Non-Toxic and Ultra-Small Biosilver Nanoclusters Trigger Apoptotic Cell Death in Fluconazole-Resistant Candida albicans via Ras Signaling
Abstract
1. Introduction
2. Materials and Methods
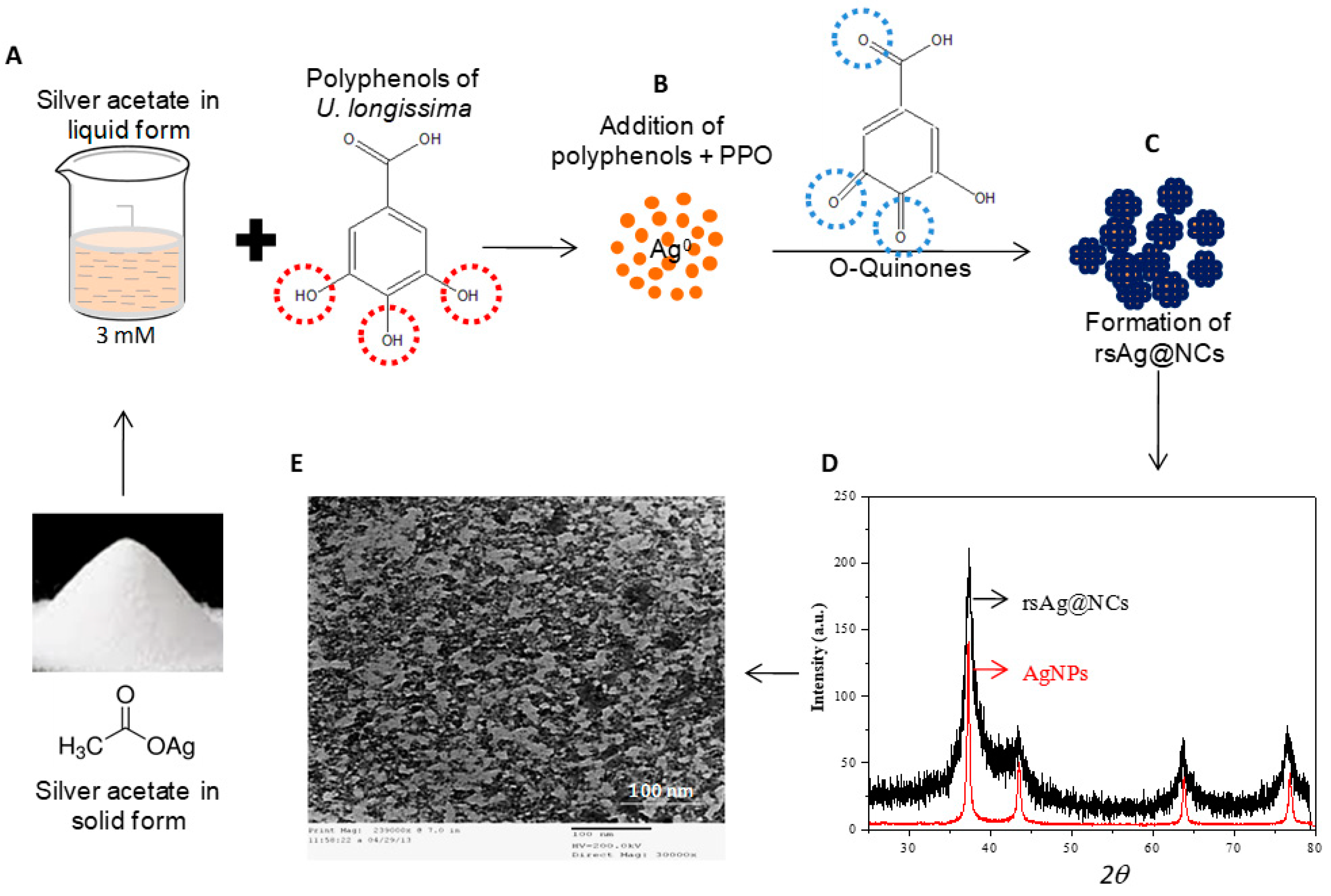
2.1. Biosynthesis and Characterization of rsAg@NCs
2.2. Assessment of Anti-Candidal Activity of rsAg@NCs
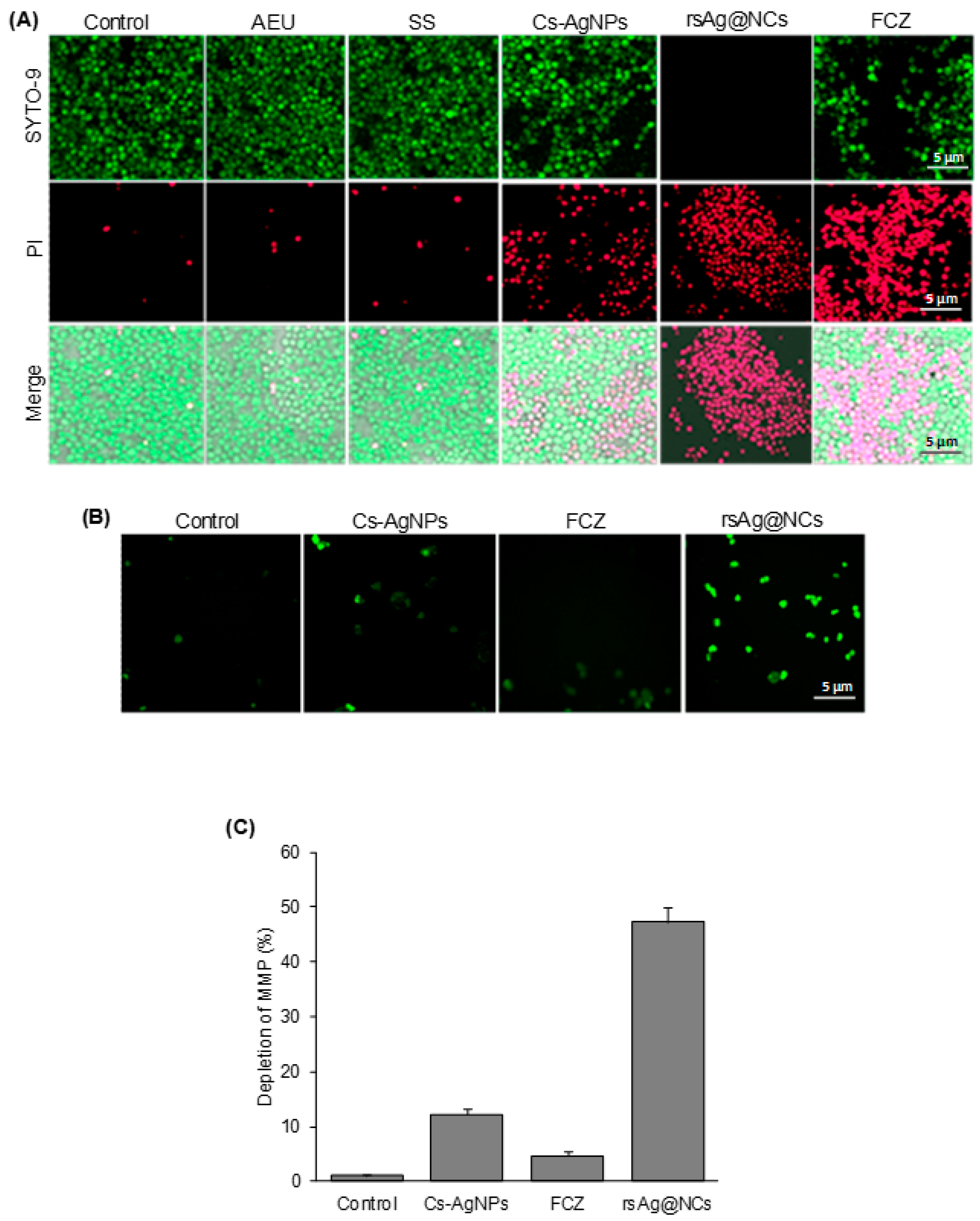
2.3. Live and Dead Cell Staining Assay
2.4. Cell Viability Assay
2.5. Mitochondrial Membrane Potential (MMP) Assay
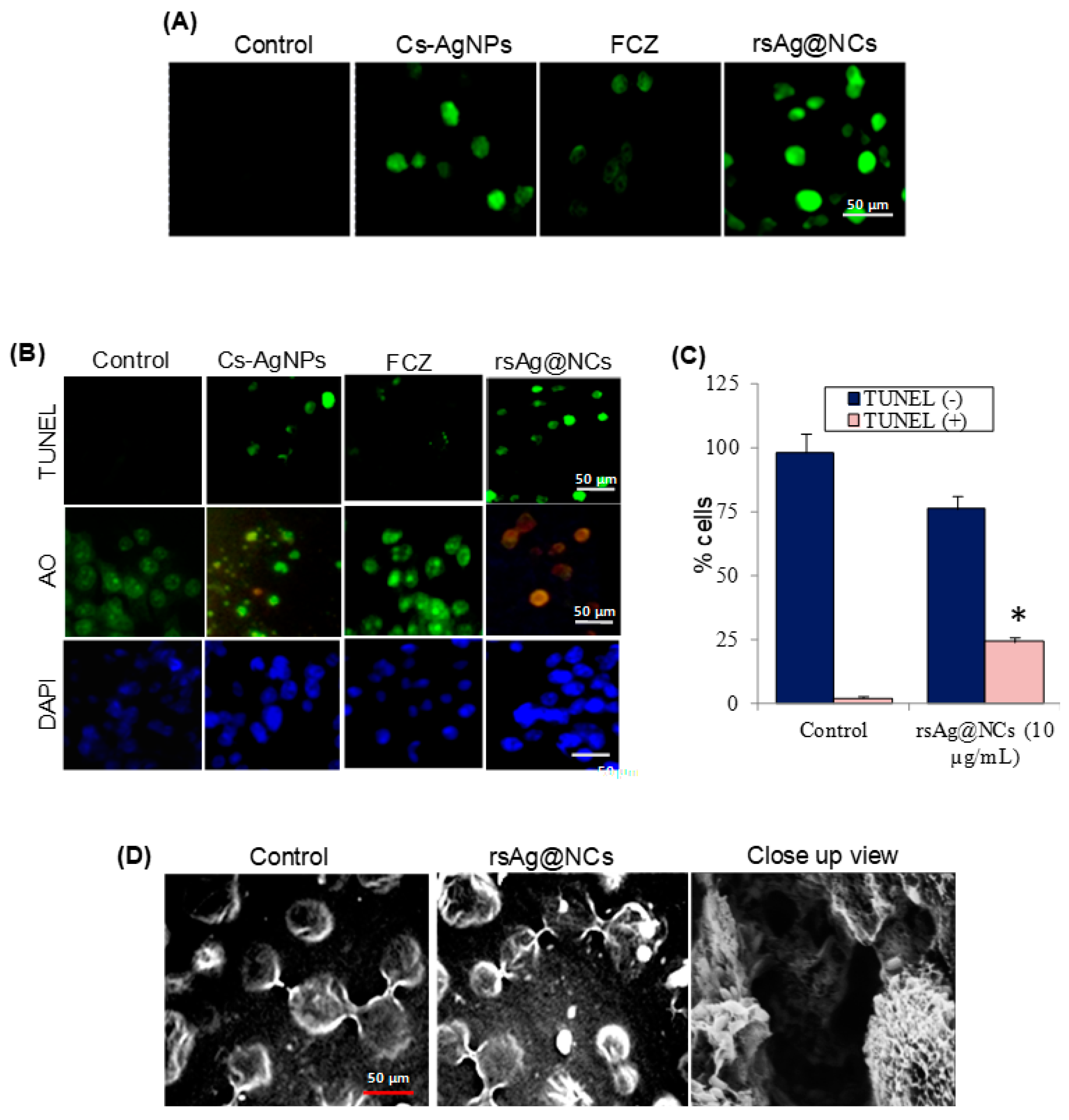
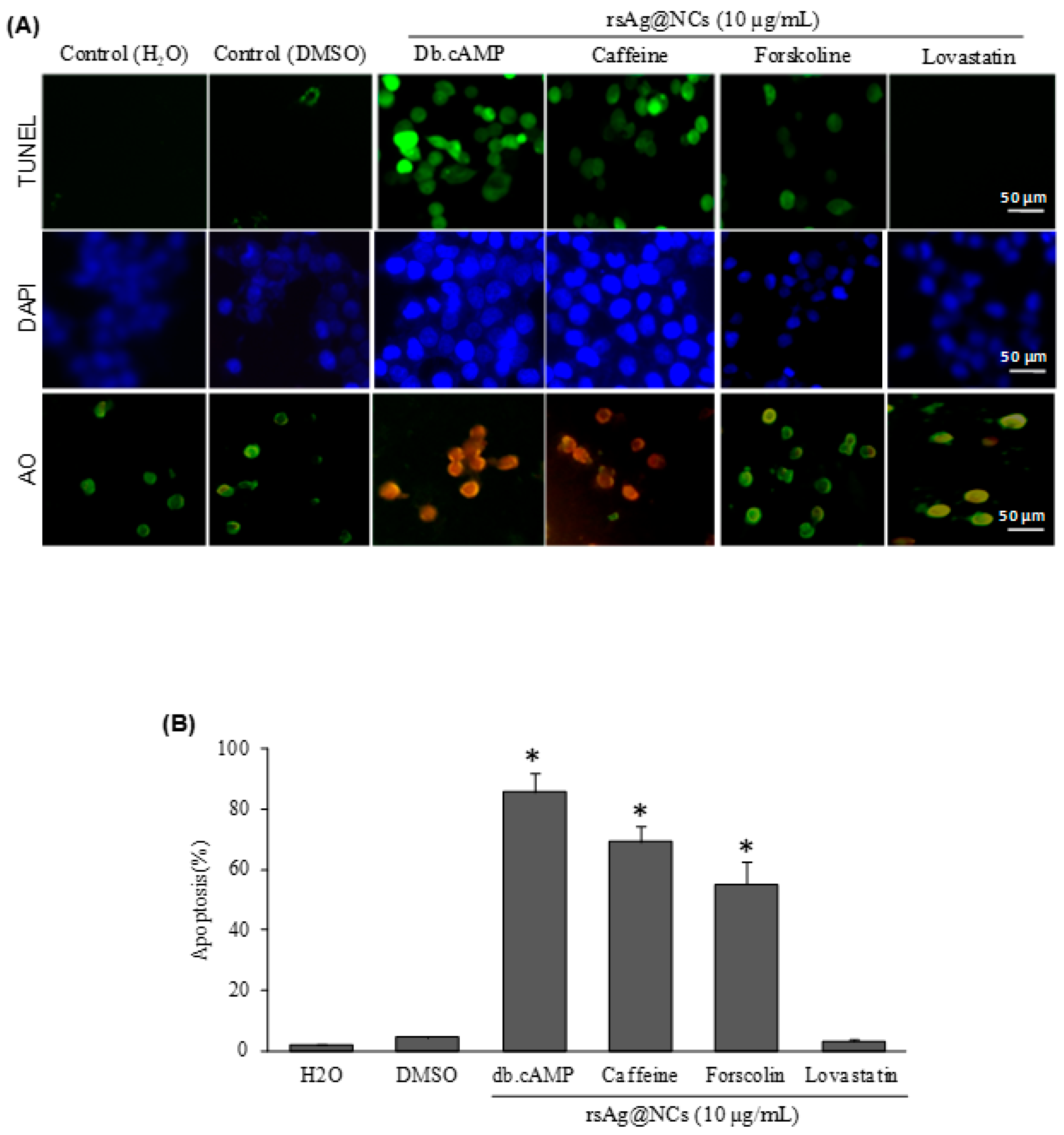
2.6. Apoptosis Assays
2.7. Analysis of Oxidative Stress Markers
2.8. Extraction and Identification of C. albicans Proteins for Proteome Analysis
2.9. ROS Detection Assay
2.10. Measurement of Endogenous Antioxidants
2.11. Detection of AhpC Expression
2.12. Statistical Analysis
3. Results
3.1. Biosynthesis of rsAg@NCs and Their Characterization
3.2. rsAg@NCs Inhibit the Cell Viability of Candida albicans
3.3. rsAg@NCs Induce Mitochondrial Dysfunction
