HIV Vaccine Mystery and Viral Shell Disorder
Abstract
1. Introduction
1.1. The Mystery of the Elusive HIV Vaccine
1.2. Protein Intrinsic Disorder
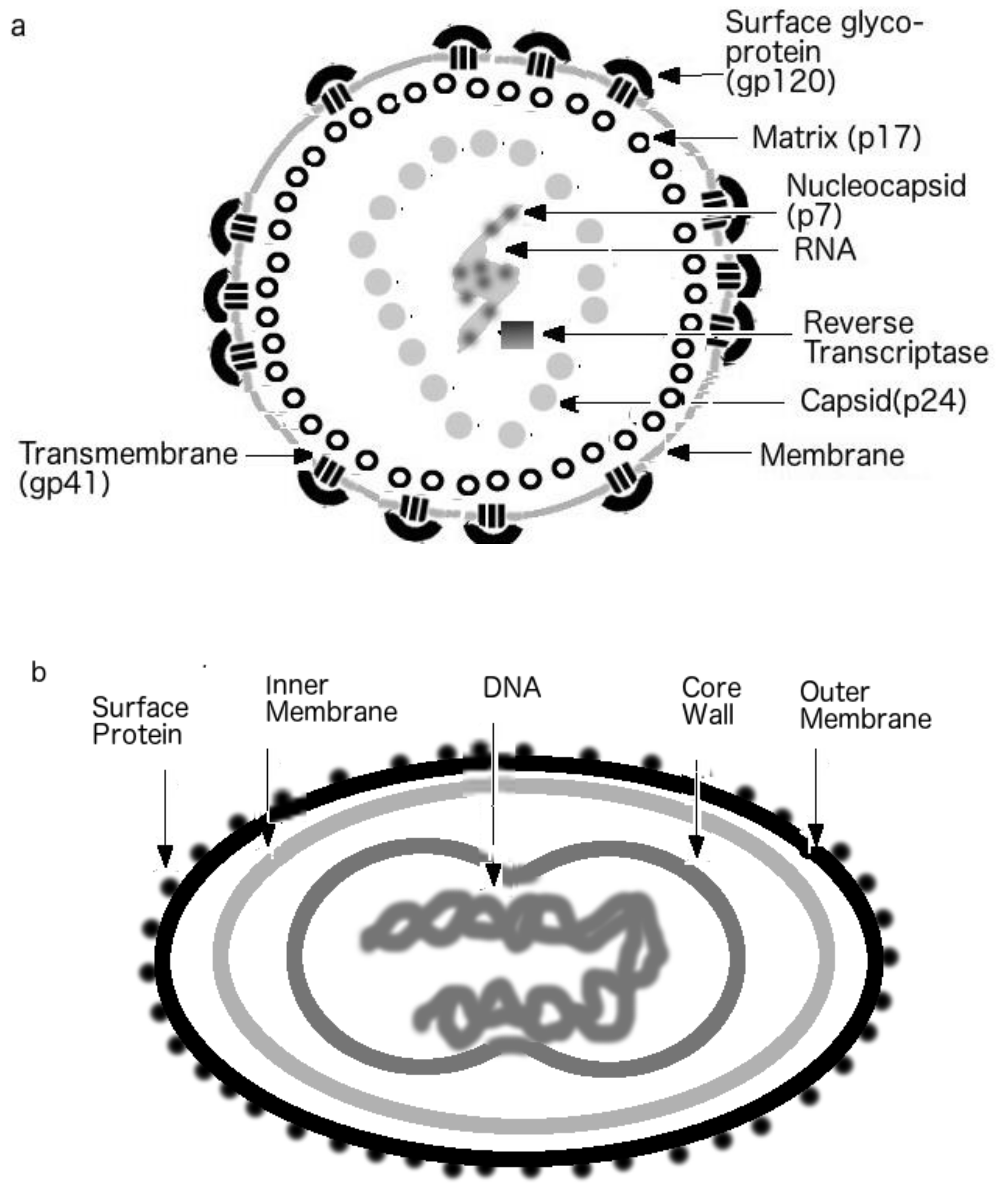
1.3. Viral Shells
2. Virus Selection: Classical Vaccine Successes vs. HIV, HSV-2 and HCV
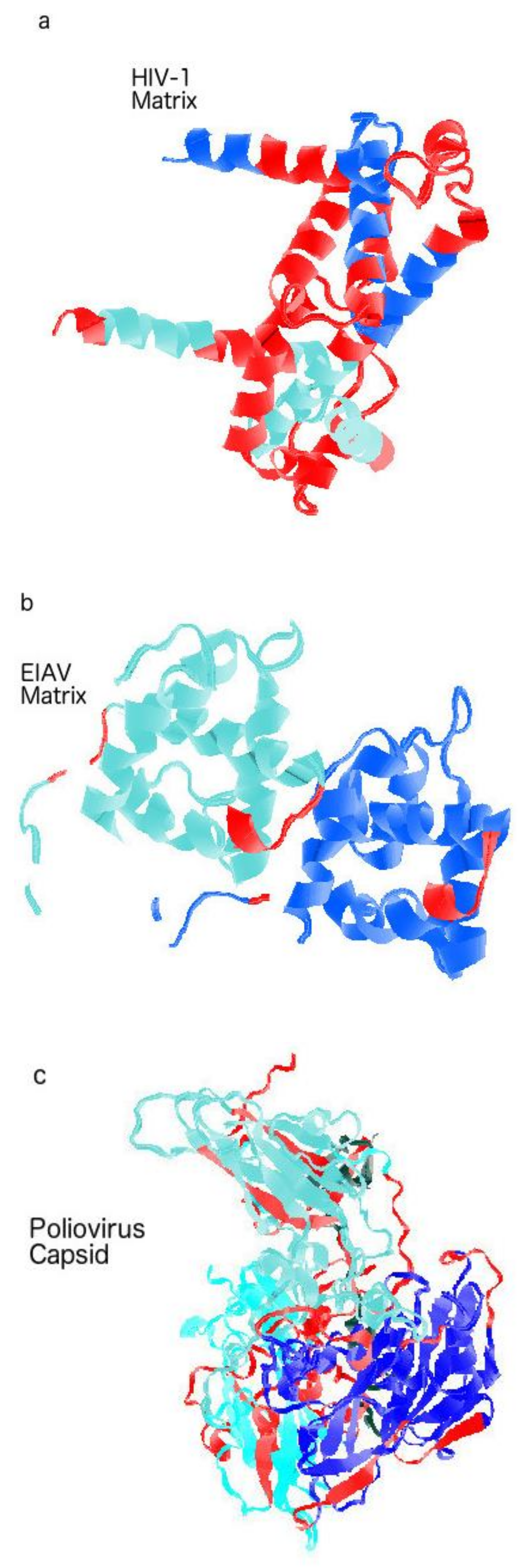
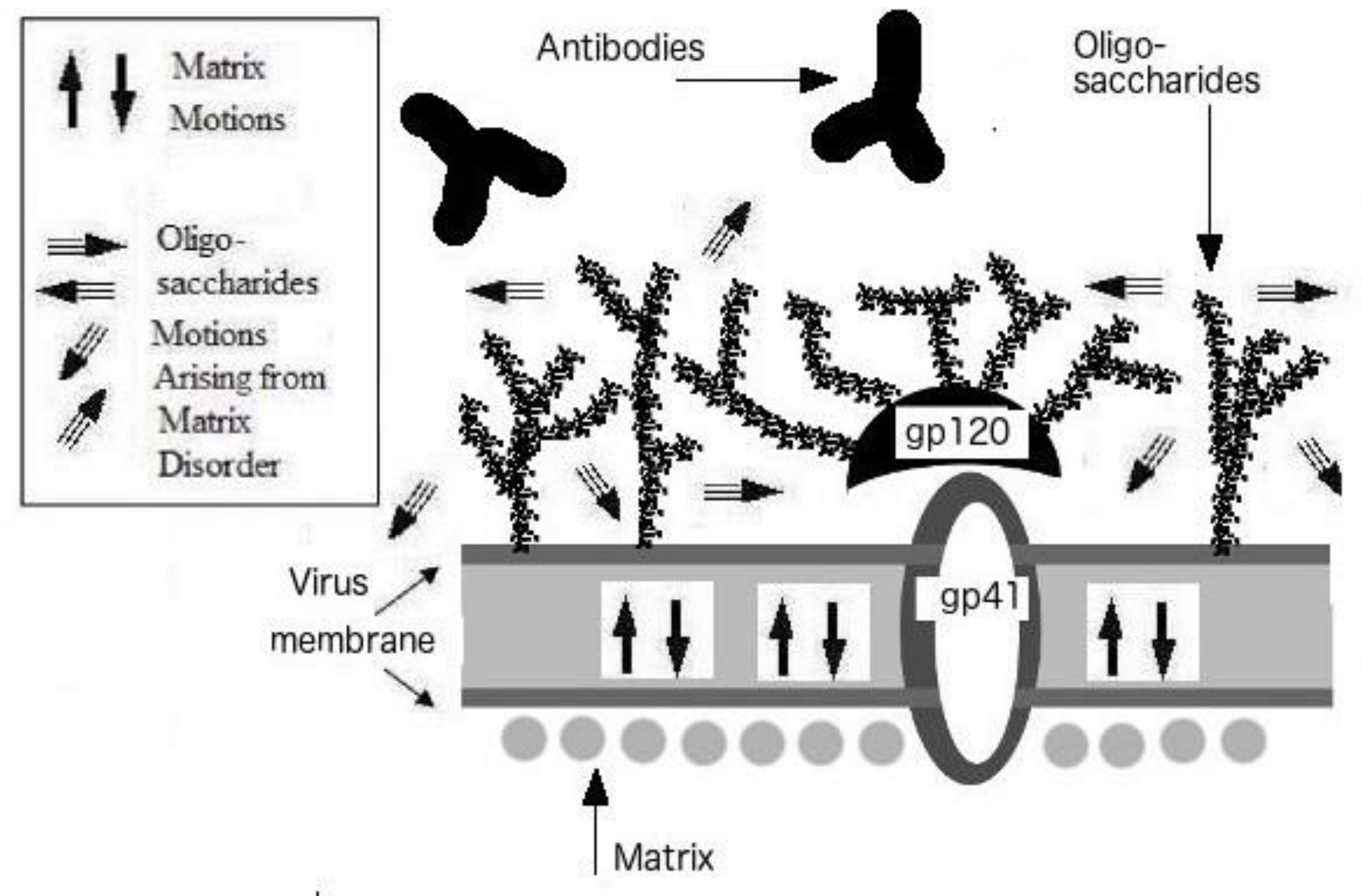
3. Protein Intrinsic Disorder of Viral Shells: Abnormality of the HIV Outer Shell
3.1. The HIV vs. EIAV Vaccine Puzzle: EIAV’s Hard Shells
3.2. Outer Shell Hardness and the Classical Vaccine Successes: Smallpox, Rabies, Polio and YFV
3.3. Influenza Virus: No Highly Disordered Outer Shell Like HIV, HSV and HCV
4. Viral Shapeshifters of Another Kind: Trojan Horse with Inner Shell Disorder
