COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Growth Conditions
2.2. Plasmid and Strain Construction
2.2.1. Construction of Cloning Vector pME4696
2.2.2. Construction of pME4701 Resulting in Strains AGB1169 and AGB1233
2.2.3. Construction of Plasmids pME4703 and pME4704 and Resulting Strains AGB1156 and AGB1157
2.2.4. Construction of Plasmid pME4706 and A. nidulans Strain AGB1159 and AGB1161
2.2.5. Construction of Plasmid pME4707 and A. nidulans Strain AGB1162 and AGB1163
2.2.6. Construction of BiFC Plasmids pME4708-4712 and the Resulting, A. nidulans Strains AGB1170-AGB1174
2.2.7. Construction of Plasmid pME4714 and the A. nidulans Strains AGB1165 and AGB1167
2.2.8. Construction of AGB1164, AGB1166, AGB1234, and AGB822
2.2.9. Construction of Plasmid pME4715
2.3. Transformations of E. coli, A. nidulans, and S. cerevisiae
2.4. Genomic DNA Extraction
2.5. Database Searches and BLAST Analyses
2.6. Phenotypical Characterization
2.7. Protein Isolation and Western Blot
2.8. RNA Extraction and Quantitative Real-Time PCR
2.9. Yeast-Two-Hybrid Assay
2.10. Fluorescence Microscopy
2.11. GFP Pull-Down
2.12. Tryptic Digestion
2.13. LC-MS Analysis
3. Results
3.1. The Repertoire of Deubiquitinating Enzymes (DUBs) in Aspergillus nidulans
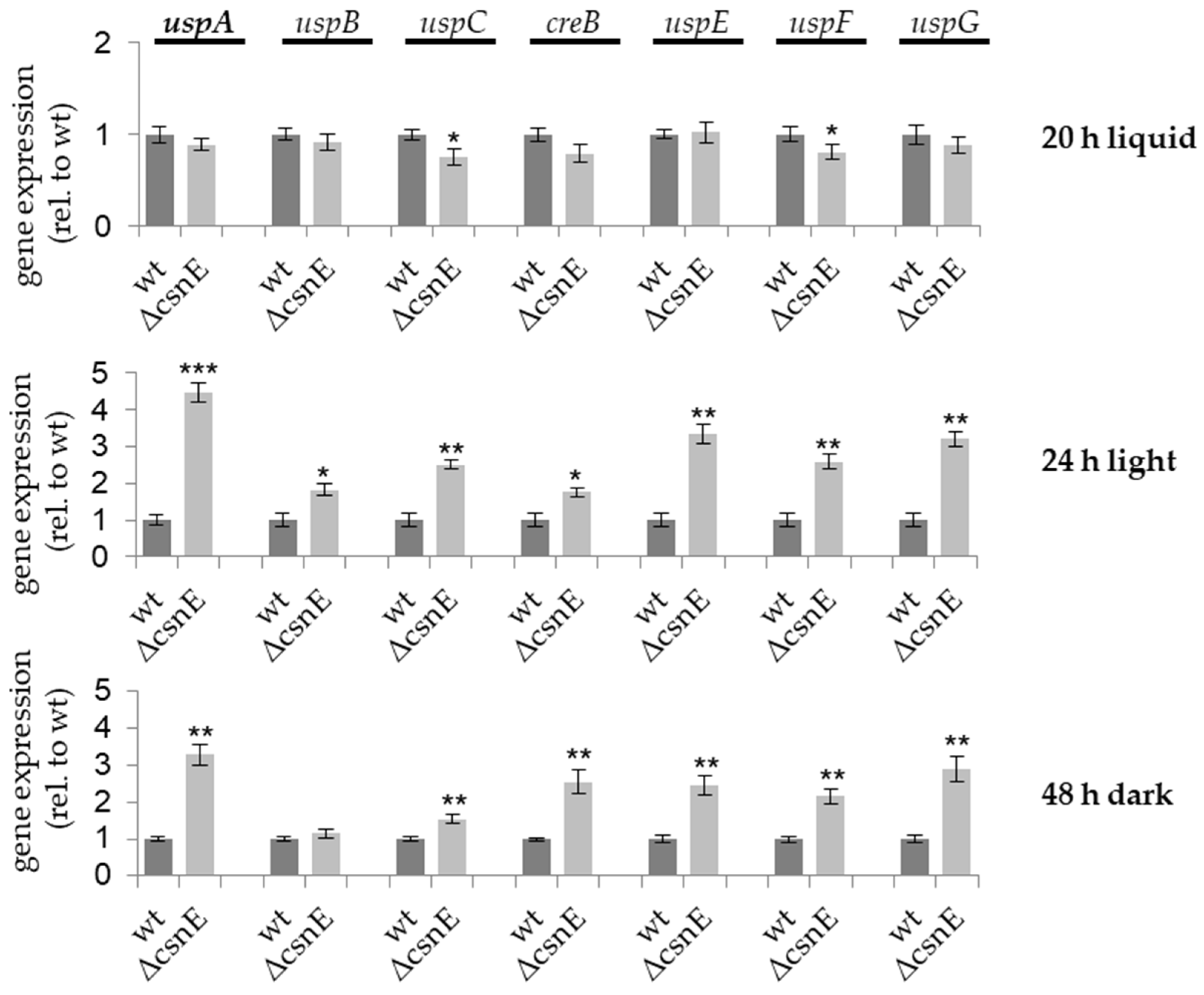
3.2. CsnE Causes the Repression of Ubiquitin-Specific Protease Encoding Genes
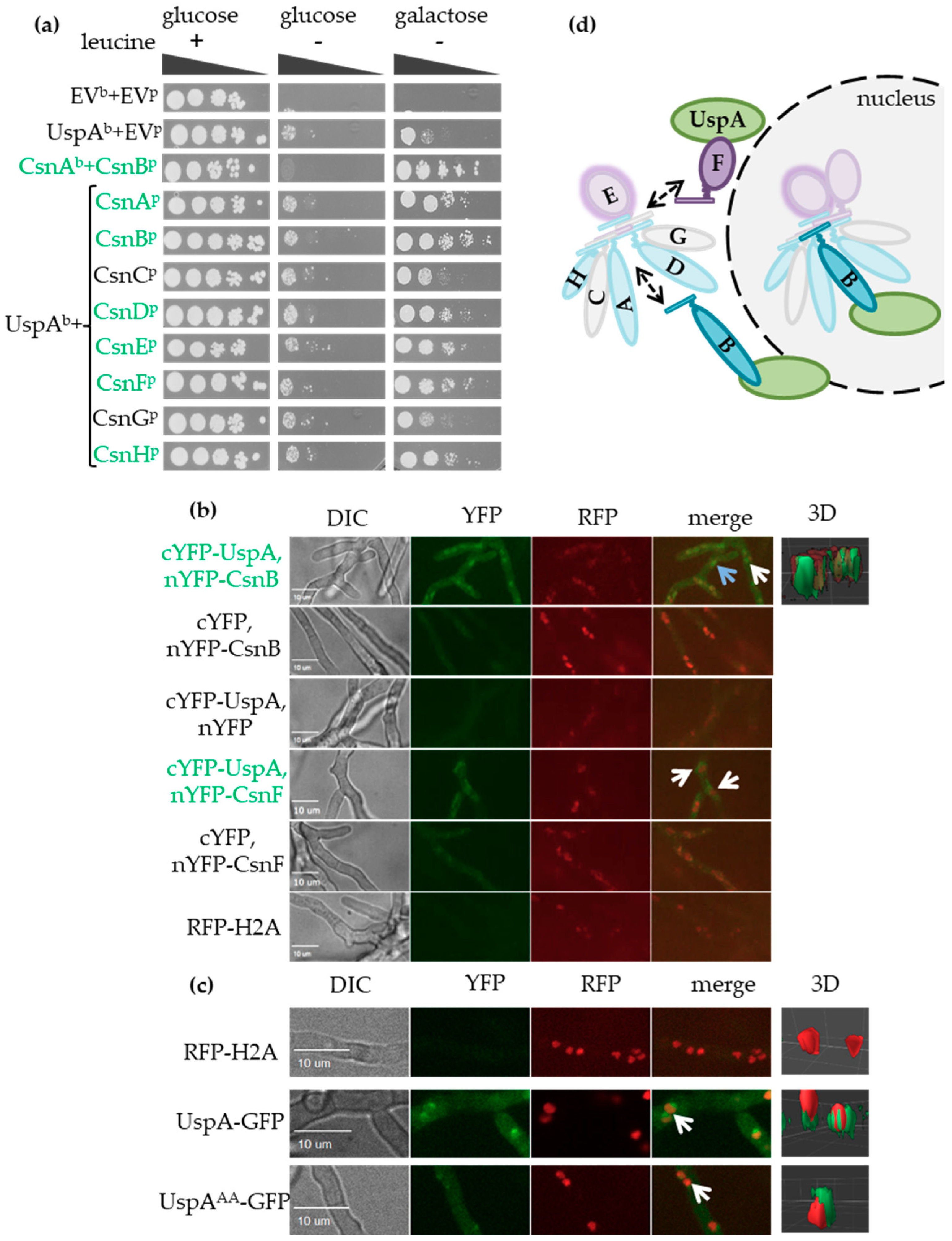
3.3. UspA-GFP Interacts with CSN Subunits In Vivo and In Vitro and is Mainly Localized in the Periphery of Nuclei
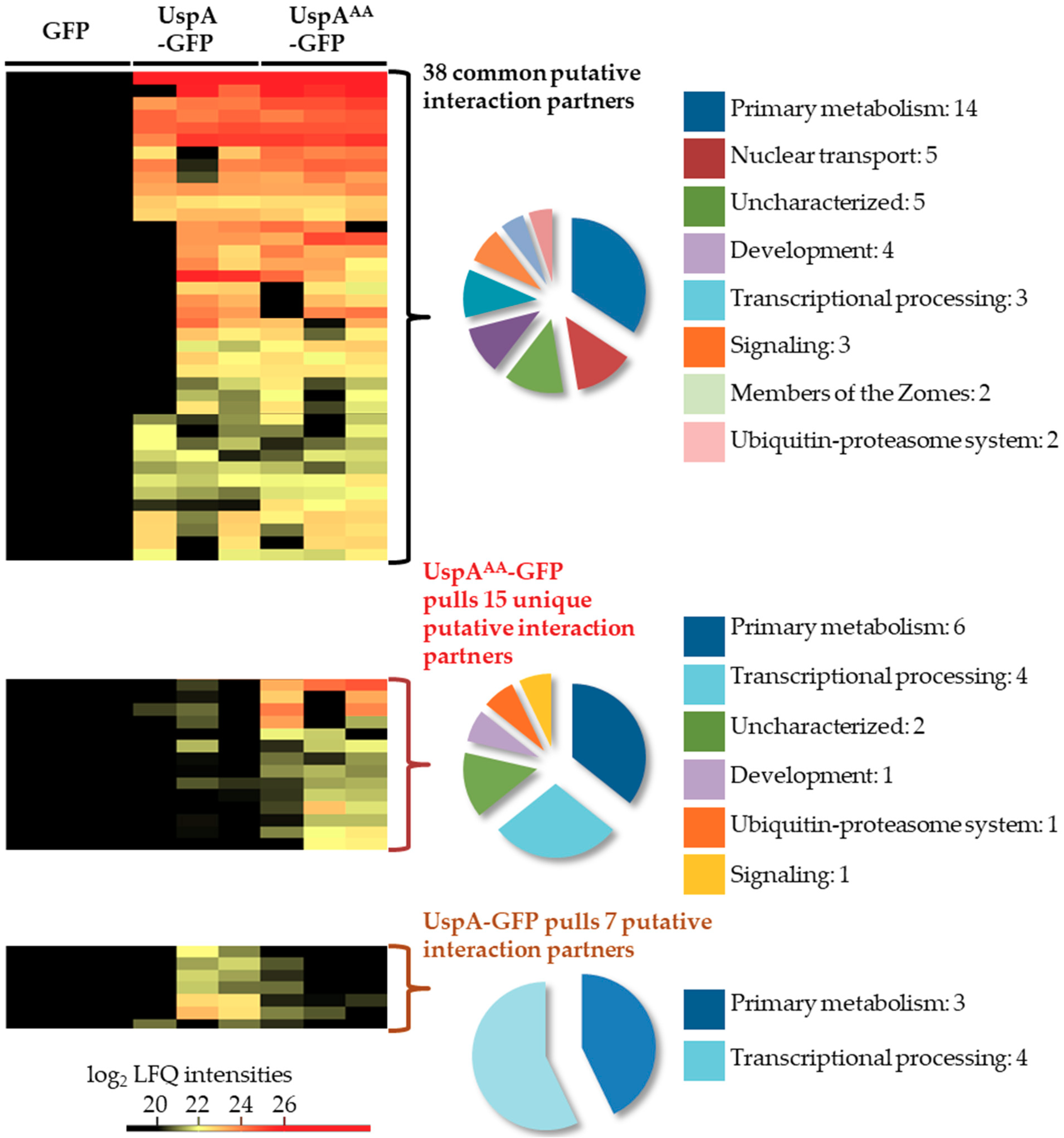
3.4. UspA-GFP Pulls Proteins Related to Nuclear Transport, Transcriptional Processing, and Ubiquitin-Proteasome System
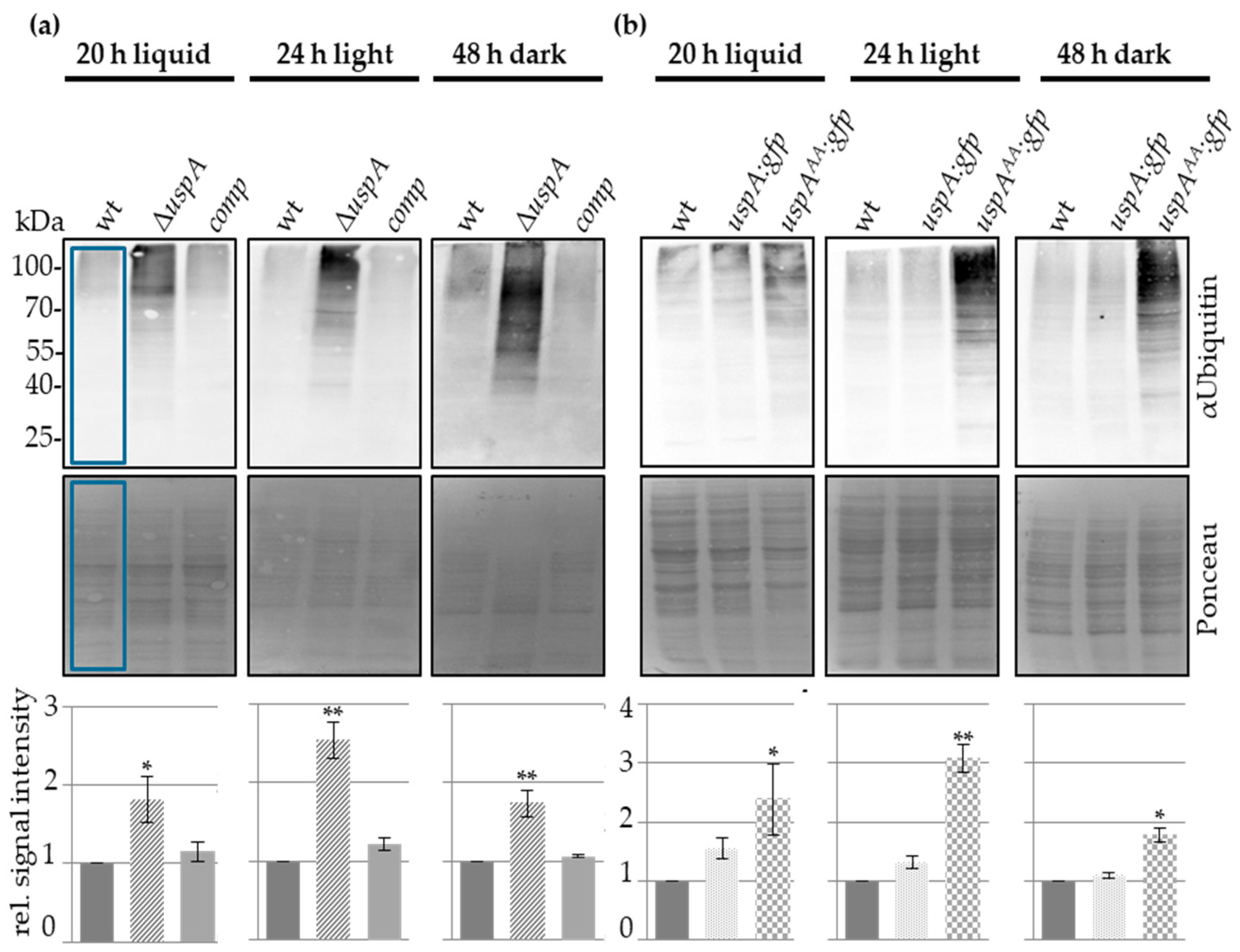
3.5. UspA Reduces the Cellular Pool of Ubiquitinated Proteins in A. nidulans
3.6. UspA Accelerates the Formation of Asexual Conidiospores and Sexual Fruiting Bodies in A. nidulans
3.7. UspA Represses the Expression of Derivative of Benzaldehyde (dba) Gene Cluster Members
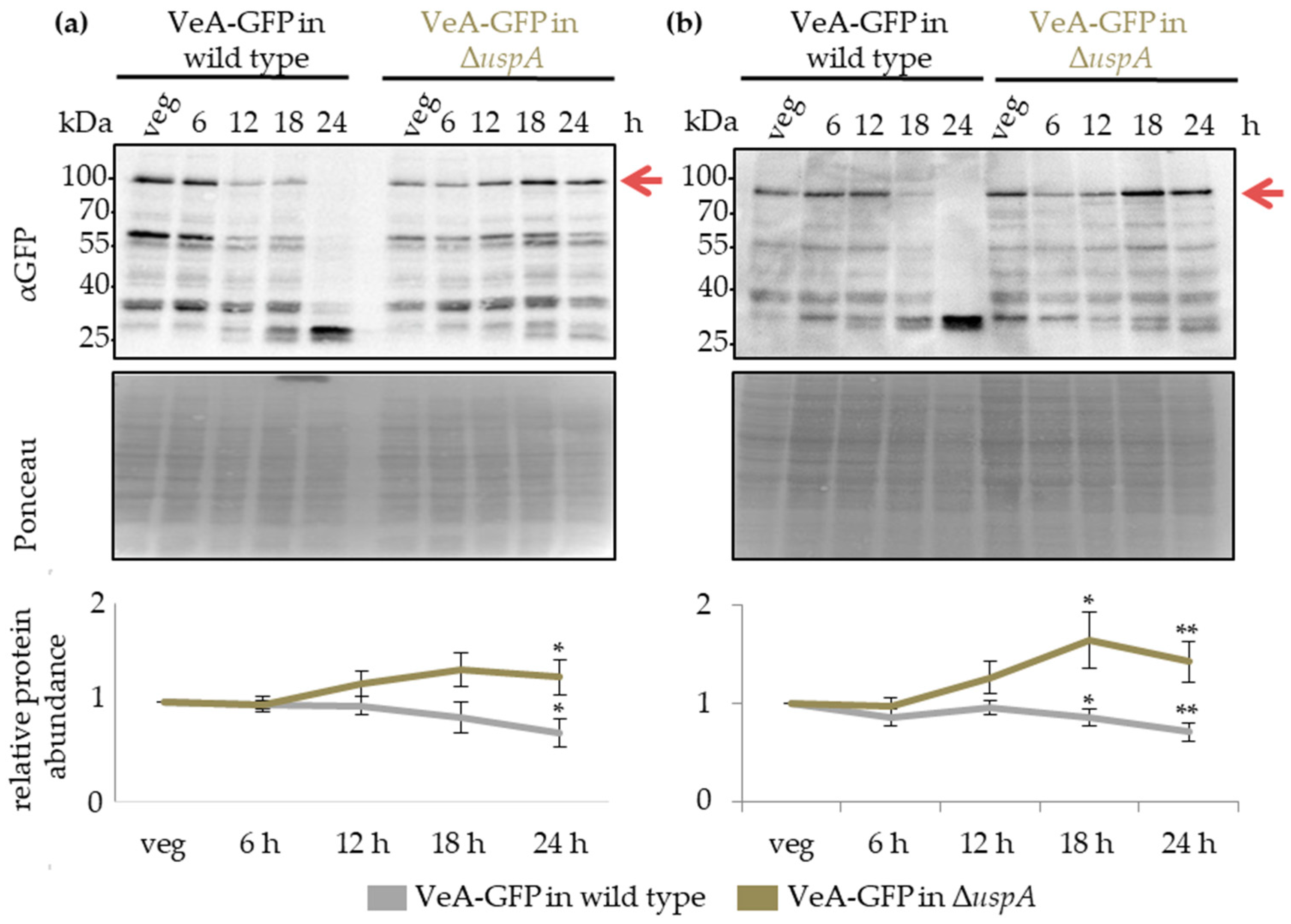
3.8. UspA Prevents Accumulation of the Velvet Domain Protein VeA During Late Asexual and Sexual Development
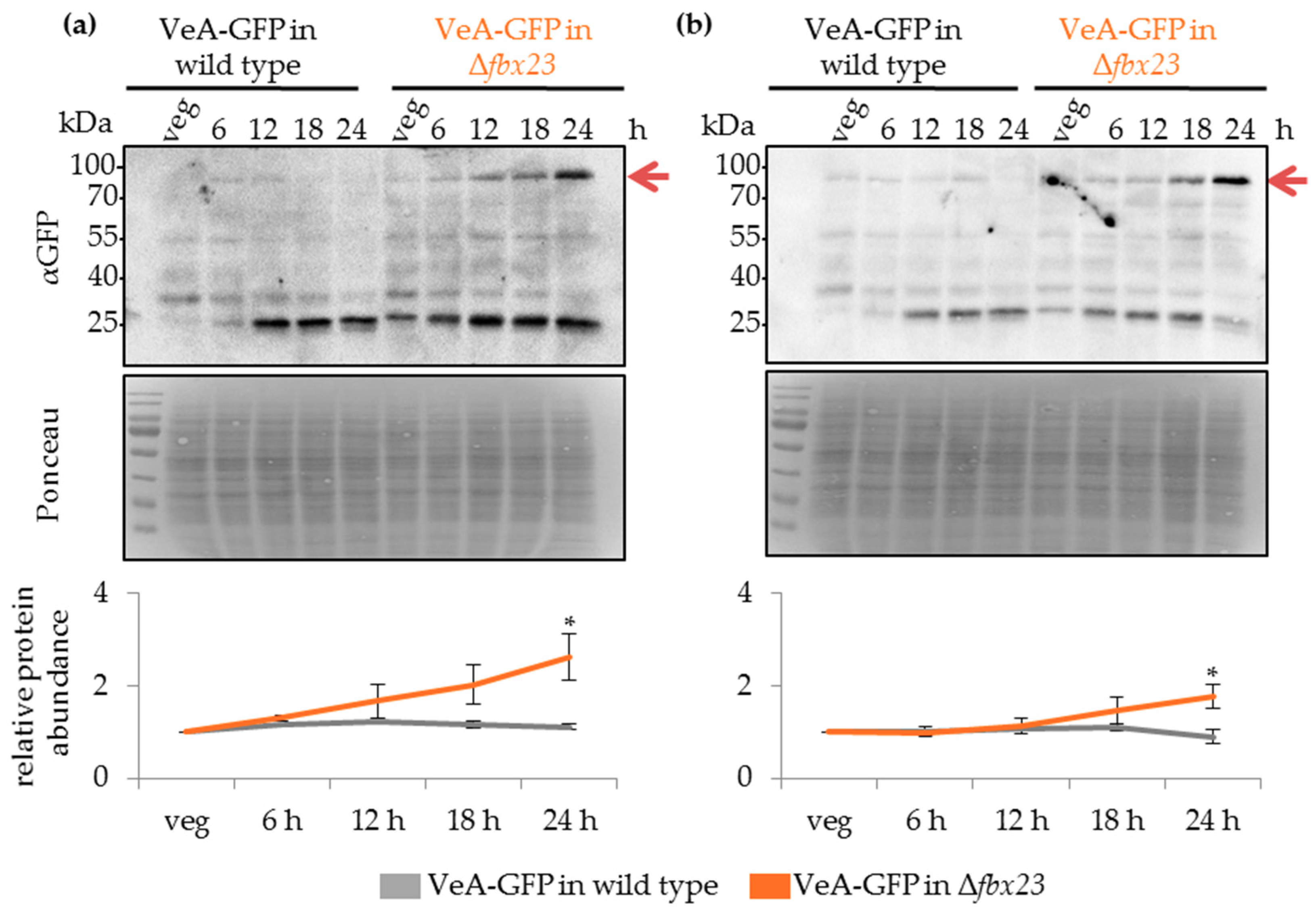
3.9. Fbx23 Prevents Accumulation of VeA-GFP in Late Developmental Time Points
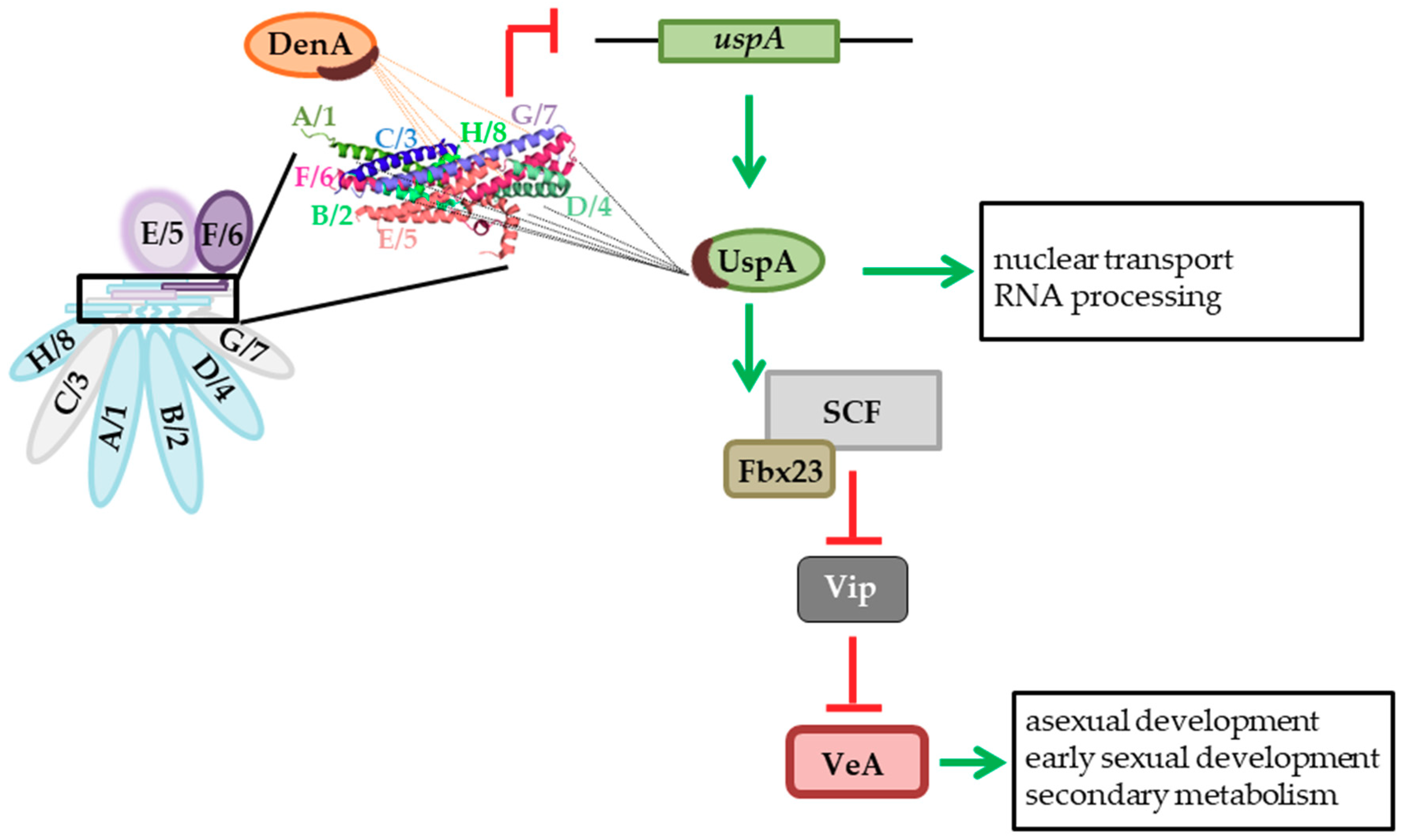
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, D.Y.; Kumar, S.; Hedges, S.B. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc Biol. Sci. 1999, 266, 163–171. [Google Scholar] [CrossRef]
- Santos, A.L.; Lindner, A.B. Protein posttranslational modifications: Roles in aging and age-related disease. Oxid. Med. Cell. Longev. 2017, 2017. [Google Scholar] [CrossRef]
