Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Genomic DNA Extraction and Genotyping
2.2.1. DNA Extraction
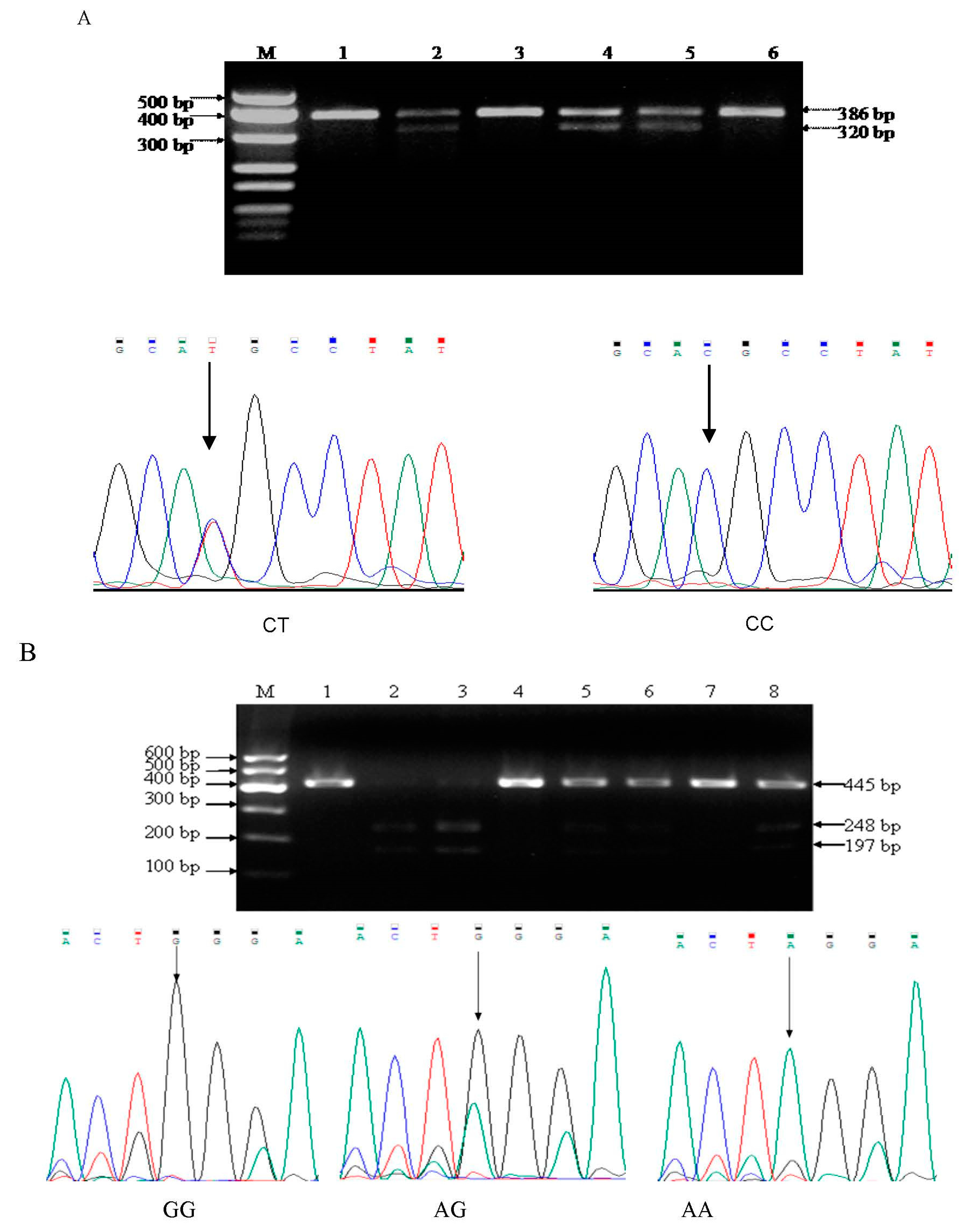
2.2.2. MMP9 -1562C/T Polymorphism
2.2.3. MMP13 -77A/G Polymorphism
2.3. Measurement of MMP-9 and MMP-13 ELISA
2.4. Statistical Analysis
3. Results
3.1. Characteristics of Study Subjects
3.2. Distribution of MMP9 and MMP13 Polymorphisms
3.3. The Association between MMP Polymorphisms and NSCLC Risk
3.4. Distribution of with Serum Levels MMP and Polymorphisms
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; ebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Bray Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer. 2015, 136, E359. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.K.; Gupta, U.; Jain, S. Epidemiology of lung cancer and approaches for its prediction: A systematic review and analysis. Chin. J. Cancer 2016, 35, 71. [Google Scholar] [CrossRef]
- Hech, S.S. Cigarette smoking and lung cancer: Chemical mechanisms and approaches to prevention. Lancet Oncol. 2002, 3, 461–469. [Google Scholar] [CrossRef]
- Li, H.; Liang, X.; Qin, X.; Cai, S.; Yu, S. Association of matrix metalloproteinase family gene polymorphisms with lung cancer risk: Logistic regression and generalized odds of published data. Sci. Rep. 2015, 5, 10056. [Google Scholar] [CrossRef] [PubMed]
- Banday, M.Z.; Sameer, A.S.; Mir, A.H.; Mokhdomi, T.A.; Chowdri, N.A.; Haq, E. Matrix metalloproteinase (MMP)-2,-7 and-9promoter polymorphisms in colorectal cancer in ethnic Kashmiri population—A case-control study and a mini review. Gene 2016, 589, 81–89. [Google Scholar] [CrossRef]
- Cui, N.; Hu, M.; Khalil, R.A. Biochemical and Biological Attributes of Matrix Metalloproteinases. Prog. Mol. Biol. Transl. Sci. 2017, 147, 71–73. [Google Scholar]
- Łukaszewicz-Zając, M.; Szmitkowski, M.; Litman-Zawadzka, A.; Mroczko, B. Matrix Metalloproteinases and their tissue inhibitors in comparison to other inflammatory proteins in Gastric Cancer (GC). Cancer Investig. 2016, 34, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.S.; Song, J.; Tu, X.Y.; Zhao, J.H.; Ye, X.Q. The association between MMP-12 82 A/Gpolymorphism and susceptibility to various malignant tumors: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 10845–10854. [Google Scholar]
- Liu, L.; Sun, J.; Li, G.; Gu, B.; Wang, X.; Chi, H.; Guo, F. Association between MMP-12-82A/G polymorphism and cancer risk: A meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 11896–11904. [Google Scholar]