3.4. rsAg@NCs Enhance Intracellular ROS Accumulation
3.5. rsAg@NCs Accelerate ROS-Mediated Apoptosis
3.6. rsAg@NCs Modulate the Expression of Metacaspases in DNA Damage
3.7. rsAg@NCs Alter Protein Expression in C. albicans
3.8. rsAg@NCs Regulate the Ras Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cohen, M.L. Changing patterns of infectious disease. Nature 2000, 406, 762–767. [Google Scholar] [CrossRef] [PubMed]
- Huh, A.J.; Kwon, Y.J. “Nanoantibiotics”: A new paradigm for treating infectious diseases using nanomaterials in the antibiotics resistant era. J. Control Release 2011, 156, 128–145. [Google Scholar] [CrossRef] [PubMed]
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad Bugs, No Drugs: No ESKAPE! An Update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Z.M.; Zhang, Q.B.; Puppala, H.L.; Colvin, V.L.; Alvarez, P.J. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano. Lett. 2012, 12, 4271–4275. [Google Scholar] [CrossRef] [PubMed]
- Atiyeh, B.S.; Costagliola, M.; Hayek, S.N.; Dibo, S.A. Effect of silver on burn wound infection control and healing: Review of the literature. Burns 2007, 33, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.R.; Singh, B.N.; Singh, A.; Khan, W.; Naqvi, A.H.; Singh, H.B. Mycofabricated biosilver nanoparticles interrupt Pseudomonas aeruginosa quorum sensing systems. Sci. Rep. 2015, 5, 13719. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Prateeksha; Pandey, G.; Jadaun, V.; Singh, S.; Bajpai, R.; Nayaka, S.; Naqvi, A.H.; Singh Rawat, A.K.; Upreti, D.K.; et al. Development and characterization of a novel Swarna-based herbo-metallic colloidal nano-formulation—Inhibitor of Streptococcus mutans quorum sensing. RSC Adv. 2015, 5, 5809–5822. [Google Scholar] [CrossRef]
- Zhang, X.; Hicks, E.M.; Zhao, J.; Schatz, G.C.; Van Duyne, R.P. Electrochemical tuning of silver nanoparticles fabricated by nanosphere lithography. Nano Lett. 2005, 5, 1503–1507. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.N.; Emori, T.G.; Culver, D.H.; Gaynes, R.P.; Jarvis, W.R.; Horan, T.; Edwards, J.R.; Tolson, J.; Henderson, T.; Martone, W.J. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. National Nosocomial Infections Surveillance System. Am. J. Med. 1991, 91, 86S–89S. [Google Scholar] [CrossRef]
- Alastruey-Izquierdo, A.; Mandelblat, M.; Ben Ami, R.; Perlin, D.S.; Segal, E. Multilocus sequence typing of Candida albicans isolates from candidemia and superficial candidiasis in Israel. Med. Mycol. 2013. [Google Scholar] [CrossRef] [PubMed]
- Mah, T.F.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Lu, D.; Liu, Q.; Zhang, T.; Cai, Y.; Yin, Y.; Jiang, G. Stable silver isotope fractionation in the natural transformation process of silver nanoparticles. Nat. Nanotechnol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Richter, A.P.; Brown, J.S.; Bharti, B.; Wang, A.; Gangwal, S.; Houck, K.; Cohen Hubal, E.A.; Paunov, V.N.; Stoyanov, S.D.; Velev, O.D. An environmentally benign antimicrobial nanoparticle based on a silver-infused lignin core. Nat. Nanotechnol. 2015, 10, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Controlled release: Tuning up silver. Nat Nano 2010.