- Karve, T.M.; Cheema, A.K. Small changes huge impact: The role of protein posttranslational modifications in cellular homeostasis and disease. J. Amin. Acids. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 1998, 67, 425–479. [Google Scholar] [CrossRef] [PubMed]
- Hutchins, A.P.; Liu, S.; Diez, D.; Miranda-Saavedra, D. The repertoires of ubiquitinating and deubiquitinating enzymes in eukaryotic genomes. Mol. Biol. Evol. 2013, 30, 1172–1187. [Google Scholar] [CrossRef]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.S.; Deng, X.W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef] [PubMed]
- Petroski, M.D.; Deshaies, R.J. Function and regulation of cullin-RING ubiquitin ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef]
- Bosu, D.R.; Kipreos, E.T. Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div. 2008, 3. [Google Scholar] [CrossRef]
- Spasser, L.; Brik, A. Chemistry and biology of the ubiquitin signal. Angew. Chemie. 2012, 51, 6840–6862. [Google Scholar] [CrossRef]
- Saeki, Y. Recent Topics in Ubiquitin-Proteasome System and Autophagy Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Thrower, J.S.; Hoffman, L.; Rechsteiner, M.; Pickart, C.M. Recognition of the polyubiquitin proteolytic signal. EMBO J. 2000, 19, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.; Nikosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol. Cell. 2016, 63, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Kwasna, D.; Rehman, S.A.A.; Natarajan, J.; Matthews, S.; Madden, R.; De Cesare, V.; Weidlich, S.; Virdee, S.; Ahel, I.; Gibbs-Seymour, I.; et al. Distinct Deubiquitinase Class Important for Genome Article Discovery and Characterization of ZUFSP / ZUP1, a Distinct Deubiquitinase Class Important for Genome Stability. Mol. Cell. 2018, 70, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Clague, M.J.; Barsukov, I.; Coulson, J.M.; Liu, H.; Rigden, D.J.; Urbé, S. Deubiquitylases from genes to organism. Physiol Rev. 2013, 93, 1289–1315. [Google Scholar] [CrossRef]
- Tsou, W.; Sheedlo, M.J.; Morrow, M.E.; Blount, J.R.; McGregor, K.M.; Das, C.; Todi, S.V. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One. 2012, 7. [Google Scholar] [CrossRef]
- Lockington, R.A.; Kelly, J.M. Carbon catabolite repression in Aspergillus nidulans. Eur. J. Biochem. 2001, 40, 1311–1321. [Google Scholar] [CrossRef]
- Lockington, R.A.; Kelly, J.M. The WD40-repeat protein CreC interacts with and stabilizes the deubiquitinating enzyme CreB in vivo in Aspergillus nidulans. Mol. Microbiol. 2002, 43, 1173–1182. [Google Scholar] [CrossRef]
- Hetfeld, B.K.J.; Helfrich, A.; Kapelari, B.; Scheel, H.; Hofmann, K.; Guterman, A.; Glickman, M.; Schade, R.; Kloetzel, P.M.; Dubiel, W. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr. Biol. 2005, 15, 1217–1221. [Google Scholar] [CrossRef]