- Moreno-Ortiz, J.M.; Gutiérrez-Angulo, M.; Partida-Pérez, M.; Peregrina-Sandoval, J.; Ramírez-Ramírez, R.; Muñiz-Mendoza, R.; Suárez-Villanueva, S.; Centeno-Flores, M.; Maciel-Gutiérrez, V.; Cabrales-Vazquez, J.E.; et al. Association of MMP7-181A/G and MMP13-77A/G polymorphisms with colorectal cancer in a Mexican population. Genet. Mol. Res. 2014, 3, 3537–3544. [Google Scholar] [CrossRef]
- Gao, P.; Yang, J.L.; Zhao, H.; You, J.H.; Hu, Y. Common polymorphism in the MMP-13 gene may contribute to the risk of human cancers: A meta-analysis. Tumor Biol. 2014, 35, 10137–10148. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Xia, J.; Xing, H.; Yang, W.; Xiong, X.; Pan, W.; Han, S.; Shang, J.; Zhou, C.; Zhou, L.; Yang, M. The Sp1-mediaded allelic regulation of MMP13 expression by an ESCC susceptibility SNP rs2252070. Sci. Rep. 2016, 6, 27103. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.; Jamerson, M.; Cabral, G.; Carlesso, A.M.; Marciano-Cabral, F. Expression of matrix metalloproteinases in Naegleria fowleri and their role in invasion of the central nervous system. Microbiology 2017, 163, 1436–1444. [Google Scholar] [CrossRef] [PubMed]
- Decock, J.; Paridaens, R.; Ye, S. Genetic polymorphisms of matrix metalloproteinases in lung, breast and colorectal cancer. Clin. Genet. 2008, 73, 197–211. [Google Scholar] [CrossRef]
- Li, Y.; Sun, D.L.; Duan, Y.N.; Zhang, X.J.; Wang, N.; Zhou, R.M.; Chen, Z.F.; Wang, S.J. Association of functional polymorphisms in MMPs genes with gastric cardia adenocarcinoma and esophageal squamous cell carcinoma in high incidence region of North China. Mol. Biol. Rep. 2010, 37, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Houghton, A.M. Matrix metalloproteinases in destructive lung disease. Matrix Biol. 2015, 44–46, 167–174. [Google Scholar] [CrossRef]
- Hughes, S.; Agbaje, O.; Bowen, R.L.; Holliday, D.L.; Shaw, J.A.; Duffy, S.; Jones, J.L. Matrix metalloproteinase single-nucleotide polymorphisms and haplotypes predict breast cancer progression. Clin. Cancer Res. 2007, 13, 6673–6680. [Google Scholar] [CrossRef]
- Chaudhary, A.K.; Singh, M.; Bharti, A.C.; Asotra, K.; Sundaram, S.; Mehrotra, R. Genetic polymorphisms of matrix metalloproteinases and their inhibitors in potentially malignant and malignant lesions of the head and neck. J. Biomed. Sci. 2010, 17, 10. [Google Scholar] [CrossRef]
- Lu, L.; Li, W.; Liu, C.; Jia, M.; Xing, F.; Jiang, P.; Tang, J.X. Association between -1562c/t polymorphisms in the matrix metalloproteinase-9 and the risk of lung cancer among south-central chinese population. J. Bionanosci. 2016, 10, 506–510. [Google Scholar] [CrossRef]
- Langers, A.M.; Verspaget, H.W.; Hommes, D.W. Single-nucleotide polymorphisms of matrix metalloproteinases and their inhibitors in gastrointestinal cancer. World J. Gastrointest. Oncol. 2011, 3, 79–98. [Google Scholar] [CrossRef]
