Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin
Abstract
1. Introduction
2. Materials and Methods
2.1. Size-Exclusion Chromatography
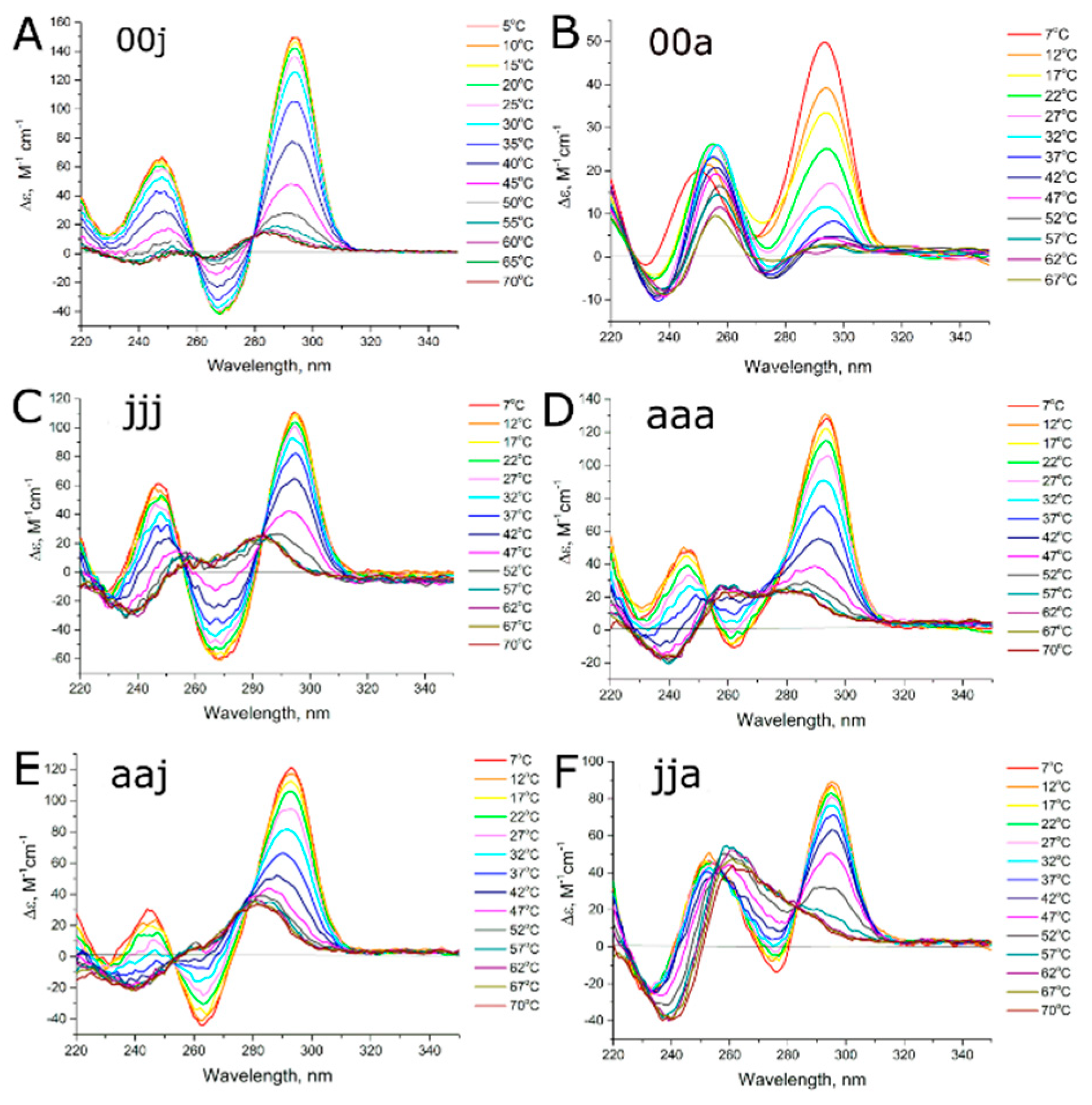
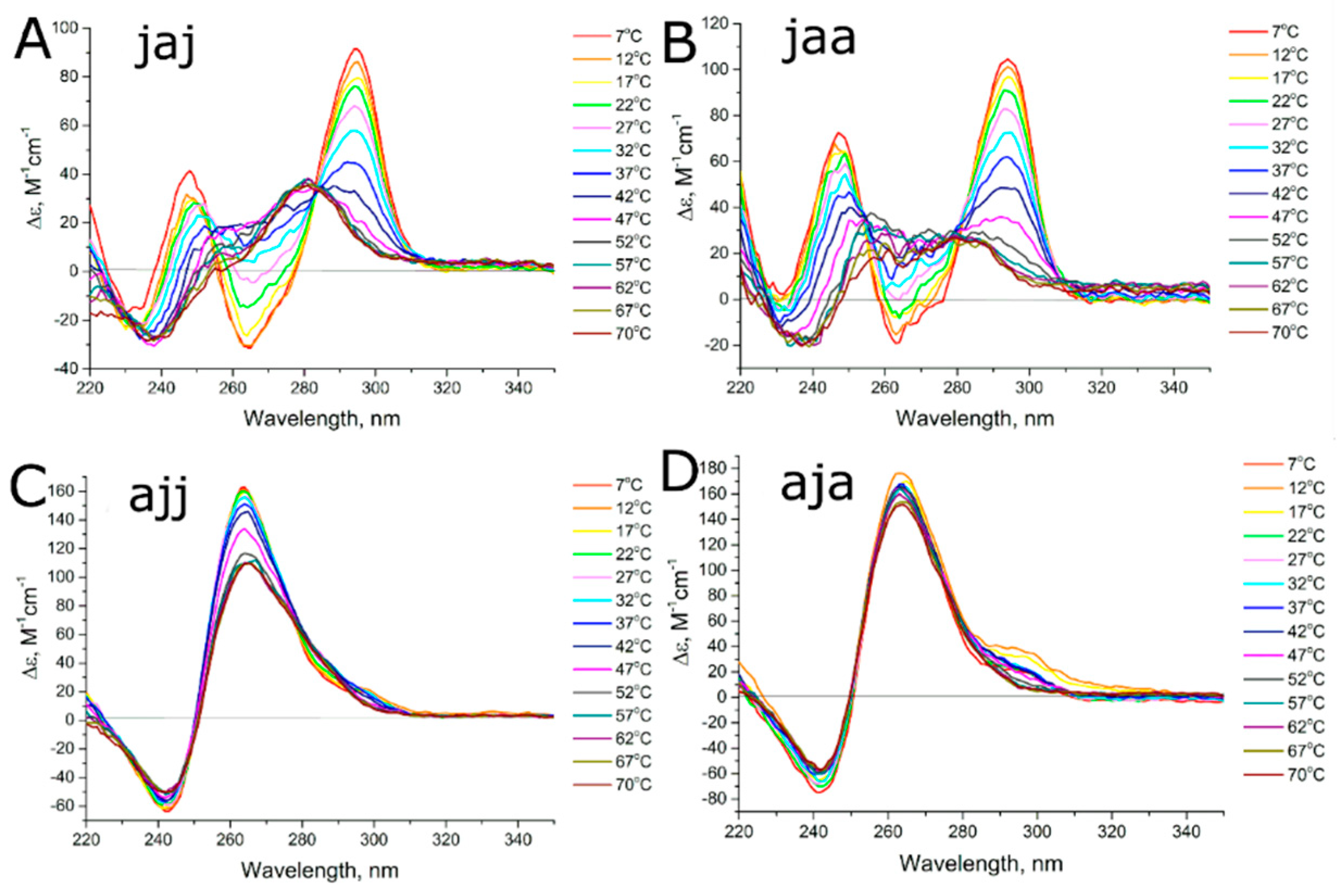
2.2. Circular Dichroism and UV Melting
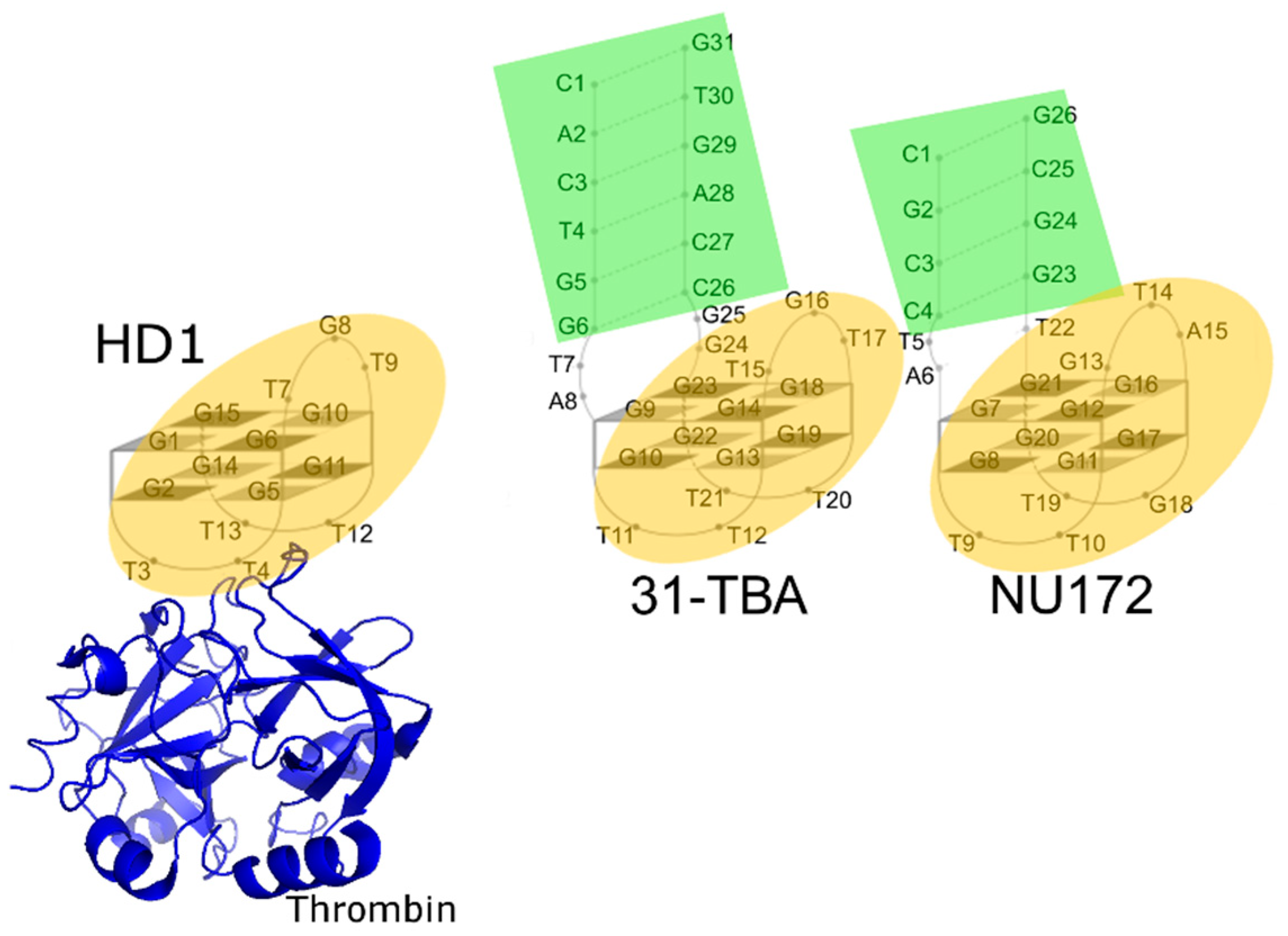
2.3. Analysis of Structures of Aptamer–Thrombin Complexes
3. Results
3.1. Testing Oligomeric Composition with Size-Exclusion Chromatography
3.2. Studying the Conformational Homogeneity of Aptamers and Stability of Their Modules with Circular Dichroism and UV Spectroscopies
3.3. Thermodynamics of Aptamer Unfolding
3.4. Comparison of the Structures of Aptamer–Thrombin Complexes
4. Discussion
4.1. Conformational Polymorphism as a Reason for Reduced Functional Activity
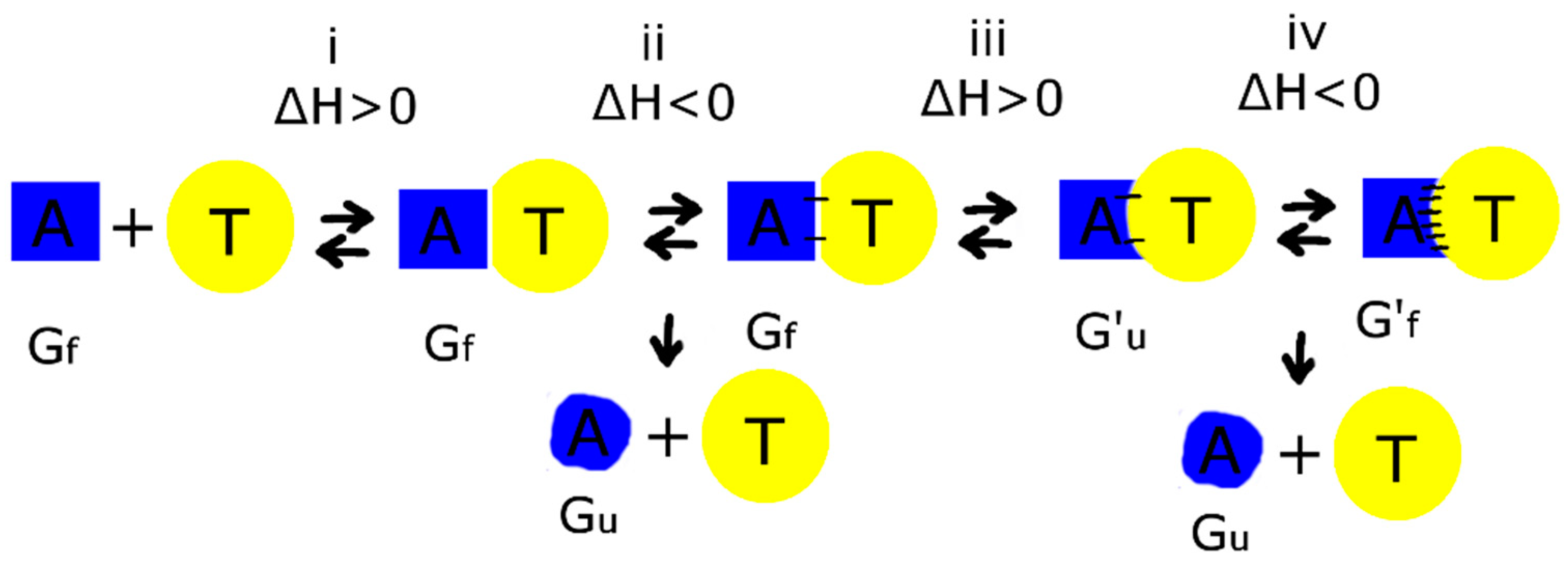
4.2. Thermodynamic Aspects of Bimodular Aptamers
4.3. Possible Mechanism for Complex Formation between Aptamer and Thrombin
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nimjee, S.M.; Rusconi, C.P.; Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005, 56, 555–583. [Google Scholar] [CrossRef] [PubMed]
- Woodruff, R.S.; Sullenger, B.A. Modulation of the coagulation cascade using aptamers. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2083–2091. [Google Scholar] [CrossRef] [PubMed]
- Nimjee, S.M.; Povsic, T.J.; Sullenger, B.A.; Becker, R.C. Translation and clinical development of antithrombotic aptamers. Nucleic Acid Ther. 2016, 26, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Golovin, A.; Pavlova, G.; Kopylov, A. Development of antithrombotic aptamers: From recognizing elements to drugs. Curr. Pharm. Des. 2016, 22, 5163–5176. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Ustinov, N.; Golovin, A.; Pavlova, G.; Kopylov, A. G-quadruplex aptamers to human thrombin versus other direct thrombin inhibitors: The focus on mechanism of action and drug efficiency as anticoagulants. Curr. Med. Chem. 2016, 23, 2230–2244. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Samoylenkova, N.; Revishchin, A.; Turashev, A.; Gordeychuk, I.; Golovin, A.; Kopylov, A.; Pavlova, G. The evaluation of pharmacodynamics and pharmacokinetics of anti-thrombin DNA aptamer RA-36. Front. Pharmacol. 2017, 8, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Abeydeera, N.D.; Egli, M.; Cox, N.; Mercier, K.; Conde, J.N.; Pallan, P.S.; Mizurini, D.M.; Sierant, M.; Hibti, F.E.; Hassell, T.; et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016, 44, 8052–8064. [Google Scholar] [CrossRef]
- Novoseltseva, A.; Zavyalova, E.; Golovin, A.; Kopylov, A. An insight into aptamer–protein complexes. Aptamers 2018, 2, 1–19. [Google Scholar]
- Zavyalova, E.; Golovin, A.; Pavlova, G.; Kopylov, A. Module-activity relationship of G-quadruplex based DNA aptamers for human thrombin. Curr. Med. Chem. 2013, 20, 4836–4843. [Google Scholar] [CrossRef]
- Zavyalova, E.; Kopylov, A. G-quadruplexes and i-motifs as scaffolds for molecular engineering of DNA aptamers. In G-Quadruplex Structures, Formation and Roles in Biology; Santos, H., Ed.; Nova Publishers: New York, NY, USA, 2016; pp. 53–80. ISBN 978-1-63485-512-9. [Google Scholar]