- Zhou, C.; Wee, S.; Rhee, E.; Naumann, M.; Dubiel, W.; Wolf, D.A. Fission yeast COP signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol. Cell. 2003, 11, 927–938. [Google Scholar] [CrossRef]
- Braus, G.H.; Irniger, S.; Bayram, Ö. Fungal development and the COP9 signalosome. Curr. Opin. Microbiol. 2010, 13, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Lingaraju, G.M.; Bunker, R.D.; Cavadini, S.; Hess, D.; Hassiepen, U.; Renatus, M.; Fischer, E.S.; Thomä, N.H. Crystal structure of the human COP9 signalosome. Nature. 2014, 512, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.W.; Dubiel, W.; Wei, N.; Hofmann, K.; Mundt, K.; Colicelli, J.; Kato, J.; Naumann, M.; Segal, D.; Seeger, M.; et al. Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet. 2000, 16, 202–203. [Google Scholar] [CrossRef]
- Busch, S.; Schwier, E.U.; Nahlik, K.; Bayram, Ö.; Helmstaedt, K.; Draht, O.W.; Krappmann, S.; Valerius, O.; Lipscomb, W.N.; Braus, G.H. An eight-subunit COP9 signalosome with an intact JAMM motif is required for fungal fruit body formation. Proc. Natl. Acad. Sci. USA. 2007, 104, 8089–8094. [Google Scholar] [CrossRef] [PubMed]
- Lyapina, S.; Cope, G.; Shevchenko, A.; Serino, G.; Tsuge, T.; Zhou, C.; Wolf, D.A.; Wei, N.; Shevchenko, A.; Deshaies, R.J. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001, 292, 1382–1385. [Google Scholar] [CrossRef]
- Beckmann, E.A.; Köhler, A.M.; Meister, C.; Christmann, M.; Draht, O.W.; Rakebrandt, N.; Valerius, O.; Braus, G.H. Integration of the catalytic subunit activates deneddylase activity in vivo as final step in fungal COP9 signalosome assembly. Mol. Microbiol. 2015, 97, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural insights into NEDD8 activation of cullin-RING ligases: Conformational control of conjugation. Cell. 2008, 134, 995–1006. [Google Scholar] [CrossRef]
- Köhler, A.M.; Harting, R.; Langeneckert, A.E.; Valerius, O.; Gerke, J.; Meister, C.; Strohdiek, A.; Braus, G.H. Integration of Fungus-Specific CandA-C1 into a Trimeric CandA Complex Allowed Splitting of the Gene for the Conserved Receptor Exchange Factor of CullinA E3 Ubiquitin Ligases in Aspergilli. MBio. 2019. [Google Scholar] [CrossRef]
- Freilich, S.; Oron, E.; Kapp, Y.; Nevo-Caspi, Y.; Orgad, S.; Segal, D.; Chamovitz, D.A. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr. Biol. 1999, 9, PS1–S4. [Google Scholar] [CrossRef]
- Lykke-Andersen, K.; Schaefer, L.; Menon, S.; Deng, X.; Miller, J.B.; Wei, N. Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol. Cell. Biol. 2003, 23, 6790–6797. [Google Scholar] [CrossRef]
