Combinatorial Use of Chitosan Nanoparticles, Reversine, and Ionising Radiation on Breast Cancer Cells Associated with Mitosis Deregulation
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Nanoparticle Synthesis
2.3. Nanoparticle Characterisation
2.4. Nanoparticles’ Intracellular Localisation
2.5. Cell Culture
2.6. Cell Irradiation
2.7. Cell Viability Assay
2.8. Comet Assay
2.9. Mitotic Analysis and Western Blot
2.10. Statistical Analyses
3. Results
3.1. Physicochemical Characterisation of Chitosan Nanoparticles
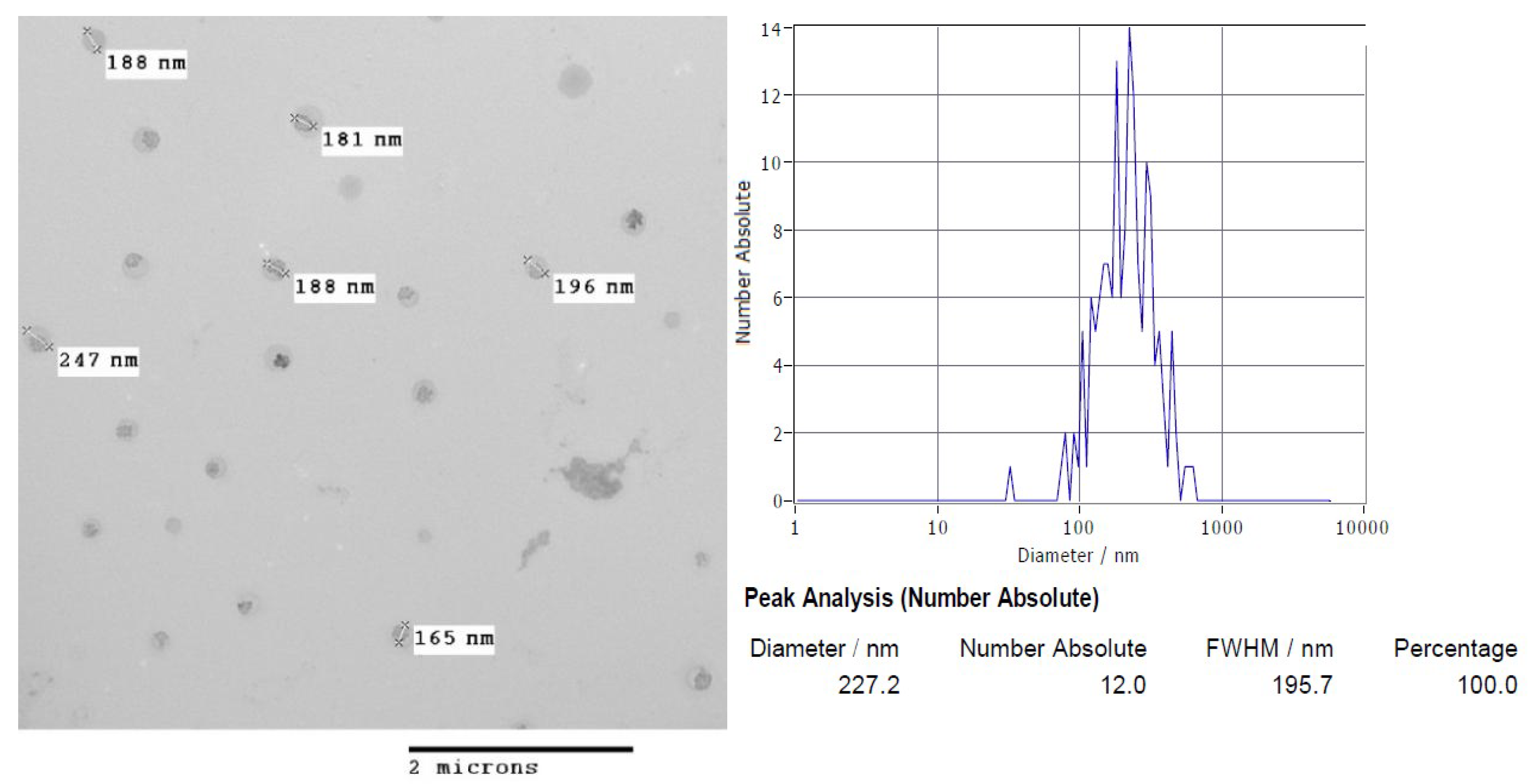
3.2. Nanoparticles Morphology
3.3. Nanoparticles Cellular Localisation
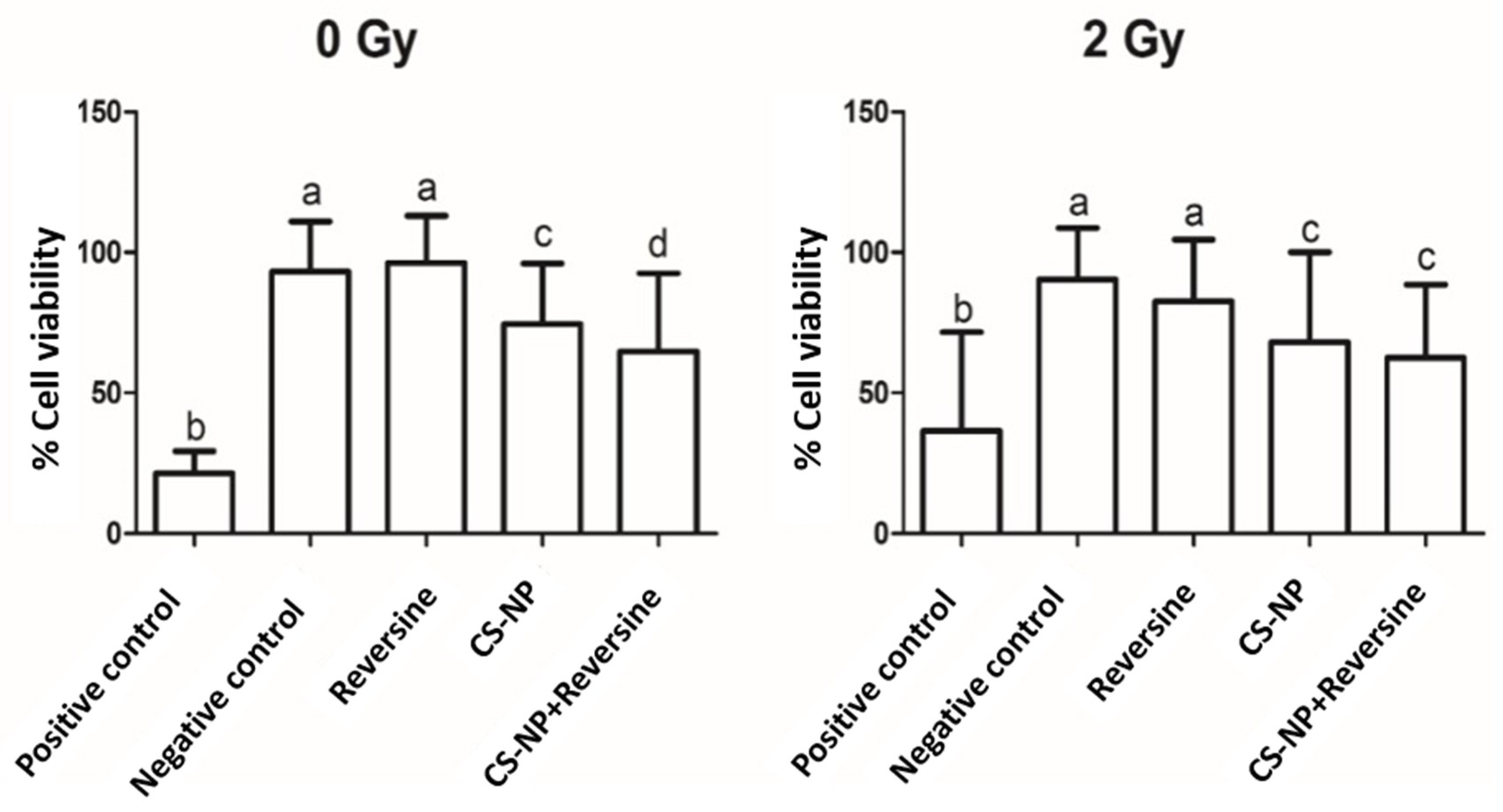
3.4. Cell Viability Assay
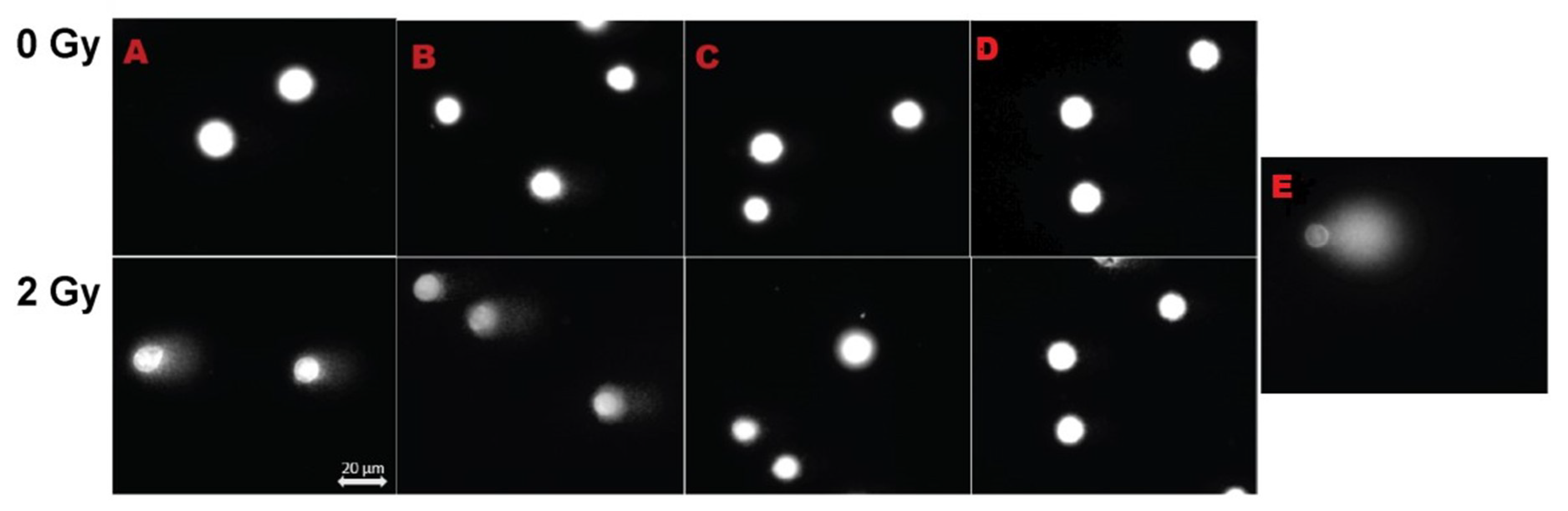
3.5. DNA Damage by Comet Assay
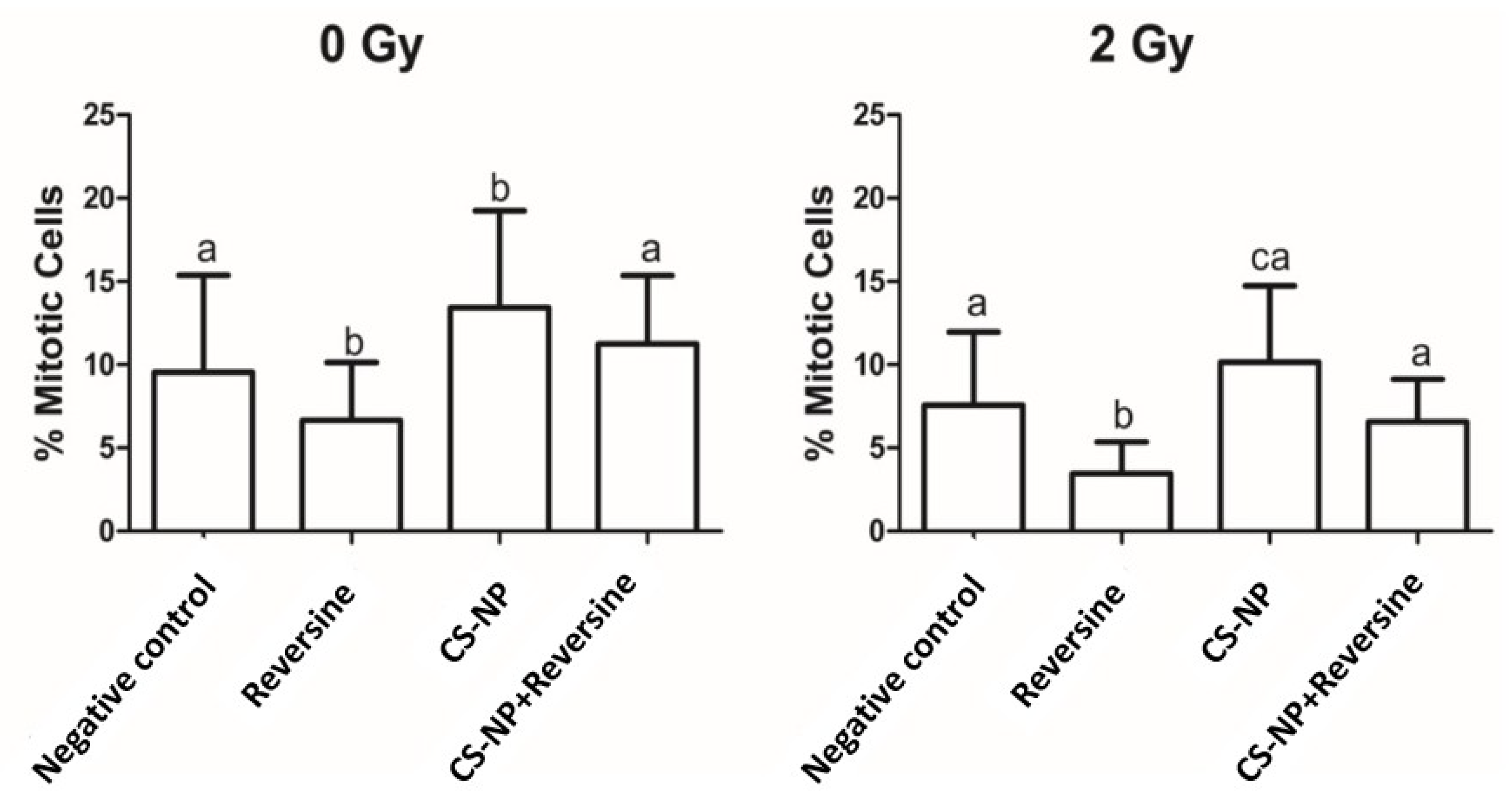
3.6. Mitotic Index
3.7. Mps1 Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Dou, Z.; Prifti, D.K.; Gui, P.; Liu, X.; Elowe, S.; Yao, X. Recent progress on the localization of the spindle assembly checkpoint machinery to kinetochores. Cells 2019, 23, 278. [Google Scholar] [CrossRef]
- Wang, H.; Mu, X.; He, H.; Zhang, X.D. Cancer Radiosensitizers. Trends Pharmacol. Sci. 2018, 39, 24–48. [Google Scholar] [CrossRef] [PubMed]
- Rancoule, C.; Magné, N.; Vallard, A.; Guy, J.B.; Rodriguez-Lafrasse, C.; Deutsch, E.; Chargari, C. Nanoparticles in radiation oncology: From bench-side to bedside. Cancer Lett. 2016, 375, 256–262. [Google Scholar] [CrossRef]