- Musumeci, D.; Montesarchio, D. Polyvalent nucleic acid aptamers and modulation of their activity: A focus on the thrombin binding aptamer. Pharmacol. Ther. 2012, 136, 202–215. [Google Scholar] [CrossRef]
- Spiridonova, V.A.; Barinova, K.V.; Glinkina, K.A.; Melnichuk, A.V.; Gainutdynov, A.A.; Safenkova, I.V.; Dzantiev, B.B. A family of DNA aptamers with varied duplex region length that forms complexes with thrombin and prothrombin. FEBS Lett. 2015, 589, 2043–2049. [Google Scholar] [CrossRef] [PubMed]
- Zavyalova, E.; Tagiltsev, G.; Reshetnikov, R.; Arutyunyan, A.; Kopylov, A. Cation coordination alters the conformation of a thrombin-binding G-quadruplex DNA aptamer that affects inhibition of thrombin. Nucleic Acid Ther. 2016, 26, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Dailey, M.M.; Miller, M.C.; Bates, P.J.; Lane, A.N.; Trent, J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010, 38, 4877–4888. [Google Scholar] [CrossRef] [PubMed]
- Vorlícková, M.; Bednárová, K.; Kejnovská, I.; Kypr, J. Intramolecular and intermolecular guanine quadruplexes of DNA in aqueous salt and ethanol solutions. Biopolymers 2007, 86, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mergny, J.L.; Lacroix, L. Analysis of thermal melting curves. Oligonucleotides 2003, 13, 515–537. [Google Scholar] [CrossRef] [PubMed]
- Vorlíčková, M.; Kejnovská, I.; Bednářová, K.; Renčiuk, D.; Kypr, J. Circular dichroism spectroscopy of DNA: From duplexes to quadruplexes. Chirality 2012, 24, 691–698. [Google Scholar] [CrossRef] [PubMed]