- Wei, N.; Deng, X.W. The COP9 signalosome. Annu. Rev. Cell Dev. Biol. 2003, 19, 261–286. [Google Scholar] [CrossRef] [PubMed]
- Busch, S.; Eckert, S.E.; Krappmann, S.; Braus, G.H. The COP9 signalosome is an essential regulator of development in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 2003, 49, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Busch, S.; Braus, G.H. How to build a fungal fruit body: from uniform cells to specialized tissue. Mol. Microbiol. 2007, 64, 873–876. [Google Scholar] [CrossRef] [PubMed]
- Nahlik, K.; Dumkow, M.; Bayram, Ö.; Helmstaedt, K.; Busch, S.; Valerius, O.; Gerke, J.; Hoppert, M.; Schwier, E.; Opitz, L.; et al. The COP9 signalosome mediates transcriptional and metabolic response to hormones, oxidative stress protection and cell wall rearrangement during fungal development. Mol. Microbiol. 2010, 78, 964–979. [Google Scholar] [CrossRef]
- Christmann, M.; Schmaler, T.; Gordon, C.; Huang, X.; Bayram, Ö.; Schinke, J.; Stumpf, S.K.; Dubiel, W.; Braus, G.H. Control of multicellular development by the physically interacting deneddylases DEN1/DenA and COP9 signalosome. PLoS Genet. 2013, 9, e1003275. [Google Scholar] [CrossRef] [PubMed]
- Mergner, J.; Heinzlmeir, S.; Kuster, B.; Schwechheimer, C. Deneddylase1 deconjugates NEDD8 from non-cullin protein substrates in Arabidopsis thaliana. Plant Cell. 2015, 27, 741–753. [Google Scholar] [CrossRef][Green Version]
- Bayram, Ö.; Braus, G.H. Coordination of secondary metabolism and development in fungi: The velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012, 36. [Google Scholar] [CrossRef]
- Ahmed, Y.L.; Gerke, J.; Park, H.S.; Bayram, Ö.; Neumann, P.; Ni, M.; Dickmanns, A.; Kim, S.C.; Yu, J.H.; Braus, G.H.; et al. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biol. 2013, 11, e1001849. [Google Scholar] [CrossRef]
- Oeckinghaus, A.; Ghosh, S. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb. Perspect. Biol. 2009, 1. [Google Scholar] [CrossRef]
- Gutierrez, H.; Davies, A.M. Regulation of neural process growth, elaboration and structural plasticity by NF-kB. Trends Neurosci. 2011, 34, 316–325. [Google Scholar] [CrossRef]
- Sarikaya-Bayram, Ö.; Bayram, Ö.; Feussner, K.; Kim, J.H.; Kim, H.S.; Kaever, A.; Feussner, I.; Chae, K.S.; Han, D.M.; Han, K.H.; et al. Membrane-bound methyltransferase complex VapA-VipC-VapB guides epigenetic control of fungal development. Dev. Cell. 2014, 29, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Bok, J.W.; Keller, N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004, 3, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Han, K.; Kim, K.; Han, D.; Jahng, K.; Chae, K. The veA gene activates sexual development in Aspergillus nidulans. Fungal Genet Biol. 2002, 37, 72–80. [Google Scholar] [CrossRef]