5. HIV, HSV-2 and HCV: Presence of Disordered Proteins at the Outer Shell and the Lack of Effective Vaccines
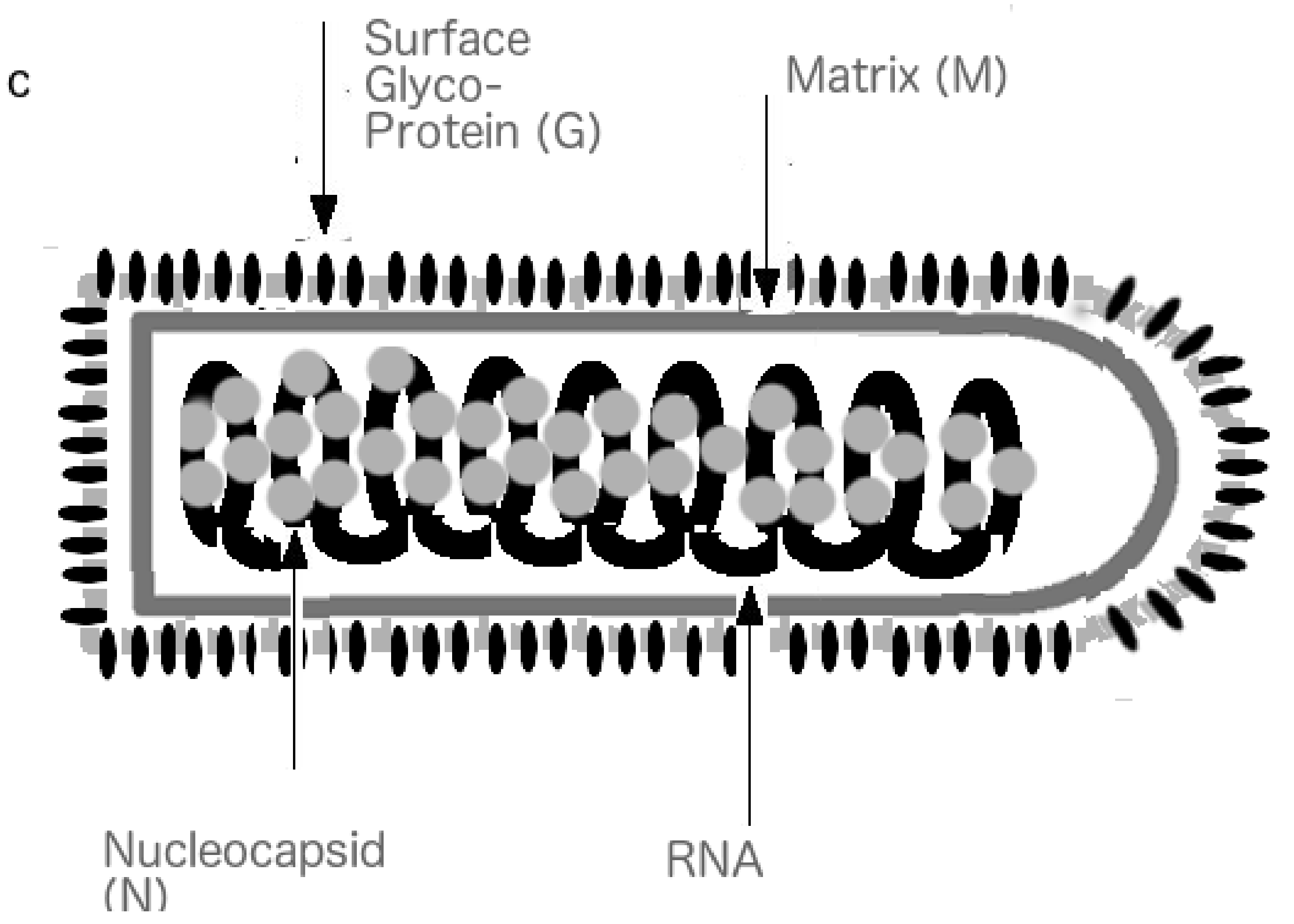
5.1. HSV, HCV and HIV: Similarities and Differences in Shell Disorder and Transmission Evolution
5.2. HIV-1 and HIV-2: Similarities and Differences in Shell Disorder and Evolution
5.3. HCV and HIV: Similarities and Differences in Shell Disorder and Evolution
5.4. HSV-1, HSV-2 and HIV: Similarities and Differences in Shell Disorder and Evolution
6. More than One Mode of Evasion: Latency
7. Experimental and Empirical Evidence Has Trickled in During the Last 10 Years
Mechanism of Immune Evasion for the True Viral Shapeshifter
8. Implications and Potential Applications of the Viral Shapeshifters: Cancer and Bacterial Virotherapy as an Example