- Dolinnaya, N.G.; Yuminova, A.V.; Spiridonova, V.A.; Arutyunyan, A.M.; Kopylov, A.M. Coexistence of G-quadruplex and duplex domains within the secondary structure of 31-mer DNA thrombin-binding aptamer. J. Biomol. Struct. Dyn. 2012, 30, 524–531. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Spiridonova, V.; Pica, A.; Napolitano, V.; Sica, F. Different duplex/quadruplex junctions determine the properties of anti-thrombin aptamers with mixed folding. Nucleic Acids Res. 2016, 44, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Troisi, R.; Napolitano, V.; Spiridonova, V.; Russo Krauss, I.; Sica, F. Several structural motifs cooperate in determining the highly effective anti-thrombin activity of NU172 aptamer. Nucleic Acids Res. 2018, gky990. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Giancola, C.; Randazzo, A.; Mazzarella, L.; Sica, F. Thrombin-aptamer recognition: A revealed ambiguity. Nucleic Acids Res. 2011, 39, 7858–7867. [Google Scholar] [CrossRef]
- Pica, A.; Russo Krauss, I.; Merlino, A.; Nagatoishi, S.; Sugimoto, N.; Sica, F. Dissecting the contribution of thrombin exosite I in the recognition of thrombin binding aptamer. FEBS J. 2013, 280, 6581–6588. [Google Scholar] [CrossRef] [PubMed]
- Nagatoishi, S.; Sugimoto, N. Interaction of water with the G-quadruplex loop contributes to the binding energy of G-quadruplex to protein. Mol Biosyst. 2012, 8, 2766–2770. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Merlino, A.; Randazzo, A.; Novellino, E.; Mazzarella, L.; Sica, F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012, 40, 8119–8128. [Google Scholar] [CrossRef]