- Kato, N.; Brooks, W.; Calvo, A.M. The expression of sterigmatocystin and penicillin genes in Aspergillus nidulans is controlled by veA, a gene required for sexual development. Am. Soc. Microbiol. 2003, 2, 1178–1186. [Google Scholar] [CrossRef]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [PubMed]
- Käfer, E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977, 19, 33–131. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Bayram, Ö.S.; Ahmed, Y.L.; Maruyama, J.I.; Valerius, O.; Rizzoli, S.O.; Ficner, R.; Irniger, S.; Braus, G.H. The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet. 2012, 8. [Google Scholar] [CrossRef]
- Thieme, K.G.; Gerke, J.; Sasse, C.; Valerius, O.; Thieme, S.; Karimi, R.; Heinrich, A.; Finkernager, F.; Smith, K.; Bode, H.B.; et al. Velvet domain protein VosA represses the zinc cluster transcription factor SclB regulatory network for Aspergillus nidulans asexual development, oxidative stress response and secondary metabolism. PLOS Genet. 2018, 14, e1007511. [Google Scholar] [CrossRef]
- Inoue, H.; Nojima, H.; Okayama, H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990, 96, 23–28. [Google Scholar] [CrossRef]
- Grant, S.G.; Jessee, J.; Bloom, F.R.; Hanahan, D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Nat. Acad. Sci. USA 1990, 87, 4645–4649. [Google Scholar] [CrossRef]
- Punt, P.; van den Hondel, C. Transformation of filamentous fungi based on hygromycin B and phleomycin resistance markers. Methods Enzymol. 1992, 216, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Southern, E.M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 1975, 98, 503–517. [Google Scholar] [CrossRef]
- Ito, H.; Fukuda, Y.; Murata, K.; Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983, 153, 163–168. [Google Scholar] [PubMed]
- Gietz, D.; St, A.; Robin, J.; Schiestl, R.H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992, 20, 1992. [Google Scholar] [CrossRef] [PubMed]
- Manian, S.; Sreenivasaprasad, S.; Mills, P.R. DNA extraction method for PCR in mycorrhizal fungi. Lett. Appl. Microbiol. 2001, 33, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, G.C.; Arnaud, M.B.; Inglis, D.O.; Skrzypek, M.S.; Binkley, G.; Simison, M.; Miyasato, S.R.; Binkley, J.; Orvis, J.; Shah, P.; et al. The Aspergillus Genome Database: Multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations. Nucleic Acids Res. 2014, 42, 705–710. [Google Scholar] [CrossRef] [PubMed]