- Schveigert, D.; Valuckas, K.P.; Kovalcis, V.; Ulys, A.; Chvatovic, G.; Didziapetriene, J. Significance of MMP-9 expression and MMP-9 polymorphism in prostate cancer. Tumori 2013, 99, 523–529. [Google Scholar] [CrossRef]
- Verma, S.; Kesh, K.; Gupta, A.; Swarnakar, S. An overview of matrix metalloproteinase 9 polymorphism and gastric cancer risk. Asian Pac. J. Cancer Prev. 2015, 16, 7393–7400. [Google Scholar] [CrossRef]
- Avcı, N.; Ture, M.; Deligonul, A.; Cubukcu, E.; Olmez, O.F.; Sahinturk, S.; Topak, A.; Kurt, E.; Evrensel, T.; Şahin, A.B.; et al. Association and prognostic significance of the functional -1562C/T polymorphism in the promoter region of MMP-9 in Turkish patients with gastric cancer. Pathol. Oncol. Res. 2015, 21, 1243–1247. [Google Scholar] [CrossRef]
- Beeghly-Fadiel, A.; Lu, W.; Shu, X.O.; Lu, W.; Long, J.; Cai, Q.; Xiang, Y.B.; Zheng, Y.; Zhao, Z.; Gu, K.; et al. Mmp 9 polymorphisms and breast cancer risk: A report from the shanghai breastcancer genetics study. Breast Cancer Res. Treat. 2011, 126, 507–513. [Google Scholar] [CrossRef]
- Matsumura, S.; Oue, N.; Nakayama, H.; Kitadai, Y.; Yoshida, K.; Yamaguchi, Y.; Imai, K.; Nakachi, K.; Matsusaki, K.; Chayama, K.; et al. A single nucleotide polymorphism in the mmp-9, promoter affects tumor progression and invasive phenotype of gastric cancer. J. Cancer Res. Clin. Oncol. 2005, 131, 19–25. [Google Scholar] [CrossRef]
- Roncevic, J.; Djoric, I.; Selemetjev, S.; Jankovic, J.; Dencic, T.I.; Bozic, V.; Cvejic, D. MMP-9-1562 C/T single nucleotide polymorphism associates with increased MMP-9 level and activity during papillary thyroid carcinoma progression. Pathology 2019, 51, 55–61. [Google Scholar] [CrossRef]
- Isaacson, K.J.; Martin Jensen, M.; Subrahmanyam, N.B.; Ghandehari, H. Matrix-metalloproteinases as targets for controlled delivery in cancer: An analysis of upregulation and expression. J. Control. Release 2017, 10, 62–75. [Google Scholar] [CrossRef]
- Yamada, T.; Oshima, T.; Yoshihara, K.; Tamura, S.; Kanazawa, A.; Inagaki, D.; Kunisaki, C. Overexpression of MMP-13 gene in colorectal cancer with liver metastasis. Anticancer Res. 2010, 30, 2693–2699. [Google Scholar]
- Borghese, B.; Chiche, J.D.; Vernerey, D.; Chenot, C.; Mir, O.; Bijaoui, G.; Bonaiti-Pellié, C. Genetic polymorphisms of matrix metalloproteinase 12 and 13 genes are implicated in endometriosis progression. Hum. Reprod. 2008, 23, 1207–1213. [Google Scholar] [CrossRef]
- Kincl, V.; Máchal, J.; Drozdová, A.; Panovský, R.; Vašků, A. The Relation between eNOS -786 C/T,4a/b,MMP-13 rs640198 G/T, Eotaxin 426C/T, -384A/G, and 67G/A polymorphisms and long-term outcome in patients with coronary artery disease. Dis. Mark. 2015, 2015, 232048. [Google Scholar]