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Esparza, J. A brief history of the global effort to develop a preventive HIV vaccine. Vaccine 2013, 31, 3502–3518. [Google Scholar] [CrossRef]
- Acheson, N.H. Fundamentals of Molecular Virology; Wiley: Hoboken, NJ, USA, 2007. [Google Scholar]
- Goh, G.K. Viral Shapeshifters: Strange Behaviors of HIV and Other Viruses, 1st ed.; Simplicity Research Institute: Singapore, 2017. [Google Scholar]
- Levine, A.J. Viruses; Scientific American Library; Henry Holt and Co.: New York, NY, USA.
- Moelling, K. Viruses: More Friends Than Foes; World Scientific: Singapore, 2015. [Google Scholar]
- Malmquist, W.A.; Barnett, D.; Becvar, C.S. Production of equine infectious anemia antigen in a persistently infected cell line. Archiv für die gesamte Virusforschung 1973, 42, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Leroux, C.; Cadoré, J.L.; Montelaro, R.C. Equine Infectious Anemia Virus (EIAV): What has HIV’s country cousin got to tell us? Vet. Res. 2004, 35, 485–512. [Google Scholar] [CrossRef] [PubMed]
- Montelaro, R.C.; Grund, C.; Raabe, M.; Woodson, B.; Cook, R.F.; Cook, S.; Issel, C.J. Characterization of protective and enhancing immune responses to equine infectious anemia virus resulting from experimental vaccines. AIDS Res. Hum. Retroviruses 1996, 12, 413–415. [Google Scholar] [CrossRef] [PubMed]
- World Organization for Animal Health (OIE). Terrestrial Manual 2013; OIE 2013:2.5.6, Equine Infectious Anemial. Available online: http://www.oie.int/standard-setting/terrestrial-manual/access-online/ (accessed on 3 May 2019).