- Stajich, J.E.; Harris, T.; Brunk, B.P.; Brestelli, J.; Fischer, S.; Harb, O.S.; Kissinger, J.C.; Li, W.; Nayak, V.; Pinney, D.F.; et al. FungiDB: An integrated functional genomics database for fungi. Nucleic Acids Res. 2012, 40, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Derbyshire, M.K.; Gonzales, N.R.; Lu, S.; Chitsaz, F.; Geer, L.Y.; Geer, R.C.; He, J.; Gwadz, M.; Hurwitz, D.I.; et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015, 43, D222–D226. [Google Scholar] [CrossRef]
- Golemis, E.A.; Serebriiskii, I.; Law, S.F. The yeast two-hybrid system: criteria for detecting physiologically significant protein-protein interactions. Curr Issues Mol Biol. 1999, 1, 31–45. [Google Scholar] [CrossRef]
- Golemis, E.A.; Serebriiskii, I.; Finley, R.L.; Kolonin, M.G.; Gyuris, J.; Brent, R. Interaction trap/two-hybrid system to identify interacting proteins. Curr Protoc Cell Biol. 2001, 17, 20.1.1–20.1.4. [Google Scholar] [CrossRef]
- Shevchenko, A.; Tomas, H.; Havliš, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2007, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Wessel, D.; Flügge, U.I. A Method for the Quantitative Recovery of Protein in Dilute Solution in the Presence of Detergents and Lipids. Anal. Chem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Rappsilber, J.; Ishihama, Y.; Mann, M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003, 75, 663–670. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Hanpude, P.; Bhattacharya, S.; Dey, A.K.; Maiti, T.K. Deubiquitinating enzymes in cellular signaling and disease regulation enzymatic mechanism of DUBs. IUBMB Life. 2015, 67, 544–555. [Google Scholar] [CrossRef]
- Haahr, P.; Borgermann, N.; Guo, X.; Typas, D.; Achuthankutty, D.; Shearer, R.; Sixma, T.K.; Mailand, N. ZUFSP Deubiquitylates K63-Linked Polyubiquitin Chains to Promote Genome Stability. Mol. Cell. 2018, 70, 165–174. [Google Scholar] [CrossRef]
- Vlasschaert, C.; Xia, X.; Coulombe, J.; Gray, D.A. Evolution of the highly networked deubiquitinating enzymes USP4, USP15, and USP11. BMC Evol. Biol. 2015, 15. [Google Scholar] [CrossRef]
- Hotton, S.K.; Callis, J. Regulation of cullin RING ligases. Annu. Rev. Plant Biol. 2008, 59, 467–489. [Google Scholar] [CrossRef]
- Morimoto, M.; Nishida, T.; Honda, R.; Yasuda, H. Modification of cullin-1 by ubiquitin-like protein Nedd8 enhances the activity of SCF(skp2) toward p27(kip1). Biochem. Biophys. Res. Commun. 2000, 270, 1093–1096. [Google Scholar] [CrossRef] [PubMed]
- Schinke, J.; Gulko, M.K.; Christmann, M.; Valerius, O.; Stumpf, S.K.; Stirz, M.; Braus, G.H. The DenA/DEN1 interacting phosphatase DipA controls septa positioning and phosphorylation-dependent stability of cytoplasmatic DenA/DEN1 during fungal development. PLoS Genet. 2016, 12, e1005949. [Google Scholar] [CrossRef] [PubMed]