- Bergs, J.W.; Wacker, M.G.; Hehlgans, S.; Piiper, A.; Multhoff, G.; Rödelm, C.; Rödel, F. The role of recent nanotechnology in enhancing the efficacy of radiation therapy. Biochim. Biophys. Acta. 2015, 1856, 130–143. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, S.; Deng, F.; Chen, X.; Wang, X.; Wang, Y.; Zhang, H.; Dai, W.; He, B.; Zhang, Q.; et al. The function and mechanism of preactivated thiomers in triggering epithelial tight junctions opening. Eur. J. Pharm. Biopharm. 2018, 133, 188–199. [Google Scholar] [CrossRef]
- Townley, H.E.; Kim, J.; Dobson, P.J. In vivo demonstration of enhanced radiotherapy using rare earth doped titania nanoparticles. Nanoscale 2012, 4, 5043–5050. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, A.; Bonengel, S.; Laffleur, F.; Ijaz, M.; Leonaviciute, G.; Bernkop-Schnürch, A. An in-vitro exploration of permeation enhancement by novel polysulfonate thiomers. Int. J. Pharm. 2015, 496, 304–313. [Google Scholar] [CrossRef]
- Garaiova, Z.; Strand, S.P.; Reitan, N.K.; Lélu, S.; Størset, S.T.; Berg, K.; Malmo, J.; Folasire, O.; Bjørkøy, A.; Davies, C.D.L. Cellular uptake of DNA-chitosan nanoparticles: The role of clathrin- and caveolae-mediated pathways. Int. J. Biol. Macromol. 2012, 51, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Subhapradha, N.; Shanmugam, V.; Shanmugam, A. Chitosan nanoparticles from marine squid protect liver cells against N-diethylnitrosoamine-induced hepatocellular carcinoma. Carbohydr. Polym. 2017, 171, 18–26. [Google Scholar] [CrossRef]