- Wang, X.F.; Lin, Y.Z.; Li, Q.; Liu, Q.; Zhao, W.W.; Du, C.; Chen, J.; Wang, X.; Zhou, J.H. Genetic Evolution during the development of an attenuated EIAV vaccine. Retrovirology 2016, 13, 9. [Google Scholar] [CrossRef] [PubMed]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. A comparative analysis of viral matrix proteins using disorder predictors. Virol. J. 2008, 5, 126. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Shell disorder, immune evasion and transmission behaviors among human and animal retroviruses. Mol. Biosyst. 2015, 11, 2312–2323. [Google Scholar] [CrossRef]
- Avery, O.T.; Goebel, W.F. Chemo-Immunological Studies on Conjugated Carbohydrate-Proteins: V. The Immunological Specifity of an Antigen Prepared by Combining the Capsular Polysaccharide of Type III Pneumococcus with Foreign Protein. J. Exp. Med. 1931, 54, 437–447. [Google Scholar] [CrossRef]
- Avery, O.T.; Goebel, W.F. Chemo-Immunological Studies on Conjugated Carbohydrate- Proteins: II. Immunological Specificity of Synthetic Sugar-Protein Antigens. J. Exp. Med. 1929, 50, 533–550. [Google Scholar] [CrossRef]
- Goebel, W.F.; Avery, O.T. Chemo-Immunological Studies on Conjugated Carbohydrate Proteins: I. The Synthesis of p-Aminophenol beta-glycoside, p-Aminophenol beta- Galactoside, and their Coupling with Serum Globulin. J. Exp. Med. 1929, 50, 521–531. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genom. 2008, 9 (Suppl. 2). [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Protein intrinsic disorder and influenza virulence: The 1918 H1N1 and H5N1 viruses. Virol. J. 2009, 6, 69. [Google Scholar] [CrossRef]
- Fonseca, A.A.; Heinemann, M.B.; Leite, R.C.; Reis, J.K. A comparative analysis of envelope and tegument proteins of suid herpesvirus 1, bovine herpesvirus 1 and bovine herpesvirus 5. Arch. Virol. 2010, 155, 1687–1692. [Google Scholar] [CrossRef]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Meng Goh, G.K.; Dunker, A.; Uversky, V.N. Viral disorder or disordered viruses: Do viral proteins possess unique features? Protein Pept. Lett. 2010, 17, 932–951. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Understanding Viral transmission behavior via protein intrinsic disorder prediction: Coronaviruses. J. Pathog. 2012, 2012, 738590. [Google Scholar] [CrossRef]
- Xue, B.; Mizianty, M.J.; Kurgan, L.; Uversky, V.N. Protein intrinsic disorder as a flexible armor and a weapon of HIV-1. Cell. Mol. Life Sci. 2012, 69, 1211–1259. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Prediction of Intrinsic Disorder in MERS CoV/HCoV-EMC Supports a High Oral-Fecal Transmission. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef]
- Fan, X.; Xue, B.; Dolan, P.T.; LaCount, D.J.; Kurgan, L.; Uversky, V.N. The intrinsic disorder status of the human hepatitis C virus proteome. Mol. Biosyst. 2014, 10, 1345–1363. [Google Scholar] [CrossRef]
- Dolan, P.T.; Roth, A.P.; Xue, B.; Sun, R.; Dunker, A.K.; Uversky, V.N.; LaCount, D.J. Intrinsic disorder mediates hepatitis C virus core-host cell protein interactions. Protein Sci. 2015, 24, 221–235. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Detection of links between Ebola nucleocapsidand virulence using disorder analysis. Mol. Biosyst. 2015, 11, 2337–2344. [Google Scholar] [CrossRef]
- Goh, G.K.; Dunker, A.K.; Uversky, V.N. Correlating Flavivirus virulence and levels of intrinsic disorder in shell proteins: Protective roles vs. immune evasion. Mol. Biosyst. 2016, 12, 1881–1891. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26–59. [Google Scholar] [CrossRef]
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002, 27, 527–533. [Google Scholar] [CrossRef]
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins 2000, 41, 415–427. [Google Scholar] [CrossRef]
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999, 293, 321–331. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38–48. [Google Scholar] [CrossRef]
- Romero, P.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E.; Dunker, A.K. Identifying disordered regions in proteins from amino acid sequence. In Proceedings of the IEEE International Conference on Neural Networks, Houston, TX, USA, 12 June 1997; pp. 90–95. [Google Scholar]