- Sharon, M.; Mao, H.; Boeri Erba, E.; Stephens, E.; Zheng, N.; Robinson, C.V. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009, 17, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Mundt, K.E.; Liu, C.; Carr, A.M. Deletion mutants in COP9 signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol. Biol. Cell. 2002, 13, 493–502. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tomoda, K.; Kubota, Y.; Arata, Y.; Mori, S.; Maeda, M.; Tanaka, T.; Yoshida, M.; Yoneda-Kato, N.; Kato, J.Y. The cytoplasmic shuttling and subsequent degradation of p27Kip1mediated by Jab1/CSN5 and the COP9 signalosome complex. J. Biol. Chem. 2002, 277, 2302–2310. [Google Scholar] [CrossRef] [PubMed]
- Staub, J.M.; Wei, N.; Deng, X. Evidence for FUSG as a Component of the Nuclear-Localized COP9 Complex in Arabidopsis. Plant Cell. 1996, 8, 2047–2056. [Google Scholar] [CrossRef] [PubMed]
- Maytal-Kivity, V.; Piran, R.; Pick, E.; Hofmann, K.; Glickman, M.H. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 2002, 3, 1215–1221. [Google Scholar] [CrossRef]
- Markina-Inarrairaegui, A.; Etxebeste, O.; Herrero-Garcia, E.; Araujo-Bazan, L.; Fernandez-Martinez, J.; Flores, J.A.; Osmani, S.A.; Espeso, E.A. Nuclear transporters in a multinucleated organism: Functional and localization analyses in Aspergillus nidulans. Mol. Biol. Cell. 2011, 22, 3874–3886. [Google Scholar] [CrossRef]
- Yang, Y.; El-Ganiny, A.M.; Bray, G.E.; Sanders, D.A.R.; Kaminskyj, S.G.W. Aspergillus nidulans hypB encodes a Sec7-domain protein important for hyphal morphogenesis. Fungal Genet. Biol. 2008, 45, 749–759. [Google Scholar] [CrossRef]
- Gerke, J.; Bayram, Ö.; Feussner, K.; Landesfeind, M.; Shelest, E.; Feussner, I.; Braus, G.H. Breaking the silence: Protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl. Environ. Microbiol. 2012, 78, 8234–8244. [Google Scholar] [CrossRef]
- Sarikaya-Bayram, Ö.; Palmer, J.M.; Keller, N.; Braus, G.H.; Bayram, Ö. One Juliet and four Romeos: VeA and its methyltransferases. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Guardavaccaro, D.; Kudo, Y.; Boulaire, J.; Barchi, M.; Busino, L.; Donzelli, M.; Margottin-Goguet, F.; Jackson, P.K.; Yamasaki, L.; Pagano, M. Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell. 2003, 4, 799–812. [Google Scholar] [CrossRef]
- von Zeska Kress, M.R.; Harting, R.; Bayram, Ö.; Christmann, M.; Irmer, H.; Valerius, O.; Schinke, J.; Goldman, G.H.; Braus, G.H. The COP9 signalosome counteracts the accumulation of cullin SCF ubiquitin E3 RING ligases during fungal development. Mol. Microbiol. 2012, 83, 1162–1177. [Google Scholar] [CrossRef] [PubMed]