- Hwang, I.S.; Lee, J.; Hwang, J.H.; Kim, K.J.; Lee, D.G. Silver nanoparticles induce apoptotic cell death in Candida albicans through the increase of hydroxyl radicals. FEBS J. 2012, 279, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Wahab, R.; Dwivedi, S.; Khan, F.; Mishra, Y.K.; Hwang, I.H.; Shin, H.S.; Musarrat, J.; Al-Khedhairy, A.A. Statistical analysis of gold nanoparticle-induced oxidative stress and apoptosis in myoblast (C2C12) cells. Colloids Surf. B Biointerfaces 2014, 123, 664–672. [Google Scholar] [CrossRef] [PubMed]
- Wahab, R.; Khan, F.; Yang, Y.B.; Hwang, I.H.; Shin, H.S.; Ahmad, J.; Dwivedi, S.; Khan, S.T.; Siddiqui, M.A.; Saquib, Q.; et al. Zinc oxide quantum dots: Multifunctional candidates for arresting C2C12 cancer cells and their role towards caspase 3 and 7 genes. RSC Adv. 2016, 6, 26111–26120. [Google Scholar] [CrossRef]
- Singh, B.N.; Prateeksha; Upreti, D.K.; Singh, B.R.; Defoirdt, T.; Gupta, V.K.; De Souza, A.O.; Singh, H.B.; Barreira, J.C.; Ferreira, I.C.; et al. Bactericidal, quorum quenching and anti-biofilm nanofactories: A new niche for nanotechnologists. Crit. Rev. Biotechnol. 2017, 37, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Vanhaecke, F. Nanoecotoxicology: Nanoparticle behaviour dissected. Nat. Nanotechnol. 2016. [Google Scholar] [CrossRef]
- Singh, S.; Saikia, J.P.; Buragohain, A.K. A novel ‘green’ synthesis of colloidal silver nanoparticles (SNP) using Dillenia indica fruit extract. Colloids Surf. B Biointerfaces 2013, 102, 83–85. [Google Scholar] [CrossRef]
- Eckhardt, S.; Brunetto, P.S.; Gagnon, J.; Priebe, M.; Giese, B.; Fromm, K.M. Nanobio Silver: Its Interactions with Peptides and Bacteria, and Its Uses in Medicine. Chem. Rev. 2013, 113, 4708–4754. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Chelliah, R.; Shanmugam, S.; Varukattu, N.B.; Oh, D.H.; Kathiresan, K.; Wang, M.H. Green synthesis and characterization of biologically active nanosilver from seed extract of Gardenia jasminoides Ellis. J. Photochem. Photobiol. B 2018, 185, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Wang, M.H. Trichoderma based synthesis of anti-pathogenic silver nanoparticles and their characterization, antioxidant and cytotoxicity properties. Microb. Pathog. 2018, 114, 269–273. [Google Scholar] [CrossRef] [PubMed]
- Saravanakumar, K.; Shanmugam, S.; Varukattu, N.B.; MubarakAli, D.; Kathiresan, K.; Wang, M.H. Biosynthesis and characterization of copper oxide nanoparticles from indigenous fungi and its effect of photothermolysis on human lung carcinoma. J. Photochem. Photobiol. B 2019, 190, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.N.; Prateeksha; Gupta, V.K.; Chen, J.Y.; Atanasov, A.G. Organic Nanoparticle-Based Combinatory Approaches for Gene Therapy. Trends Biotechnol. 2017, 35, 1121–1124. [Google Scholar] [CrossRef]
- Singh, B.N.; Singh, B.R.; Gupta, V.K.; Kharwar, R.N.; Pecoraro, L. Coating with Microbial Hydrophobins: A Novel Approach to Develop Smart Drug Nanoparticles. Trends Biotechnol. 2018, 36, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, D.R.; Silva, S.; Negri, M.; Gorup, L.F.; de Camargo, E.R.; Oliveira, R.; Barbosa, D.B.; Henriques, M. Silver nanoparticles: Influence of stabilizing agent and diameter on antifungal activity against Candida albicans and Candida glabrata biofilms. Lett. Appl. Microbiol. 2012, 54, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Panacek, A.; Kolar, M.; Vecerova, R.; Prucek, R.; Soukupova, J.; Krystof, V.; Hamal, P.; Zboril, R.; Kvitek, L. Antifungal activity of silver nanoparticles against Candida spp. Biomaterials 2009, 30, 6333–6340. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.R.; Singh, B.N.; Khan, W.; Singh, H.B.; Naqvi, A.H. ROS-mediated apoptotic cell death in prostate cancer LNCaP cells induced by biosurfactant stabilized CdS quantum dots. Biomaterials 2012, 33, 5753–5767. [Google Scholar] [CrossRef] [PubMed]