- Garner, E.; Romero, P.; Dunker, A.K.; Brown, C.; Obradovic, Z. Predicting Binding Regions within Disordered Proteins. Genome Inf. 1999, 10, 41–50. [Google Scholar]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005, 44, 12454–12470. [Google Scholar] [CrossRef]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468–13477. [Google Scholar] [CrossRef] [PubMed]
- Tagmyer, T.L.; Craigo, J.K.; Cook, S.J.; Even, D.L.; Issel, C.J.; Montelaro, R.C. Envelope determinants of equine infectious anemia virus vaccine protection and the effects of sequence variation on immune recognition. J. Virol. 2008, 82, 4052–4063. [Google Scholar] [CrossRef]
- Gelderblom, H.R. Medical Microbiology, 4th ed.; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996. [Google Scholar]
- Walpole, G.A. The New British Traveller; Alexander Hogg: London, UK, 1784; p. 365. [Google Scholar]
- Ambrose, C.T. Osler and the infected letter. Emerg. Infect. Dis. 2005, 11, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Razzell, P. Smallpox extinction-a note of caution. New Sci. 1976, 71, 35. [Google Scholar]
- Belshe, R.B.; Leone, P.A.; Bernstein, D.I.; Wald, A.; Levin, M.J.; Stapleton, J.T.; Gorfinkel, I.; Ashley Morrow, R.L.; Ewell, M.G.; Stokes-Riner, A.; et al. Efficacy results of a trial of a herpes simplex vaccine. N. Engl. J. Med. 2012, 366, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Shen, S.; Wang, L.; Deng, H. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 2010, 1, 987–998. [Google Scholar] [CrossRef]
- Lambers, F.A.; Prins, M.; Thomas, X.; Molenkamp, R.; Kwa, D.; Brinkman, K.; Van der Meer, J.T.; Schinkel, J.; MOSAIC (MSM Observational Study of Acute Infection with Hepatitis C) Study Group. Alarming incidence of hepatitis C virus re-infection after treatment of sexually acquired acute hepatitis C virus infection in HIV-infected MSM. AIDS 2011, 25, F21–F27. [Google Scholar] [CrossRef]
- Alencar, W.K.; Duarte, P.S.; Waldman, E.A. Survival analysis of acquired immune deficiency syndrome patients with and without hepatitis C virus infection at a reference center for sexually transmitted diseases/acquired immune deficiency syndrome in Sao Paulo, Brazil. Braz. J. Infect. Dis. 2013, 18, 150–157. [Google Scholar] [CrossRef]
- Chan, D.P.; Sun, H.Y.; Wong, H.T.; Lee, S.S.; Hung, C.C. Sexually acquired hepatitis C virus infection: A review. Int. J. Infect. Dis. 2016, 49, 47–58. [Google Scholar] [CrossRef]
- Bloch, H. Edward Jenner (1749–1823). The history and effects of smallpox, inoculation, and vaccination. Am. J. Dis. Child. 1993, 147, 772–774. [Google Scholar] [CrossRef]
- Riedel, S. Edward Jenner and the history of smallpox and vaccination. In Baylor University Medical Center Proceedings; Taylor & Francis: Abingdon, UK, 2005; Volume 18, pp. 21–25. [Google Scholar]
- Berche, P. Louis Pasteur, from crystals of life to vaccination. Clin. Microbiol. Infect. 2012, 18 (Suppl. 5), 1–6. [Google Scholar] [CrossRef]
- John, T.J. The golden jubilee of vaccination against poliomyelitis. Indian J. Med. Res. 2004, 119, 1–17. [Google Scholar]
- Norrby, E. Yellow fever and Max Theiler: The only Nobel Prize for a virus vaccine. J. Exp. Med. 2007, 204, 2779–2784. [Google Scholar] [CrossRef]
- Du, J.; Wang, X.; Ma, J.; Wang, J.; Qin, Y.; Zhu, C.; Liu, F.; Shao, Y.; Zhou, J.; Qiao, W.; et al. Structural and biochemical insights into the V/I505T mutation found in the EIAV gp45 vaccine strain. Retrovirology 2014, 11, 26. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, Y.; Stone, S.; Harty, R.N. Characterization of filovirus protein-protein interactions in mammalian cells using bimolecular complementation. J. Infect. Dis. 2011, 204 (Suppl. 3), S817–S824. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Liu, Z.; Minowa, A.; Imai, T.; Tanaka, M.; Sugimoto, K.; Nishiyama, Y.; Arii, J.; Kawaguchi, Y. Herpes simplex virus 1 protein kinase Us3 and major tegument protein UL47 reciprocally regulate their subcellular localization in infected cells. J. Virol. 2011, 85, 9599–9613. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Khandkar, M. Endocytosis of human immunodeficiency virus 1 (HIV-1) in astrocytes: A fiery path to its destination. Microb. Pathog. 2015, 78, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Goudsmit, J. Viral Sex: The Nature of AIDS; Oxford University Press: New York, NY, USA, 1997. [Google Scholar]