- de Assis, L.J.; Ulas, M.; Ries, L.N.A.; El Ramli, N.A.M.; Sarikaya-Bayram, Ö.; Braus, G.H.; Bayram, Ö.; Goldman, G.H. Regulation of Aspergillus nidulans CreA-mediated catabolite repression by the F-Box proteins Fbx23 and Fbx47. MBio. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Uhle, S.; Medalia, O.; Waldron, R.; Dumdey, R.; Henklein, P.; Bech-otschir, D.; Huang, X.; Berse, M.; Sperling, J.; Schade, R.; et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 2003, 22, 1302–1312. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wilson, M.P.; Majerus, P.W. Inositol 1,3,4-triphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J. Biol. Chem. 2002, 277, 45759–45764. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Feussner, K.; Dumkow, M.; Herrfurth, C.; Feussner, I.; Braus, G.H. Changes of global gene expression and secondary metabolite accumulation during light-dependent Aspergillus nidulans development. Fungal Genet. Biol. 2016, 87, 30–53. [Google Scholar] [CrossRef] [PubMed]
- Bayram, Ö.; Krappmann, S.; Ni, M.; Bok, J.W.; Helmstaedt, K.; Valerius, O.; Braus-Stromeyer, S.; Kwon, N.J.; Keller, N.P.; Yu, J.H.; et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008, 320, 1504–1506. [Google Scholar] [CrossRef]
- Stinnett, S.M.; Espeso, E.A.; Cobeño, L.; Araújo-bazán, L.; Calvo, A.M. Aspergillus nidulans VeA subcellular localization is dependent on the importin a carrier and on light. Mol. Microbiol. 2007, 63, 242–255. [Google Scholar] [CrossRef]
- Schweitzer, K.; Bozko, P.M.; Dubiel, W.; Naumann, M. CSN controls NF-kB by deubiquitinylation of IkBα. EMBO J. 2007, 26, 1532–1541. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-kB activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef] [PubMed]



 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meister, C.; Thieme, K.G.; Thieme, S.; Köhler, A.M.; Schmitt, K.; Valerius, O.; Braus, G.H. COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development. Biomolecules 2019, 9, 238. https://doi.org/10.3390/biom9060238
Meister C, Thieme KG, Thieme S, Köhler AM, Schmitt K, Valerius O, Braus GH. COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development. Biomolecules. 2019; 9(6):238. https://doi.org/10.3390/biom9060238
Chicago/Turabian StyleMeister, Cindy, Karl G. Thieme, Sabine Thieme, Anna M. Köhler, Kerstin Schmitt, Oliver Valerius, and Gerhard H. Braus. 2019. "COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development" Biomolecules 9, no. 6: 238. https://doi.org/10.3390/biom9060238
APA StyleMeister, C., Thieme, K. G., Thieme, S., Köhler, A. M., Schmitt, K., Valerius, O., & Braus, G. H. (2019). COP9 Signalosome Interaction with UspA/Usp15 Deubiquitinase Controls VeA-Mediated Fungal Multicellular Development. Biomolecules, 9(6), 238. https://doi.org/10.3390/biom9060238