- Moretti, M.L.; Trabasso, P.; Lyra, L.; Fagnani, R.; Resende, M.R.; de Oliveira Cardoso, L.G.; Schreiber, A.Z. Is the incidence of candidemia caused by Candida glabrata increasing in Brazil? Five-year surveillance of Candida bloodstream infection in a university reference hospital in southeast Brazil. Med. Mycol. 2013, 51, 225–230. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Deeba, F.; Pandey, A.K.; Pandey, V. Organ Specific Proteomic Dissection of Selaginella bryopteris Undergoing Dehydration and Rehydration. Front. Plant. Sci. 2016, 7, 425. [Google Scholar] [CrossRef] [PubMed]
- Deeba, F.; Pandey, A.K.; Ranjan, S.; Mishra, A.; Singh, R.; Sharma, Y.K.; Shirke, P.A.; Pandey, V. Physiological and proteomic responses of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol. Biochem. 2012, 53, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, H.; Ohishi, N.; Yagi, K. Assay for lipid peroxides in animal tissues by thiobarbituric acid reaction. Anal. Biochem. 1979, 95, 351–358. [Google Scholar] [CrossRef]
- Sinha, A.K. Colorimetric assay of catalase. Anal. Biochem. 1972, 47, 389–394. [Google Scholar] [CrossRef]
- Kakkar, P.; Das, B.; Viswanathan, P.N. A modified spectrophotometric assay of superoxide dismutase. Indian J. Biochem. Biophys. 1984, 21, 130–132. [Google Scholar]
- Tietze, F. Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: Applications to mammalian blood and other tissues. Anal. Biochem. 1969, 27, 502–522. [Google Scholar] [CrossRef]
- Mongkolsuk, S.; Loprasert, S.; Whangsuk, W.; Fuangthong, M.; Atichartpongkun, S. Characterization of transcription organization and analysis of unique expression patterns of an alkyl hydroperoxide reductase C gene (ahpC) and the peroxide regulator operon ahpF-oxyR-orfX from Xanthomonas campestris pv. phaseoli. J. Bacteriol. 1997, 179, 3950–3955. [Google Scholar] [CrossRef] [PubMed]
- Arunachalam, K.D.; Annamalai, S.K.; Hari, S. One-step green synthesis and characterization of leaf extract-mediated biocompatible silver and gold nanoparticles from Memecylon umbellatum. Int. J. Nanomedicine 2013, 8, 307–1315. [Google Scholar] [CrossRef]
- Link, S.; El-Sayed, M.A. Optical properties and ultrafast dynamics of metallic nanocrystals. Annu. Rev. Phys Chem 2003, 54, 331–366. [Google Scholar] [CrossRef]
- Choi, Y.J.; Luo, T.J.M. Electrochemical Properties of Silver Nanoparticle Doped Aminosilica Nanocomposite. Int. J. Electrochem. Sci. 2011, 2011. [Google Scholar] [CrossRef]
- Wu, Y.N.; Yang, L.X.; Shi, X.Y.; Li, I.C.; Biazik, J.M.; Ratinac, K.R.; Chen, D.H.; Thordarson, P.; Shieh, D.B.; Braet, F. The selective growth inhibition of oral cancer by iron core-gold shell nanoparticles through mitochondria-mediated autophagy. Biomaterials 2011, 32, 4565–4573. [Google Scholar] [CrossRef] [PubMed]
- Adrain, C.; Martin, S.J. The mitochondrial apoptosome: A killer unleashed by the cytochrome seas. Trends Biochem Sci 2001, 26, 390–397. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, Y.; Tian, Y.; Zhang, W.; Lu, X.; Jiang, X. The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli. Biomaterials 2012, 33, 2327–2333. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Sudbery, I.; Ramsdale, M. Apoptosis induced by environmental stresses and amphotericin B in Candida albicans. Proc. Natl. Acad. Sci. USA 2003, 100, 14327–14332. [Google Scholar] [CrossRef] [PubMed]