- Gibot, L.; Chabaud, S.; Bouhout, S.; Bolduc, S.; Auger, F.A.; Moulin, V.J. Anticancer properties of chitosan on human melanoma are cell line dependent. Int. J. Biol. Macromol. 2015, 72, 370–379. [Google Scholar] [CrossRef]
- Naskar, S.; Sharma, S.; Koutsu, K. Chitosan-based nanoparticles: An overview of biomedical applications and its preparation. J. Drug Deliv. Sci. Technol. 2018, 49, 66–81. [Google Scholar] [CrossRef]
- Bai, X.; Liu, F.; Liu, Y.; Li, C.; Wang, S.; Zhou, H.; Wang, W.; Zhu, H.; Winkler, D.A.; Yan, B. Toward a systematic exploration of nano-bio interactions. Toxicol. Appl. Pharmacol. 2017, 323, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.A.; Syeda, J.T.M.; Wasan, K.M.; Wasan, E.K. An overview of chitosan nanoparticles and its application in non-parenteral drug delivery. Pharmaceutics 2017, 9, 53. [Google Scholar] [CrossRef] [PubMed]
- Alonso, M.J.; Calvo, P.; Remun, C. Novel hydrophilic chitosan—Polyethylene oxide nanoparticles as protein carriers. J. Appl. Polym. Sci. 1998, 63, 125–132. [Google Scholar] [CrossRef]
- Shin, S.; Song, I.; Um, S. Role of Physicochemical Properties in Nanoparticle Toxicity. Nanomaterials 2015, 5, 1351–1365. [Google Scholar] [CrossRef]
- Kiilll, C.P.; Barud, H.D.S.; Santagneli, S.H.; Ribeiro, S.J.L.; Silva, A.M.; Tercjak, A.; Gutierrez, J.; Pironi, A.M.; Gremião, M.P.D. Synthesis and factorial design applied to a novel chitosan/sodium polyphosphate nanoparticles via ionotropic gelation as an RGD delivery system. Carbohydr. Polym. 2017, 157, 1695–1702. [Google Scholar] [CrossRef]