- Vairaktaris, E.; Yapijakis, C.; Nkenke, E.; Serefoglou, Z.C.; Chatzitheofylaktou, A.; Vassiliou, S.; Derka, S.; Vylliotis, A.; Perrea, D.; Neukam, F.W.; et al. A metalloproteinase-13 polymorphism affecting its gene expression is associated with advanced stages of oral cancer. Anticancer Res. 2007, 27, 4027–4030. [Google Scholar]
- Martin, G.; Asensi, V.; Montes, A.H.; Collazos, J.; Alvarez, V.; Carton, J.A.; Taboada, F.; Valle-Garay, E. Role of plasma matrix-metalloproteases (MMPs) and their polymorphisms (SNPs) in sepsis development and outcome in ICU patients. Sci. Rep. 2014, 4, 5002. [Google Scholar] [CrossRef]
| Variable | Cases (n = 245) Number (%) | Controls (n = 258) Number (%) | p-Value | χ2 |
|---|---|---|---|---|
| Age | 0.32 | 0.97 | ||
| <55 | 100 (40.82) | 118 (45.74) | ||
| ≥55 | 145 (59.18) | 140(54.36) | ||
| Sex | 0.44 | 0.61 | ||
| Male | 180 (73.47) | 165 (63.95) | ||
| Female | 65(27.53) | 93 (36.05) | ||
| Smoking status | <0.001 | 38.61 | ||
| No | 85 (34.69) | 161 (62.40) | ||
| Yes | 160 (65.31) | 97 (37.60) | ||
| Histological type | ||||
| Squamous carcinomas | 135 (55.11) | |||
| Adenocarcinomas | 89(36.32) | |||
| Other carcinomas a | 21 (8.57) | |||
| Clinical stage I + II | 80 | |||
| III + IV | 165 | |||
| Lymph node metastasis | ||||
| Yes | 182 | |||
| No | 63 |
| Variable | Genotype | Cases (n = 245) Number (%) | Controls (n = 258) Number (%) | p | χ2 | OR (95% CI) b |
|---|---|---|---|---|---|---|
| Age | ||||||
| <55 | CC | 81 (81.00) | 80 (67.80) | 0.03 | 4.89 | 2.03 (1.08–3.81) c |
| TT+CT | 19 (19.00) | 38 (32.20) | 1 (reference) | |||
| ≥55 | CC | 120 (82.75) | 93 (66.43) | 0.002 | 10.06 | 2.43 (1.39–4.23) c |
| TT+CT | 25 (17.25) | 47 (33.57) | 1 (reference) | |||
| Sex | ||||||
| Male | CC | 148 (82.22) | 104 (63.03) | 0.00 | 16.10 | 2.71 (1.65–4.45) c |
| TT+CT | 32 (17.78) | 61 (36.97) | 1 (reference) | |||
| Female | CC | 53 (81.54) | 69 (74.19) | 0.28 | 1.17 | 1.54 (0.70–3.35) |
| TT+CT | 12 (18.56) | 24 (26.81) | 1 (reference) | |||
| Smoking status | ||||||
| Yes | CC | 132 (82.50) | 66 (68.04) | 0.01 | 7.14 | 2.21 (1.23–4.00) c |
| TT+CT | 28 (17.50) | 31 (31.96) | 1 (reference) | |||
| No | CC | 69 (81.18) | 107 (66.46) | 0.02 | 5.92 | 2.18 (1.15–4.11) c |
| TT+CT | 16 (18.82) | 54 (33.54) | 1 (reference) |
| Variable | Genotype | Cases (n = 245) Number (%) | Controls (n = 258) Number (%) | p | χ2 | OR (95% CI) b |
|---|---|---|---|---|---|---|
| Age | ||||||
| <55 | GG | 25 (17.24) | 17 (12.14) | 1 (reference) | ||