- Urdaneta, V.; Casadesús, J. Interactions between bacteria and bile Salts in the gastrointestinal and hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Nieuwenhuis, R.F.; Van Doornum, G.J.; Mulder, P.G.; Neumann, H.A.; Van der Meijden, W.I. Importance of herpes simplex virus type-1 (HSV-1) in primary genital herpes. Acta Derm. Venereol. 2006, 86, 129–134. [Google Scholar] [PubMed]
- Koelle, D.M.; Norberg, P.; Fitzgibbon, M.P.; Russell, R.M.; Greninger, A.L.; Huang, M.; Stensland, L.; Jing, L.; Magaret, A.S.; Diem, K.; et al. Worldwide circulation of HSV-2 × HSV-1 recombinant strains. Sci. Rep. 2017, 7, 44084. [Google Scholar] [CrossRef]
- Ohori, Y.; Okazaki, H.; Watanabe, S.; Tochio, N.; Arai, M.; Kigawa, T.; Nishimura, C. Flexible and rigid structures in HIV-1 p17 matrix protein monitored by relaxation and amide proton exchange with NMR. Biochim. Biophys. Acta 2014, 1844, 520–526. [Google Scholar] [CrossRef]
- Caccuri, F.; Giagulli, C.; Reichelt, J.; Martorelli, D.; Marsico, S.; Bugatti, A.; Barone, I.; Rusnati, M.; Guzman, C.A.; Dolcetti, R.; et al. Simian immunodeficiency virus and human immunodeficiency virus type 1 matrix proteins specify different capabilities to modulate B cell growth. J. Virol. 2014, 88, 5706–5717. [Google Scholar] [CrossRef]
- Giagulli, C.; D’Ursi, P.; He, W.; Zorzan, S.; Caccuri, F.; Varney, K.; Orro, A.; Marsico, S.; Otjacques, B.; Laudanna, C.; et al. A single amino acid substitution confers B-cell clonogenic activity to the HIV-1 matrix protein p17. Sci. Rep. 2017, 7, 6555. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.; Renner, D.W.; Thompson, R.L.; Szpara, M.L.; Sawtell, N.M. Inferred father-to-son transmission of herpes simplex virus results in near-perfect preservation of viral genome identity and in vivo phenotypes. Sci. Rep. 2017, 7, 13666. [Google Scholar] [CrossRef]
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: Disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012, 30, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, W.L.; Krivushin, K.; Gilichinsky, D.; Schuerger, A.C. Growth of Carnobacterium spp. from permafrost under low pressure, temperature, and anoxic atmosphere has implications for Earth microbes on Mars. Proc. Natl. Acad. Sci. USA 2013, 110, 666–671. [Google Scholar] [CrossRef]
- Burton, D.R. Antibodies, viruses and vaccines. Nat. Rev. Immunol. 2002, 2, 706–713. [Google Scholar] [CrossRef]
- Vigerust, D.J.; Shepherd, V.L. Virus glycosylation: Role in virulence and immune interactions. Trends Microbiol. 2007, 15, 211–218. [Google Scholar] [CrossRef]
- Keen, E.C. A century of phage research: Bacteriophages and the shaping of modern biology. Bioessays 2015, 37, 6–9. [Google Scholar] [CrossRef]
- Trigo, G.; Martins, T.G.; Fraga, A.G.; Longatto-Filho, A.; Castro, A.G.; Azeredo, J.; Pedrosa, J. Phage therapy is effective against infection by Mycobacterium ulcerans in a murine footpad model. PLoS Negl. Trop. Dis. 2013, 7, e2183. [Google Scholar] [CrossRef]
- Chernajovsky, Y.; Layward, L.; Lemoine, N. Fighting cancer with oncolytic viruses. BMJ 2006, 332, 170–172. [Google Scholar] [CrossRef]
- Wei, D.; Xu, J.; Liu, X.; Chen, Z.N.; Bian, H. Fighting cancer with viruses: Oncolytic virus therapy in China. Hum. Gene Ther. 2017. [Google Scholar] [CrossRef]
- Chaurasiya, S.; Chen, N.G.; Warner, S.G. Oncolytic virotherapy versus cancer stem cells: A review of approaches and mechanisms. Cancers 2018, 10, E124. [Google Scholar] [CrossRef]