- Zorzi, G.K.; Fecker, T.; El Gueddari, N.E.; Moerschbacher, B.M.; Goycoolea, F.M. A rational approach towards the design of chitosan-based nanoparticles obtained by ionotropic gelation. Colloids Surf. B Biointerfaces 2015, 135, 99–108. [Google Scholar]
- Hejjaji, E.M.A.; Smith, A.M.; Morris, G.A. Evaluation of the mucoadhesive properties of chitosan nanoparticles prepared using different chitosan to tripolyphosphate (CS:TPP) ratios. Int. J. Biol. Macromol. 2018, 120, 1610–1617. [Google Scholar] [CrossRef]
- Musumeci, T.; Pellitteri, R.; Spatuzza, M.; Puglisi, G. Nose-to-brain delivery: Evaluation of polymeric nanoparticles on olfactory ensheathing cells uptake. J. Pharm. Sci. 2014, 103, 628–635. [Google Scholar] [CrossRef]
- Fazil, M.; Md, S.; Haque, S.; Kumar, M.; Baboota, S.; Sahni, J.K.; Ali, J. Development and evaluation of rivastigmine loaded chitosan nanoparticles for brain targeting. Eur. J. Pharm. Sci. 2012, 47, 6–15. [Google Scholar] [CrossRef]
- D’Alise, A.M.; Amabile, G.; Iovino, M.; Di Giorgio, F.P.; Bartiromo, M.; Sessa, F.; Villa, F.; Musacchio, A.; Cortese, R. Reversine, a novel Aurora kinases inhibitor, inhibits colony formation of human acute myeloid leukemia cells. Mol. Cancer Ther. 2008, 7, 1140–1149. [Google Scholar] [CrossRef]
- Santaguida, S.; Tighe, A.; D’Alise, A.M.; Taylor, S.S.; Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010, 190, 73–87. [Google Scholar] [CrossRef]