| AA + AG | 120 (82.76) | 123 (87.86) | 0.23 | 1.47 | 1.51 (0.78-2.93) | |
| ≥55 | GG | 18 (18.00) | 14 (11.86) | 1 (reference) | ||
| AA + AG | 82 (82.00) | 104 (88.14) | 0.20 | 1.63 | 1.63 (0.77–3.47) | |
| Sex | ||||||
| Male | GG | 31 (17.22) | 20 (12.12) | 1 (reference) | ||
| AA + AG | 149 (82.78) | 145 (87.82) | 0.18 | 1.78 | 1.51 (0.82–2.77) | |
| Female | GG | 12 (18.46) | 11 (11.83) | 1 (reference) | ||
| AA + AG | 53 (81.54) | 82 (88.17) | 0.25 | 1.35 | 1.69 (0.69–4.10) | |
| Smoking status | ||||||
| Yes | GG | 30 (18.75) | 13 (13.40) | 1 (reference) | ||
| AA + AG | 130 (81.25) | 84 (86.60) | 0.26 | 1.24 | 1.49 (0.74–3.02) | |
| No | GG | 13 (15.29) | 18 (11.18) | 1 (reference) | ||
| AA + AG | 72 (84.71) | 143 (88.82) | 0.36 | 0.86 | 1.43 (0.67–3.09) |
| Genotype | Cases (n = 245) | Controls (n = 258) | p-Value | χ2 | OR (95% CI) b |
|---|---|---|---|---|---|
| MMP-9 | |||||
| CC/CC | 201 (82.04) | 173 (67.05) | - | - | 1 (reference) |
| CT/CT | 44 (17.96) | 85 (32.95) | 0.00 | 14.80 | 0.45 (0.29–0.68) c |
| TT/TT | 0 | 0 | |||
| T allele frequency | 44 (9.98) | 85 (16.47) | - | - | 1 (reference) |
| C allele frequency | 446 (91.02) | 431 (83.53) | 0.00 | 12.62 | 2.0 (1.36–2.95) c |
| MMP-13 | |||||
| GG/ GG | 43 (17.55) | 31 (12.02) | - | - | 1 (reference) |
| AG/ GG | 94 (38.37) | 87 (33.72) | 0.37 | 0.81 | 0.78 (0.45–1.35) |
| AA/ AA | 108 (44.08) | 140 (54.26) | 0.03 | 4.85 | 0.56 (0.33–0.94) c |
| G allele frequency | 180 (36.73) | 149 (28.88) | - | - | 1 (reference) |
| A allele frequency | 310 (63.26) | 367 (71.12) | 0.00 | 7.05 | 0.70 (0.54–0.91) c |
| Gene | Group | Alleles | p-Value (HWE Test) | χ2 | |
|---|---|---|---|---|---|
| T | C | ||||
| MMP9 | Case | 0.09 | 0.91 | 0.33 | 2.2 |
| control | 0.18 | 0.82 | 0.01 | 8.50 | |
| MMP13 | G | A | |||
| Case | 0.42 | 0.66 | 0.12 | 4.18 | |
| control | 0.35 | 0.74 | 0.06 | 5.81 | |
| Variable | N | Genotype | p | χ2 | ||
|---|---|---|---|---|---|---|
| CC | CT | TT | ||||
| Histological type | 0.82 | 0.66 | ||||
| Squamous carcinomas | 135 | 113 (82.22) | 22 (17.78) | 0 | ||
| Adenocarcinomas | 89 | 72 (80.90) | 17 (19.10) | 0 | ||
| Other carcinomas a | 21 | 16 (76.19) | 5 (23.81) | 0 | ||
| Clinical stage | ||||||
| I + II | 80 | 70 (87.50) | 10 (12.50) | 0 | 0.12 | 2.40 |
| III + IV | 165 | 131 (79.39) | 34 (20.61) | 0 | ||
| Lymph node metastasis | ||||||
| Yes | 182 | 150 (82.42) | 32 (17.58) | 0 | 0.79 | 0.068 |
| No | 63 | 51 (80.95) | 12 (19.05) | 0 | ||
| Variable | N | Genotype | p | χ2 | ||
|---|---|---|---|---|---|---|
| GG | AG | AA | ||||
| Histological type | 0.00 | 22.70 | ||||
| Squamous carcinomas | 135 | 11 (8.15) | 59 (43.70) | 65 (48.15) | ||