- Pagano, B.; Martino, L.; Randazzo, A.; Giancola, C. Stability and binding properties of a modified thrombin binding aptamer. Biophys J. 2008, 94, 562–569. [Google Scholar] [CrossRef]
- Dolot, R.; Lam, C.H.; Sierant, M.; Zhao, Q.; Liu, F.W.; Nawrot, B.; Egli, M.; Yang, X. Crystal structures of thrombin in complex with chemically modified thrombin DNA aptamers reveal the origins of enhanced affinity. Nucleic Acids Res. 2018, 46, 4819–4830. [Google Scholar] [CrossRef] [PubMed]
- Segura, J.; Fernandez-Fuentes, N. PCRPi-DB: A database of computationally annotated hot spots in protein interfaces. Nucleic Acids Res. 2011, 39, D755–D760. [Google Scholar] [CrossRef]
- Schultze, P.; Macaya, R.F.; Feigon, J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J. Mol. Biol. 1994, 235, 1532–1547. [Google Scholar] [CrossRef] [PubMed]
- Reshetnikov, R.V.; Sponer, J.; Rassokhina, O.I.; Kopylov, A.M.; Tsvetkov, P.O.; Makarov, A.A.; Golovin, A.V. Cation binding to 15-TBA quadruplex DNA is a multiple-pathway cation-dependent process. Nucleic Acids Res. 2011, 39, 9789–9802. [Google Scholar] [CrossRef] [PubMed]
- Russo Krauss, I.; Napolitano, V.; Petraccone, L.; Troisi, R.; Spiridonova, V.; Mattia, C.A.; Sica, F. Duplex/quadruplex oligonucleotides: Role of the duplex domain in the stabilization of a new generation of highly effective anti-thrombin aptamers. Int. J. Biol. Macromol. 2018, 107 Pt B, 1697–1705. [Google Scholar] [CrossRef]
- Chiorcea-Paquim, A.M.; Santos, P.V.; Eritja, R.; Oliveira-Brett, A.M. Self-assembled G-quadruplex nanostructures: AFM and voltammetric characterization. Phys. Chem. Chem. Phys. 2013, 15, 9117–9124. [Google Scholar] [CrossRef] [PubMed]