- Hill, M.A.; Thompson, J.; Kavanagh, A.; Tullis, I.D.C.; Newman, R.G.; Prentice, J.; Beech, J.; Gilchrist, S.; Smart, S.; Fokas, E.; et al. The Development of technology for effective respiratory-gated irradiation using an image-guided small animal irradiator. Radiat. Res. 2017, 188, RR14753.1. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, D.W.; Olive, P.L.; O’Neill, K.L. The comet assay: A comprehensive review. Mutat. Res. Genet. Toxicol. 1995, 339, 37–59. [Google Scholar] [CrossRef]
- Singh, N.P.; Stephens, R.E. Microgel electrophoresis: Sensitivity, mechanisms, and DNA electrostretching. Mutat. Res. DNA Repair 1997, 383, 167–175. [Google Scholar] [CrossRef]
- Lim, H.K.; Gurung, R.L.; Hande, M.P. DNA-dependent protein kinase modulates the anti-cancer properties of silver nanoparticles in human cancer cells. Mutat. Res. 2017, 824, 32–41. [Google Scholar] [CrossRef]
- Carriere, M.; Sauvaigo, S.; Douki, T.; Ravanat, J.L. Impact of nanoparticles on DNA repair processes: Current knowledge and working hypotheses. Mutagenesis 2017, 32, 203–213. [Google Scholar] [CrossRef]
- Cui, Y.; Melby, E.S.; Mensch, A.C.; Laudadio, E.D.; Hung, M.N.; Dohnalkova, A.; Hu, D.; Hamers, R.J.; Orr, G. Quantitative mapping of oxidative stress response to lithium cobalt oxide nanoparticles in single cells using multiplexed in situ gene expression analysis. Nano. Lett. 2019, 19, 1990–1997. [Google Scholar] [CrossRef] [PubMed]
- Safar, R.; Doumandji, Z.; Saidou, T.; Ferrari, L.; Nahle, S.; Rihn, B.H.; Joubert, O. Cytotoxicity and global transcriptional responses induced by zinc oxide nanoparticles NM 110 in PMA-differentiated THP-1 cells. Toxicol. Lett. 2019, 308, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Decan, N.; Wu, D.; Williams, A.; Bernatchez, S.; Johnston, M.; Hill, M.; Halappanavar, S. Characterization of in vitro genotoxic, cytotoxic and transcriptomic responses following exposures to amorphous silica of different sizes. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2016, 796, 8–22. [Google Scholar] [CrossRef]
- Daniel, J.; Coulter, J.; Woo, J.H.; Wilsbach, K.; Gabrielson, E. High levels of the MPS1 checkpoint protein are protective of aneuploidy in breast cancer cells. Proc. Natl. Acad. Sci. USA. 2011, 108, 5384–5389. [Google Scholar] [CrossRef]
- Jeynes, J.C.; Jeynes, C.; Palitsin, V.; Townley, H.E. Direct quantification of rare earth doped titania nanoparticles in individual human cells. Nanotechnology 2016, 27, 285103. [Google Scholar] [CrossRef] [PubMed]
- Richardson, S.W.; Kolbe, H.J.; Duncan, R. Potential of low molecular mass chitosan as a DNA delivery system: Biocompatibility, body distribution and ability to complex and protect DNA. Int. J. Pharm. 1999, 178, 231–243. [Google Scholar] [CrossRef]
- Huang, H.C.; Shi, J.; Orth, J.D.; Mitchison, T.J. Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 2009, 16, 347–358. [Google Scholar] [CrossRef]
- Janssen, A.; Kops, G.J.; Medema, R.H. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc. Natl. Acad. Sci. USA 2009, 106, 19108–19113. [Google Scholar] [CrossRef]


| Nanoparticles | Physicochemical Properties | ||
|---|---|---|---|
| Z-Potential (z-Average) mV ± SD | Size (Hydrodynamic Ratio) nm ± SD | Concentration Particles/mL ± SD | |
| CS-NP | 29.6 ± 9.5 | 224 ± 31 | 5.9 × 1010 ± 2 × 1010 |