- Phillips, A.J.; Crowe, J.D.; Ramsdale, M. Ras pathway signaling accelerates programmed cell death in the pathogenic fungus Candida albicans. Proc. Natl. Acad. Sci. USA 2006, 103, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Mollinedo, F. Lipid raft involvement in yeast cell growth and death. Front. Oncol. 2012, 2, 140. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Park, T.S.; Chio, L.C.; Fischl, A.S.; Ye, X.S. Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol. Cell Biol. 2003, 23, 163–177. [Google Scholar] [CrossRef] [PubMed]
- Madeo, F.; Herker, E.; Maldener, C.; Wissing, S.; Lachelt, S.; Herlan, M.; Fehr, M.; Lauber, K.; Sigrist, S.J.; Wesselborg, S.; et al. A caspase-related protease regulates apoptosis in yeast. Mol. Cell 2002, 9, 911–917. [Google Scholar] [CrossRef]
- Cabezon, V.; Vialas, V.; Gil-Bona, A.; Reales-Calderon, J.A.; Martinez-Gomariz, M.; Gutierrez-Blazquez, D.; Monteoliva, L.; Molero, G.; Ramsdale, M.; Gil, C. Apoptosis of Candida albicans during the Interaction with Murine Macrophages: Proteomics and Cell-Death Marker Monitoring. J. Proteome. Res. 2016, 15, 1418–1434. [Google Scholar] [CrossRef]
- Leger, T.; Garcia, C.; Ounissi, M.; Lelandais, G.; Camadro, J.M. The Metacaspase (Mca1p) has a Dual Role in Farnesol-induced Apoptosis in Candida albicans. Mol. Cell Proteomics 2015, 14, 93–108. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.; Dagher, M.C.; Shacter, E. Rho GDP dissociation inhibitor protects cancer cells against drug-induced apoptosis. Cancer Res. 2005, 65, 6054–6062. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Dang, C.V. Multifaceted roles of glycolytic enzymes. Trends Biochem. Sci. 2005, 30, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Tajima, H.; Tsuchiya, K.; Yamada, M.; Kondo, K.; Katsube, N.; Ishitani, R. Over-expression of GAPDH induces apoptosis in COS-7 cells transfected with cloned GAPDH cDNAs. Neuroreport 1999, 10, 2029–2033. [Google Scholar] [CrossRef] [PubMed]
- Shashidharan, P.; Chalmers-Redman, R.M.; Carlile, G.W.; Rodic, V.; Gurvich, N.; Yuen, T.; Tatton, W.G.; Sealfon, S.C. Nuclear translocation of GAPDH-GFP fusion protein during apoptosis. Neuroreport 1999, 10, 1149–1153. [Google Scholar] [CrossRef]
- Magherini, F.; Tani, C.; Gamberi, T.; Caselli, A.; Bianchi, L.; Bini, L.; Modesti, A. Protein expression profiles in Saccharomyces cerevisiae during apoptosis induced by H2O2. Proteomics 2007, 7, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Tarze, A.; Deniaud, A.; Le Bras, M.; Maillier, E.; Molle, D.; Larochette, N.; Zamzami, N.; Jan, G.; Kroemer, G.; Brenner, C. GAPDH, a novel regulator of the pro-apoptotic mitochondrial membrane permeabilization. Oncogene 2007, 26, 2606–2620. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Proestou, G.; Bourbeau, D.; Wang, E. Rapid up-regulation of peptide elongation factor EF-1 α protein levels is an immediate early event during oxidative stress-induced apoptosis. Exp. Cell Res. 2000, 259, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Nie, X.Y.; Ding, Y.F.; Chen, J.Y. Asc1, a WD-repeat protein, is required for hyphal development and virulence in Candida albicans. Acta Bioch. Bioph. Sin. 2010, 42, 793–800. [Google Scholar] [CrossRef] [PubMed]
- Alberti-Segui, C.; Morales, A.J.; Xing, H.; Kessler, M.M.; Willins, D.A.; Weinstock, K.G.; Cottarel, G.; Fechtel, K.; Rogers, B. Identification of potential cell-surface proteins in Candida albicans and investigation of the role of a putative cell-surface glycosidase in adhesion and virulence. Yeast 2004, 21, 285–302. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef] [PubMed]