- Spindler, L.; Rigler, M.; Drevensek-Olenik, I.; Ma’ani Hessari, N.; Webba da Silva, M. Effect of base sequence on G-wire formation in solution. J. Nucleic Acids 2010, 431651. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, H.; Taira, K.I.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors (Basel) 2008, 8, 1090–1098. [Google Scholar] [CrossRef] [PubMed]
- Pica, A.; Russo Krauss, I.; Parente, V.; Tateishi-Karimata, H.; Nagatoishi, S.; Tsumoto, K.; Sugimoto, N.; Sica, F. Through-bond effects in the ternary complexes of thrombin sandwiched by two DNA aptamers. Nucleic Acids Res. 2017, 45, 461–469. [Google Scholar] [CrossRef] [PubMed]
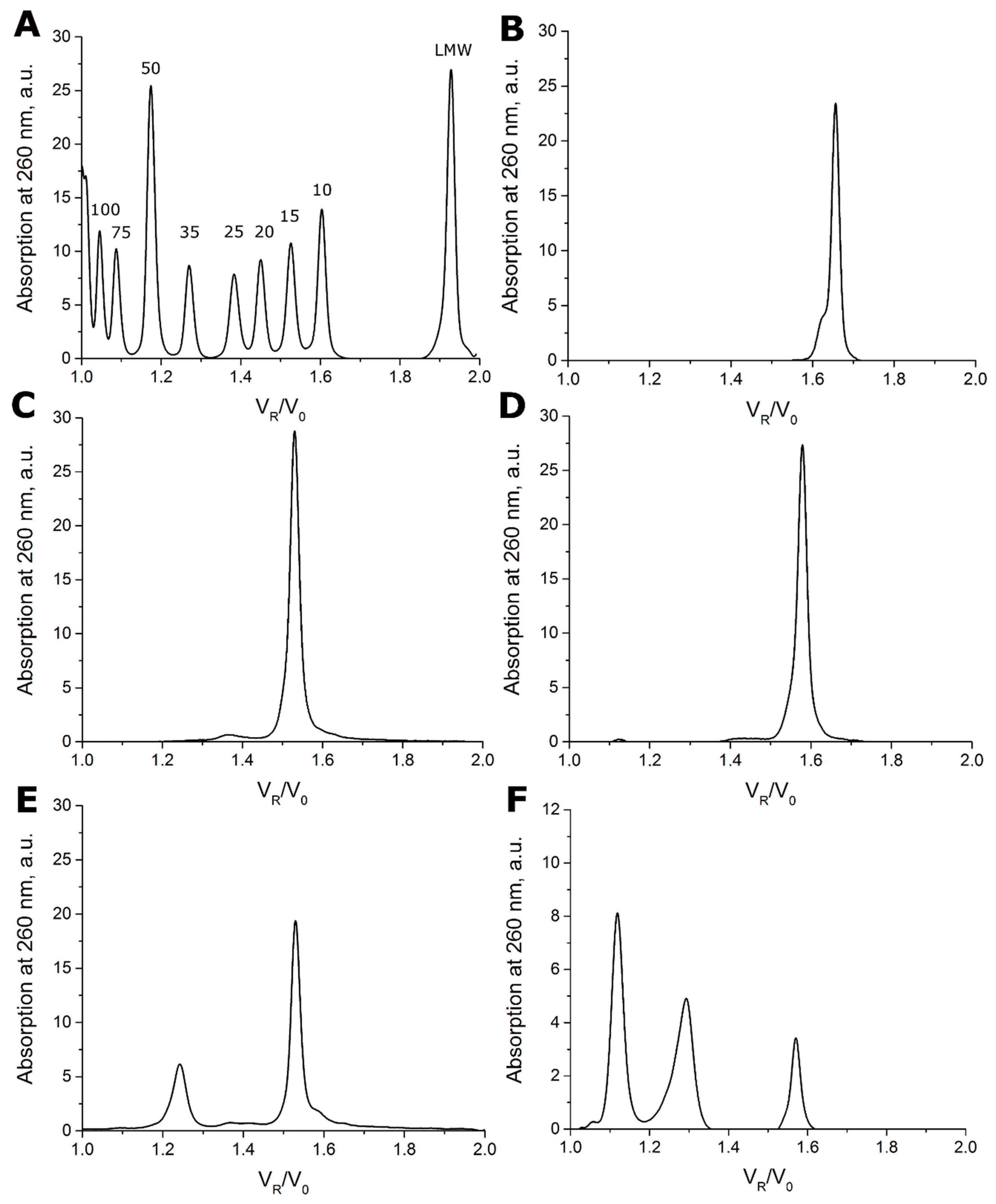
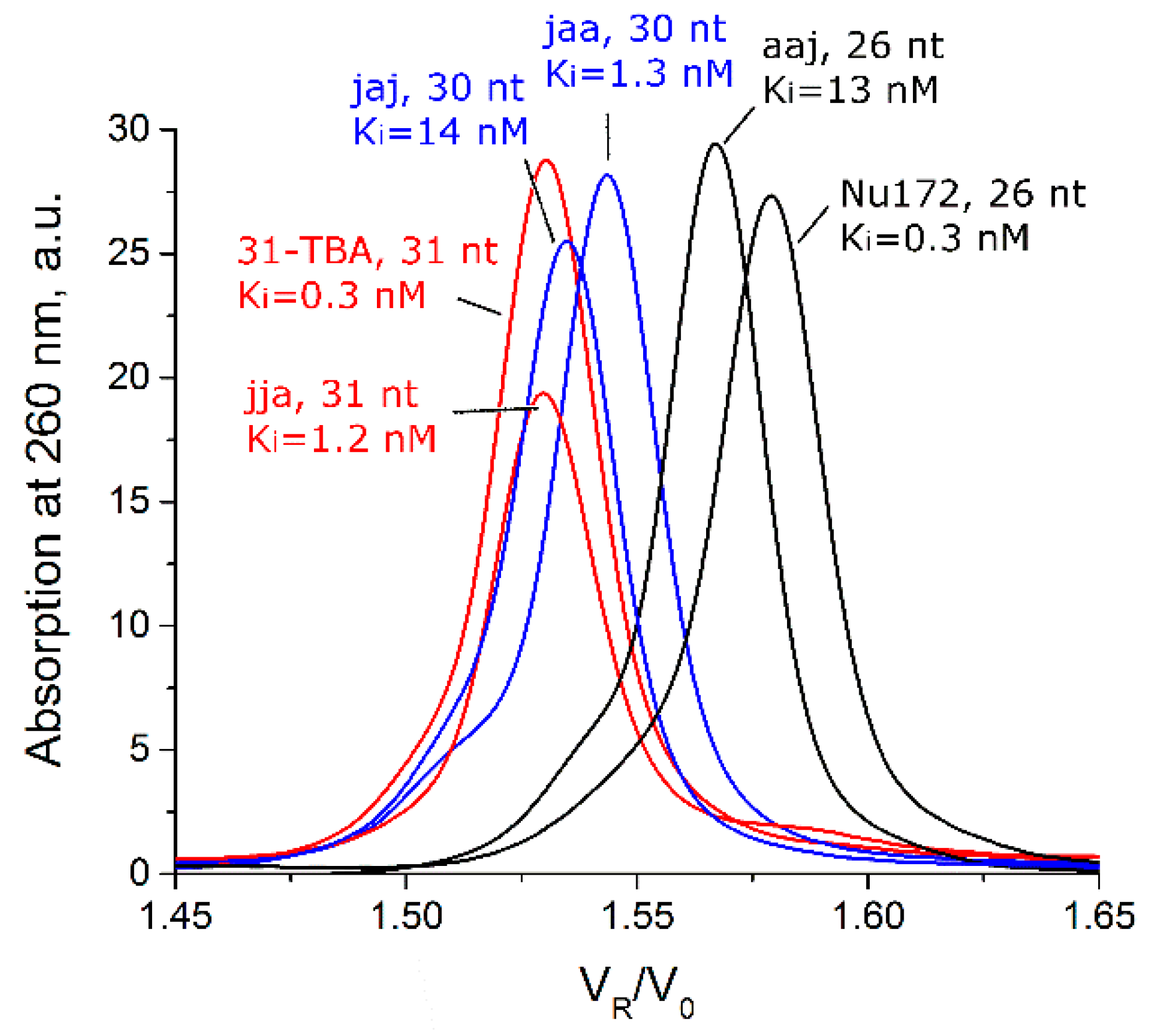
| Aptamer | Monomeric Form | Dimeric Form | Tetrameric Form | ||||||
|---|---|---|---|---|---|---|---|---|---|
| VR/ V0 | Mcalculated/ Mmonomer | Quantity (%) | VR/V0 | Mcalculated/ Mmonomer | Quantity (%) | VR/V0 | Mcalculated/ Mmonomer | Quantity (%) | |
| 31-TBA (jjj) | 1.53 | 0.96 | 100 | - | - | - | - | - | - |
| jaa | 1.54 | 0.91 | 100 | - | - | - | - | - | - |
| jja | 1.53 | 0.93 | 71 | 1.24 | 2.81 | 29 | - | - | - |
| NU172 (aaa) | 1.58 | 0.92 | 100 | - | - | - | - | - | - |
| aaj | 1.57 | 0.97 | 100 | - | - | - | - | - | - |
| jaj | 1.53 | 0.95 | 100 | - | - | - | - | - | - |
| aja | 1.57 | 0.91 | 26 | 1.29 | 2.65 | 59 | 1.12 | 5.16 | 14 |
| ajj | 1.56 | 0.95 | 16 | 1.29 | 2.67 | 41 | 1.12 | 5.29 | 42 |
| Aptamer | G-Pattern | G-Quadruplex Topology | Transition | Melting Temperature ± SD, °C | aKi ± SD, nM [9,10] | |
|---|---|---|---|---|---|---|