| CS-NP [R123] | 38 ± 0.16 | 227 ± 97 | 4.3 × 107 ± 2 × 1010 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piña Olmos, S.; Díaz Torres, R.; Elbakrawy, E.; Hughes, L.; Mckenna, J.; Hill, M.A.; Kadhim, M.; Ramírez Noguera, P.; Bolanos-Garcia, V.M. Combinatorial Use of Chitosan Nanoparticles, Reversine, and Ionising Radiation on Breast Cancer Cells Associated with Mitosis Deregulation. Biomolecules 2019, 9, 186. https://doi.org/10.3390/biom9050186
Piña Olmos S, Díaz Torres R, Elbakrawy E, Hughes L, Mckenna J, Hill MA, Kadhim M, Ramírez Noguera P, Bolanos-Garcia VM. Combinatorial Use of Chitosan Nanoparticles, Reversine, and Ionising Radiation on Breast Cancer Cells Associated with Mitosis Deregulation. Biomolecules. 2019; 9(5):186. https://doi.org/10.3390/biom9050186
Chicago/Turabian StylePiña Olmos, Sofia, Roberto Díaz Torres, Eman Elbakrawy, Louise Hughes, Joseph Mckenna, Mark A. Hill, Munira Kadhim, Patricia Ramírez Noguera, and Victor M. Bolanos-Garcia. 2019. "Combinatorial Use of Chitosan Nanoparticles, Reversine, and Ionising Radiation on Breast Cancer Cells Associated with Mitosis Deregulation" Biomolecules 9, no. 5: 186. https://doi.org/10.3390/biom9050186
APA StylePiña Olmos, S., Díaz Torres, R., Elbakrawy, E., Hughes, L., Mckenna, J., Hill, M. A., Kadhim, M., Ramírez Noguera, P., & Bolanos-Garcia, V. M. (2019). Combinatorial Use of Chitosan Nanoparticles, Reversine, and Ionising Radiation on Breast Cancer Cells Associated with Mitosis Deregulation. Biomolecules, 9(5), 186. https://doi.org/10.3390/biom9050186