- Moller, I.; Jung, M.; Beatrix, B.; Levy, R.; Kreibich, G.; Zimmermann, R.; Wiedmann, M.; Lauring, B. A general mechanism for regulation of access to the translocon: Competition for a membrane attachment site on ribosomes. Proc. Natl. Acad. Sci. USA 1998, 95, 13425–13430. [Google Scholar] [CrossRef] [PubMed]
- Funfschilling, U.; Rospert, S. Nascent polypeptide-associated complex stimulates protein import into yeast mitochondria. Mol. Biol. Cell 1999, 10, 3289–3299. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Yu, X.; Wang, B.; Zhou, H.; Ouyang, H.; Ming, J.; Jin, C. Characterization of the Aspergillus fumigatus phosphomannose isomerase Pmi1 and its impact on cell wall synthesis and morphogenesis. Microbiology 2009, 155, 3281–3293. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, S.; Tang, Z. Polyoxometalate-based functional nanostructured films: Current progress and future prospects. Nano Today 2010, 5, 267–281. [Google Scholar] [CrossRef]
- Prouzet-Mauleon, V.; Monribot-Espagne, C.; Boucherie, H.; Lagniel, G.; Lopez, S.; Labarre, J.; Garin, J.; Lauquin, G.J. Identification in Saccharomyces cerevisiae of a new stable variant of alkyl hydroperoxide reductase 1 (Ahp1) induced by oxidative stress. J. Biol. Chem. 2002, 277, 4823–4830. [Google Scholar] [CrossRef] [PubMed]
- Azam, A.; Ahmed, A.S.; Oves, M.; Khan, M.S.; Habib, S.S.; Memic, A. Antimicrobial activity of metal oxide nanoparticles against Gram-positive and Gram-negative bacteria: A comparative study. Int. J. Nanomedicine 2012, 7, 6003–6009. [Google Scholar] [CrossRef] [PubMed]
- Rockenfeller, P.; Madeo, F. Apoptotic death of ageing yeast. Exp. Gerontol. 2008, 43, 876–881. [Google Scholar] [CrossRef] [PubMed]
- Ahamed, M.; Posgai, R.; Gorey, T.J.; Nielsen, M.; Hussain, S.M.; Rowe, J.J. Silver nanoparticles induced heat shock protein 70, oxidative stress and apoptosis in Drosophila melanogaster. Toxicol. Appl. Pharmacol. 2010, 242, 263–269. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Prateeksha; Singh, B.R.; Gupta, V.K.; Deeba, F.; Bajpai, R.; Pandey, V.; Naqvi, A.H.; Upreti, D.K.; Gathergood, N.; Jiang, Y.; et al. Non-Toxic and Ultra-Small Biosilver Nanoclusters Trigger Apoptotic Cell Death in Fluconazole-Resistant Candida albicans via Ras Signaling. Biomolecules 2019, 9, 47. https://doi.org/10.3390/biom9020047
Prateeksha, Singh BR, Gupta VK, Deeba F, Bajpai R, Pandey V, Naqvi AH, Upreti DK, Gathergood N, Jiang Y, et al. Non-Toxic and Ultra-Small Biosilver Nanoclusters Trigger Apoptotic Cell Death in Fluconazole-Resistant Candida albicans via Ras Signaling. Biomolecules. 2019; 9(2):47. https://doi.org/10.3390/biom9020047
Chicago/Turabian StylePrateeksha, Braj Raj Singh, Vijai Kumar Gupta, Farah Deeba, Rajesh Bajpai, Vivek Pandey, Alim H. Naqvi, Dalip Kumar Upreti, Nicholas Gathergood, Yueming Jiang, and et al. 2019. "Non-Toxic and Ultra-Small Biosilver Nanoclusters Trigger Apoptotic Cell Death in Fluconazole-Resistant Candida albicans via Ras Signaling" Biomolecules 9, no. 2: 47. https://doi.org/10.3390/biom9020047
APA StylePrateeksha, Singh, B. R., Gupta, V. K., Deeba, F., Bajpai, R., Pandey, V., Naqvi, A. H., Upreti, D. K., Gathergood, N., Jiang, Y., El Enshasy, H. A., Sholkamy, E. N., Mostafa, A. A., Hesham, A. E.-L., & Singh, B. N. (2019). Non-Toxic and Ultra-Small Biosilver Nanoclusters Trigger Apoptotic Cell Death in Fluconazole-Resistant Candida albicans via Ras Signaling. Biomolecules, 9(2), 47. https://doi.org/10.3390/biom9020047