| G-Quadruplex Module | Duplex Module | |||||
| 31-TBA (jjj) | 2-2-2-2-4 | A | A → U | 42.7 ± 1.0 | 54.4 ± 1.0 | 0.34 ± 0.10 |
| jaa | 2-2-3-3-2 | A | A → U + P | 41 ± 4 | 54.2 ± 0.8 | 1.34 ± 0.05 |
| jja | 2-2-3-3-4 | A | A → P | 49 ± 2 | 58.9 ± 1.5 | 1.2 ± 0.3 |
| NU172 (aaa) | 2-3-3-2-2 | A | A → U | 37.1 ± 1.0 | 57 ± 3 | 0.29 ± 0.06 |
| HD1 (00j) | 2-2-2-2 | A | A → U | 39.2 ± 0.8 | - | 14.7 ± 1.0 |
| aaj | 2-2-2-2-2 | A | A → U | 32 ± 3 | 58 ± 3 | 13.4 ± 1.8 |
| jaj | 2-2-2-2-2 | A | A → U + P | 34.4 ± 1.0 | 50.8 ± 1.2 | 14.2 ± 0.7 |
| aja | 2-3-3-6 | P + A | A → ? | 49.3 ± 0.8 | n.d. | 13.2 ± 1.6 |
| ajj | 2-2-2-6 | P | P → P | 47 ± 2 | n.d. | 48.6 ± 0.2 |
| NU (00a) | 2-3-3-2 | A | A → U + P | 17 ± 2 | - | 46 ± 3 |
| Aptamer | Transition | ΔHo, kJ/mol | ΔSo, J/mol | Melting Temperature, °C | aKi, nM [9] | ||
|---|---|---|---|---|---|---|---|
| G-Quadruplex Module | ΔT | Duplex Module | |||||
| 31-TBA (jjj) | A → U | −136 ± 12 | −430 ± 40 | 42.7 ± 1.0 | +3.5 | 54.4 ± 1.0 | 0.34 ± 0.10 |
| NU172 (aaa) | A → U | −122 ± 3 | −393 ± 11 | 37.1 ± 1.0 | −2.1 | 57 ± 3 | 0.29 ± 0.06 |
| jja | A → P | −121 ± 11 | −380 ± 30 | 49 ± 2 | +9.8 | 58.9 ± 1.5 | 1.2 ± 0.3 |
| HD1 (00j) | A → U | −138 ± 3 | −443 ±10 | 39.2 ± 0.8 | 0 | - | 14.7 ± 1.0 |
| aaj | A → U | −107 ± 4 | −347 ± 13 | 32 ± 3 | −7.2 | 58 ± 3 | 13.4 ± 1.8 |
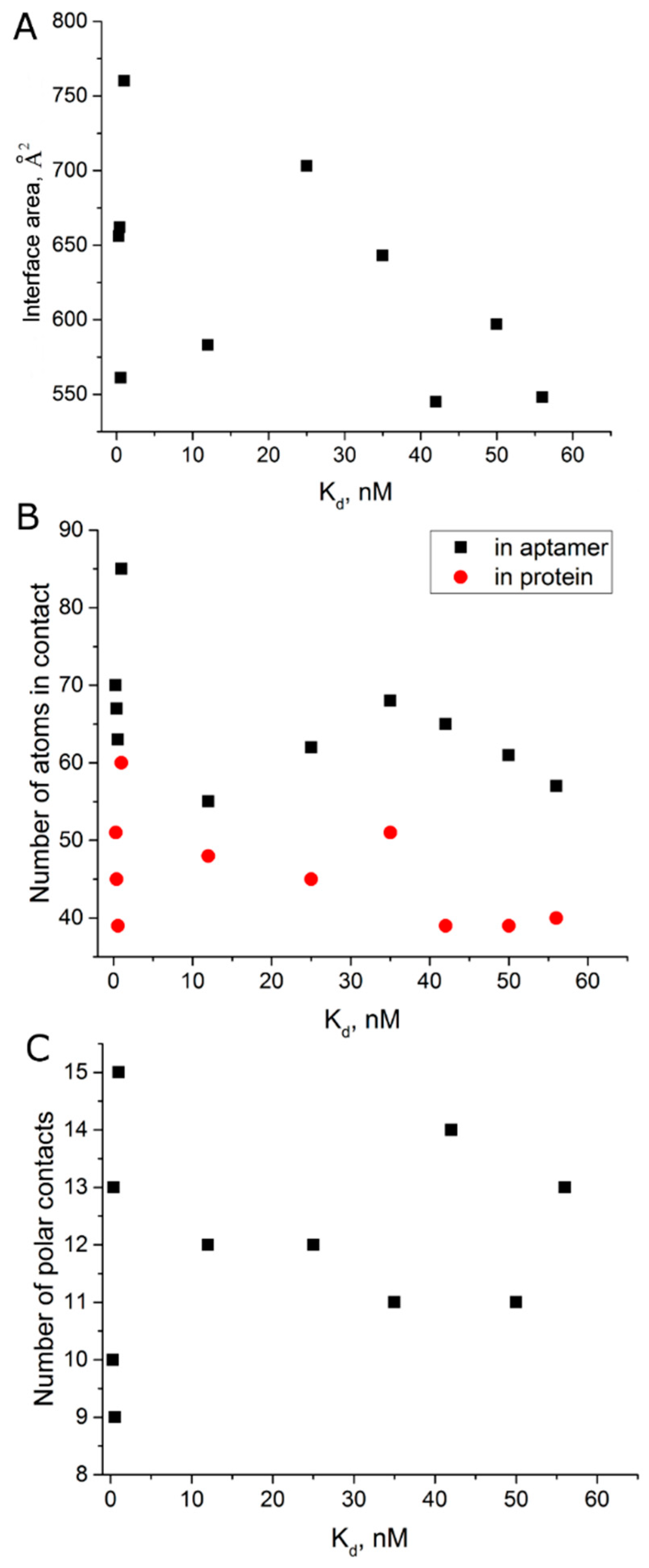
| Aptamer | PDB ID | Kd, nM | Interface Area, Å2 [21] | Number of Polar Contacts | Number of Atoms in 4Å Proximity | |
|---|---|---|---|---|---|---|
| Aptamer | Protein | |||||
| ΔT3 | 4lz4 [22] | 56 [23] | 548 | 13 | 57 | 40 |
