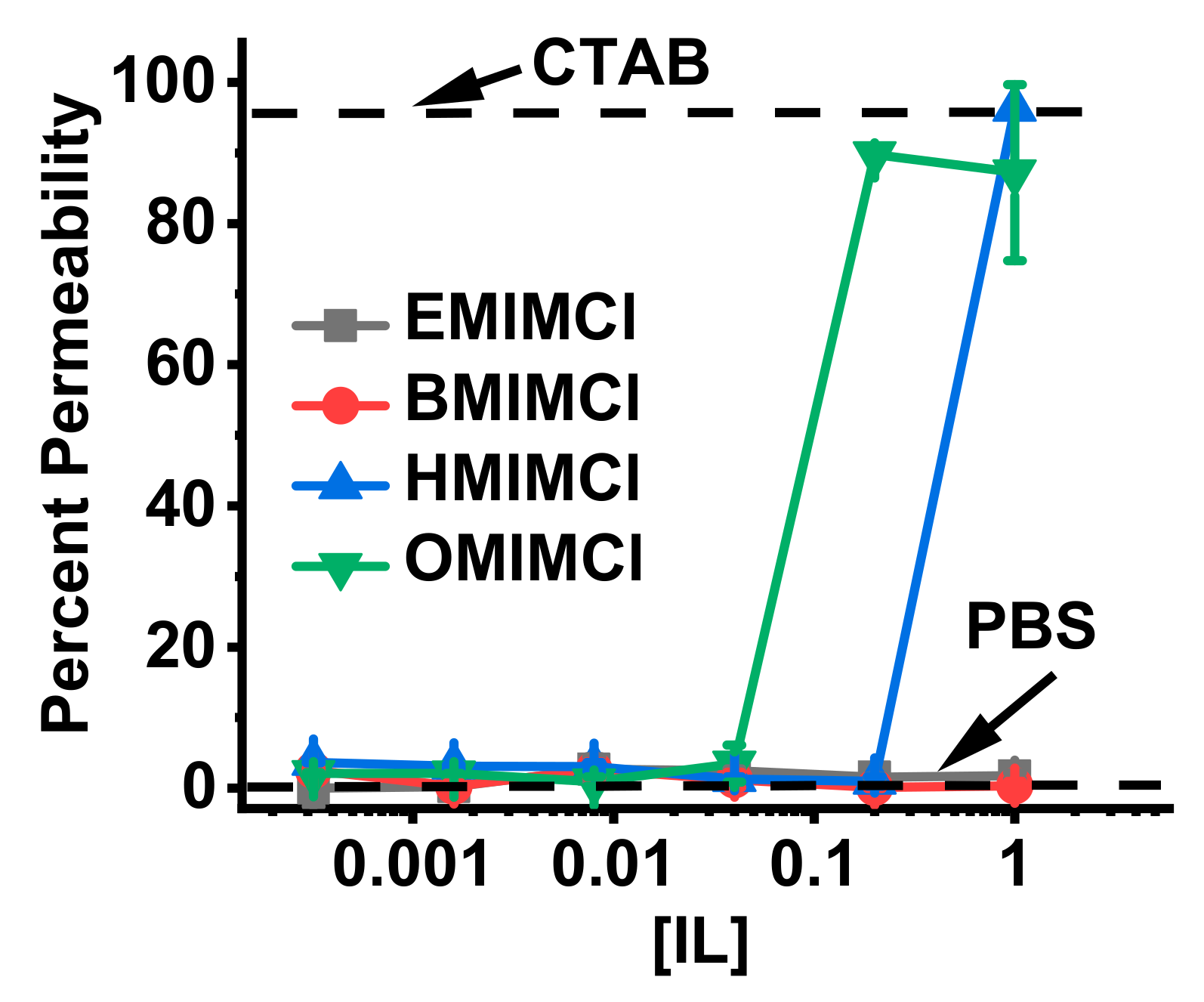
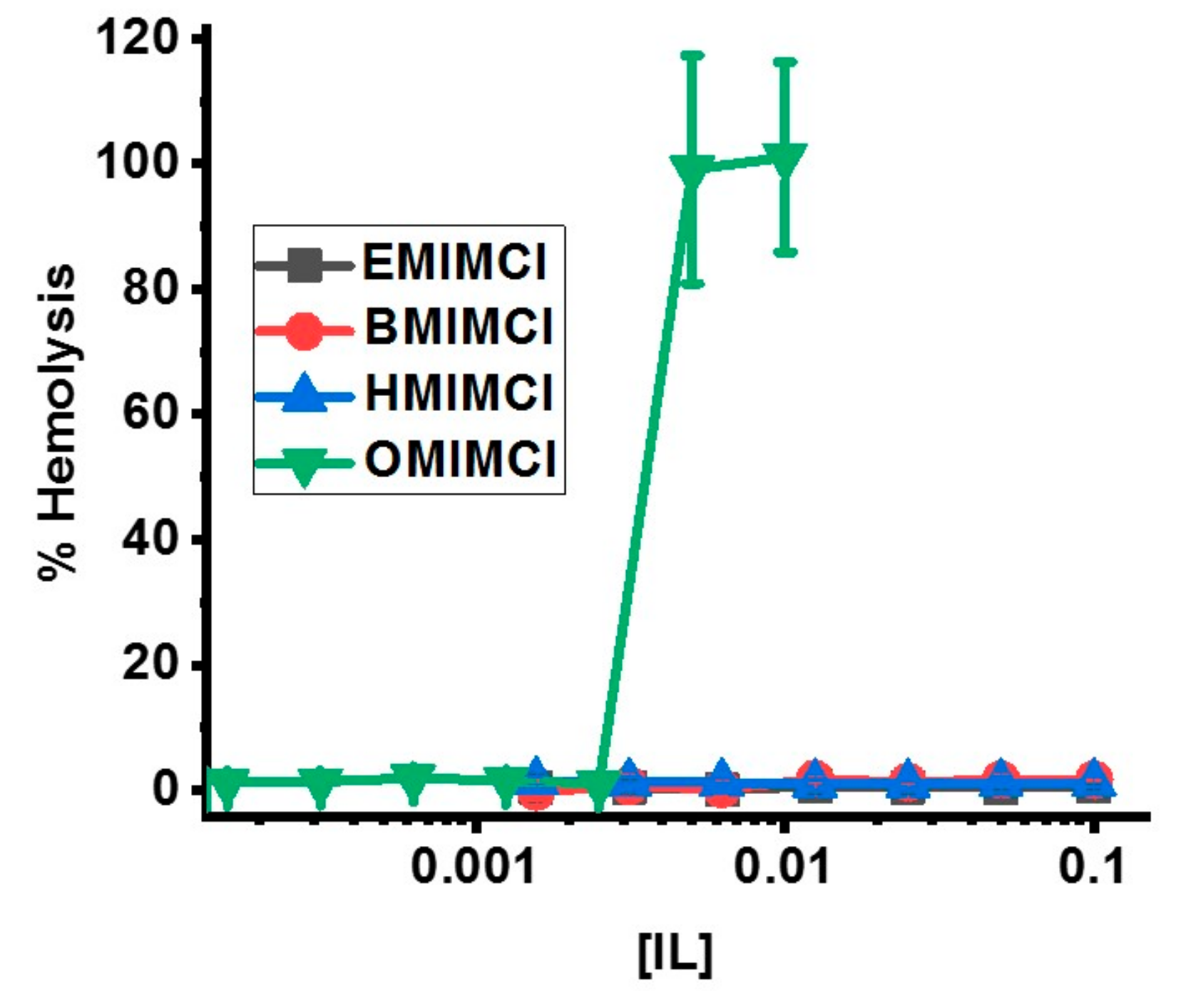
| IL | S. aureus | E. coli | P. aeruginosa |
|---|---|---|---|
| [EMIM]Cl | >200 | >200 | >100 |
| [BMIM]Cl | 25 | 50 | 100 |
| [HMIM]Cl | 6.25 | 6.25 | 25 |
| [OMIM]Cl | 2.5 | 1.25 | 10 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cook, K.; Tarnawsky, K.; Swinton, A.J.; Yang, D.D.; Senetra, A.S.; Caputo, G.A.; Carone, B.R.; Vaden, T.D. Correlating Lipid Membrane Permeabilities of Imidazolium Ionic Liquids with Their Cytotoxicities on Yeast, Bacterial, and Mammalian Cells. Biomolecules 2019, 9, 251. https://doi.org/10.3390/biom9060251
Cook K, Tarnawsky K, Swinton AJ, Yang DD, Senetra AS, Caputo GA, Carone BR, Vaden TD. Correlating Lipid Membrane Permeabilities of Imidazolium Ionic Liquids with Their Cytotoxicities on Yeast, Bacterial, and Mammalian Cells. Biomolecules. 2019; 9(6):251. https://doi.org/10.3390/biom9060251
Chicago/Turabian StyleCook, Kendall, Katharine Tarnawsky, Alana J. Swinton, Daniel D. Yang, Alexandria S. Senetra, Gregory A. Caputo, Benjamin R. Carone, and Timothy D. Vaden. 2019. "Correlating Lipid Membrane Permeabilities of Imidazolium Ionic Liquids with Their Cytotoxicities on Yeast, Bacterial, and Mammalian Cells" Biomolecules 9, no. 6: 251. https://doi.org/10.3390/biom9060251
APA StyleCook, K., Tarnawsky, K., Swinton, A. J., Yang, D. D., Senetra, A. S., Caputo, G. A., Carone, B. R., & Vaden, T. D. (2019). Correlating Lipid Membrane Permeabilities of Imidazolium Ionic Liquids with Their Cytotoxicities on Yeast, Bacterial, and Mammalian Cells. Biomolecules, 9(6), 251. https://doi.org/10.3390/biom9060251