| Adenocarcinomas | 89 | 23 (25.84) | 28 (31.46) | 38 (42.70) | ||
| Other carcinomas a | 21 | 9 (42.86) | 7 (33.33) | 5 (23.81) | ||
| Clinical stage | 0.126 | 4.15 | ||||
| I + II | 80 | 17 (21.25) | 31 (38.75) | 32 (40.00) | ||
| III + IV | 165 | 26 (15.76) | 63 (38.18) | 76 (46.06) | ||
| Lymph node metastasis | 0.00 | 64.8 | ||||
| Yes | 182 | 28 (15.38) | 72 (39.56) | 82 (45.06) | ||
| No | 63 | 15 (23.81) | 22 (34.92) | 26 (41.27) | ||
| Genotype | Cases n (%) n = 245 | Controls n (%) n = 258 | p | χ2 | OR (95% CI) | |
|---|---|---|---|---|---|---|
| MMP9 | MMP13 | |||||
| TT+CT | AA+AG | 35 (14.29) | 66 (25.58) | - | 1 (reference) | |
| TT+CT | GG | 9 (3.67) | 19 (7.36) | 0.80 | 0.06 | 0.89 (0.37–2.18) |
| CC | AA+AG | 167 (68.16) | 161 (62.41) | 0.00 | 8.20 | 1.96 (1.23–3.11) |
| CC | GG | 34 (13.88) | 12 (4.65) | 0.00 | 19.56 | 5.34 (2.46–11.60) |
| Genotype | Cases (n = 67) | Controls (n = 21) | p-Value | χ2 | OR (95% CI) b |
|---|---|---|---|---|---|
| MMP-9 | |||||
| CC/CC | 61 (198.36 ± 25.21) | 14 (46.55 ± 15.78) | - | - | 1 (reference) |
| CT/CT | 6 (154.74 ± 22.35) | 7 (42.53 ± 14.26) | 0.51 | 0.43 | 0.86 (0.54–1.36) |
| TT/TT | 0 | 0 | |||
| T allele | 6 (154.74 ± 21.71) | 7 (42.53 ± 14.79) | - | - | 1 (reference) |
| C allele | 128 (196.58 ± 25.71) | 35 (45.75 ± 15.36) | 0.468 | 0.526 | 1.19 (0.75–1.89) |
| MMP-13 | |||||
| GG/ GG | 31 (343.41 ± 37.28) | 10 (56.68 ± 18.17) | - | - | 1 (reference) |
| AG/ GG | 29 (227.29 ± 35.27) | 7 (55.96 ± 17.85) | 0.06 | 3.68 | 0.67 (0.45–1.01) |
| AA/ AA | 7 (295.64 ± 41.63) | 4 (62.61 ± 19.32) | 0.25 | 1.34 | 0.79 (0.53–1.17) |
| G allele | 91 (306.41 ± 45.39) | 27 (56.49 ± 18.29) | - | - | 1 (reference) |
| A allele | 43 (249.54 ± 19.57) | 15 (59.50 ± 18.55) | 0.18 | 1.77 | 0.76 (0.51–1.14) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Jia, M.X.; Wang, J.H.; Lu, J.L.; Deng, J.; Tang, J.X.; Liu, C. Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population. Biomolecules 2019, 9, 107. https://doi.org/10.3390/biom9030107
Li W, Jia MX, Wang JH, Lu JL, Deng J, Tang JX, Liu C. Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population. Biomolecules. 2019; 9(3):107. https://doi.org/10.3390/biom9030107
Chicago/Turabian StyleLi, Wen, Ming Xi Jia, Jian Hui Wang, Jie Li Lu, Jing Deng, Jian Xin Tang, and Cun Liu. 2019. "Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population" Biomolecules 9, no. 3: 107. https://doi.org/10.3390/biom9030107
APA StyleLi, W., Jia, M. X., Wang, J. H., Lu, J. L., Deng, J., Tang, J. X., & Liu, C. (2019). Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population. Biomolecules, 9(3), 107. https://doi.org/10.3390/biom9030107