| HD1 (Na+) | 4dih [24] | 50 [10] | 597 | 11 | 61 | 39 |
| ΔT12 | 4lz1 [22] | 42 [23] | 545 | 14 | 65 | 39 |
| NU172 (Na+) | 6gn7 [20] | 35 [13] | 643 | 11 | 68 | 51 |
| mTBA | 3qlp [21] | 25 [25] | 703 | 12 | 62 | 45 |
| HD1 | 4dii [24] | 12 [10] | 583 | 12 | 55 | 48 |
| T4W | 6eo6 [26] | 1 [26] | 760 | 15 | 85 | 60 |
| RE31 | 5cmx [19] | 0.56 [12] | 561 | 9 | 63 | 39 |
| NU172 | 6evv [20] | 0.29 [9] | 656 | 10 | 70 | 51 |
| T4K | 6eo7 [26] | 0.39 [26] | 662 | 13 | 67 | 45 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zavyalova, E.G.; Legatova, V.A.; Alieva, R.S.; Zalevsky, A.O.; Tashlitsky, V.N.; Arutyunyan, A.M.; Kopylov, A.M. Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin. Biomolecules 2019, 9, 41. https://doi.org/10.3390/biom9020041
Zavyalova EG, Legatova VA, Alieva RS, Zalevsky AO, Tashlitsky VN, Arutyunyan AM, Kopylov AM. Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin. Biomolecules. 2019; 9(2):41. https://doi.org/10.3390/biom9020041
Chicago/Turabian StyleZavyalova, Elena G., Valeriia A. Legatova, Rugiya Sh. Alieva, Arthur O. Zalevsky, Vadim N. Tashlitsky, Alexander M. Arutyunyan, and Alexey M. Kopylov. 2019. "Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin" Biomolecules 9, no. 2: 41. https://doi.org/10.3390/biom9020041
APA StyleZavyalova, E. G., Legatova, V. A., Alieva, R. S., Zalevsky, A. O., Tashlitsky, V. N., Arutyunyan, A. M., & Kopylov, A. M. (2019). Putative Mechanisms Underlying High Inhibitory Activities of Bimodular DNA Aptamers to Thrombin. Biomolecules, 9(2), 41. https://doi.org/10.3390/biom9020041