- Grigg, C.; Blake, Z.; Gartrell, R.; Sacher, A.; Taback, B.; Saenger, Y. Talimogene laherparepvec (T-Vec) for the treatment of melanoma and other cancers. Semin. Oncol. 2016, 43, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Penkett, C.J.; Redfield, C.; Dodd, I.; Hubbard, J.; McBay, D.L.; Mossakowska, D.E.; Smith, R.A.; Dobson, C.M.; Smith, L.J. NMR analysis of main-chain conformational preferences in an unfolded fibronectin-binding protein. J. Mol. Biol. 1997, 274, 152–159. [Google Scholar] [CrossRef]
- Penkett, C.J.; Redfield, C.; Jones, J.A.; Dodd, I.; Hubbardett, J.; Smith, R.A.; Smith, L.J.; Dobson, C.M. Structural and dynamical characterization of a biologically active unfolded fibronectin-binding protein from Staphylococcus aureus. Biochemistry 1998, 37, 17054–17067. [Google Scholar] [CrossRef]
- House-Pompeo, K.; Xu, Y.; Joh, D.; Speziale, P.; Hook, M.J. Conformational Changes in the Fibronectin Binding MSCRAMMs Are Induced by Ligand Binding. J. Biol. Chem. 1996, 271, 1379–1384. [Google Scholar] [CrossRef]
- Dunker, A.K.; Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573–6582. [Google Scholar] [CrossRef]

| Virus | Virus Type, Tansmission | Outer Shell, Proteins (UniProt Accession)+ | Intermediate Shell | Inner Shell |
|---|---|---|---|---|
| EIAV | Retroviridae (RNA) Insect | Matrix, p15 (P69732) | Capsid, p26 (P69732) | Nucleocapsid, p11 (P69732) |
| Influenza | Orthomyxo-viridae (RNA) Resspiratory | Matrix, m1 (P05755) | Nucleoprotein NP (P21433) | |
| HIV-1 | Retroviridae (RNA) Sexual | Matrix, p17 (P03348) | Capsid, p24 (P03348) | Nucleocapsid, p7 (P04594) |
| HIV-2 | Retroviridae (RNA) Sexual, Bite | Matrix, p17 (P04584) | Capsid, p24 (P04584) | Nucleocapsid, p7 (P04584) |
| Variola/Smallpox* | Poxviridae (DNA) Inhalation | Membrane, C9L (Q76U97), A14 (P33839), F5 (P33865) | Core, VP8 (Q0N570), 4A (Q0N532), 4B (Q0N539) | |
| Rabies | Rhabdoviridae (RNA) Bites | Matrix, M (P25224) | Nucleocapsid, N (P151979) | |
| Poliovirus | Picornaviridae (RNA) Fecal-Oral | Capsid, VP1-4 (P03302) | ||
| Yellow Fever (YFV) | Flaviviridae (RNA) Insect | Membrane, M (P03314) | Capsid, C (P03314) | |
| Hepatitis C (HCV) | Flaviviridae (RNA) Sexual, Blood Contamination | Core, p19, p21 (P26663) | ||
| Herpes Simplex Virus-2 (HSV-2)* | Herpesviridae (DNA) Oral-Oral Contacts | Tegument, VP22-UL49 (D3YPK7), VP1/2-UL36 (I1UYK0), VP13/14-UL47 (P10231), VP16-UL48 (P06492) | Capsid, VP5 (P06491) | |
| Herpes Simplex Virus-2 (HSV-2)* | Herpesviridae (DNA) Sexual | Tegument, VP22-UL49 (A7LK33), VP1/2-UL36 (G9I258), VP13/14-UL47 (A7LK25), VP16-UL48 (P68335) | Capsid, VP5 (P89442) |
| Virus | PID (%) of Outer Shell | PID (%) of Intermediate Shell | PID (%) of Inner Shell | Vaccine Available + |
|---|---|---|---|---|
| EIAV | 13 ± 0.1 | 29 ± 0.1 | 26 ± 0.1 | Yes |
| HIV-1, SIVcpz # | 56.5 ± 10.8 | 44.5 ± 2.6 | 39.5 ± 3.0 | No |
| HIV-2, SIVmac + | 51.5 ± 2.5 | 26.6 ± 2.9 | 46.5 ± 0.1 | No |
| Influenza | 35.2 ± 1.6 | 44.7 ± 5.4 | Yes | |
| Smallpox | 13 ± 0.1 | 19 ± 0.1 | Yes | |
| 8 ± 0.1 | 4 ± 0.1 | |||
| 15 ± 0.1 | 12 ± 0.1 | |||
| Rabies | 25.8 ± 1.4 | 21.5 ± 0.8 | Yes | |
| Poliovirus | 34 ± 3.8 | Yes | ||
| 15.12 ± 6.1 | ||||
| 31.3 ± 3.6 | ||||
| 27 ± 0.1 | ||||
| Yellow Fever (YFV) | 35.2 ± 0.9 | 74.3 ± 0.9 | Yes | |
| HCV | 52.5 ± 0.5 48.5 ± 0.5 | No | ||
| HSV-1 | 58.0 ± 1.4 | 18 ± 0.1 | No ^ | |
| 50.5 ± 0.5 | ||||
| 36.3 ± 0.5 | ||||
| 37 ± 0.7 | ||||
| HSV-2 | 61 ± 1.6 | 18 ± 0.1 | No | |
| 50 ± 0.1 | ||||
| 38 ± 1.1 | ||||
| 39.3 ± 0.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goh, G.K.-M.; Dunker, A.K.; Foster, J.A.; Uversky, V.N. HIV Vaccine Mystery and Viral Shell Disorder. Biomolecules 2019, 9, 178. https://doi.org/10.3390/biom9050178
Goh GK-M, Dunker AK, Foster JA, Uversky VN. HIV Vaccine Mystery and Viral Shell Disorder. Biomolecules. 2019; 9(5):178. https://doi.org/10.3390/biom9050178
Chicago/Turabian StyleGoh, Gerard Kian-Meng, A. Keith Dunker, James A. Foster, and Vladimir N. Uversky. 2019. "HIV Vaccine Mystery and Viral Shell Disorder" Biomolecules 9, no. 5: 178. https://doi.org/10.3390/biom9050178
APA StyleGoh, G. K.-M., Dunker, A. K., Foster, J. A., & Uversky, V. N. (2019). HIV Vaccine Mystery and Viral Shell Disorder. Biomolecules, 9(5), 178. https://doi.org/10.3390/biom9050178







