Molecular Dynamics Simulation of Transmembrane Transport of Chloride Ions in Mutants of Channelrhodopsin
Abstract
1. Introduction
2. Materials and Methods
2.1. Simulation-System Preparation
2.2. Molecular Dynamics Simulations
2.3. Steered Molecular Dynamics
2.4. Determining the Potential of Mean Force (PMF) with Umbrella Sampling
3. Results
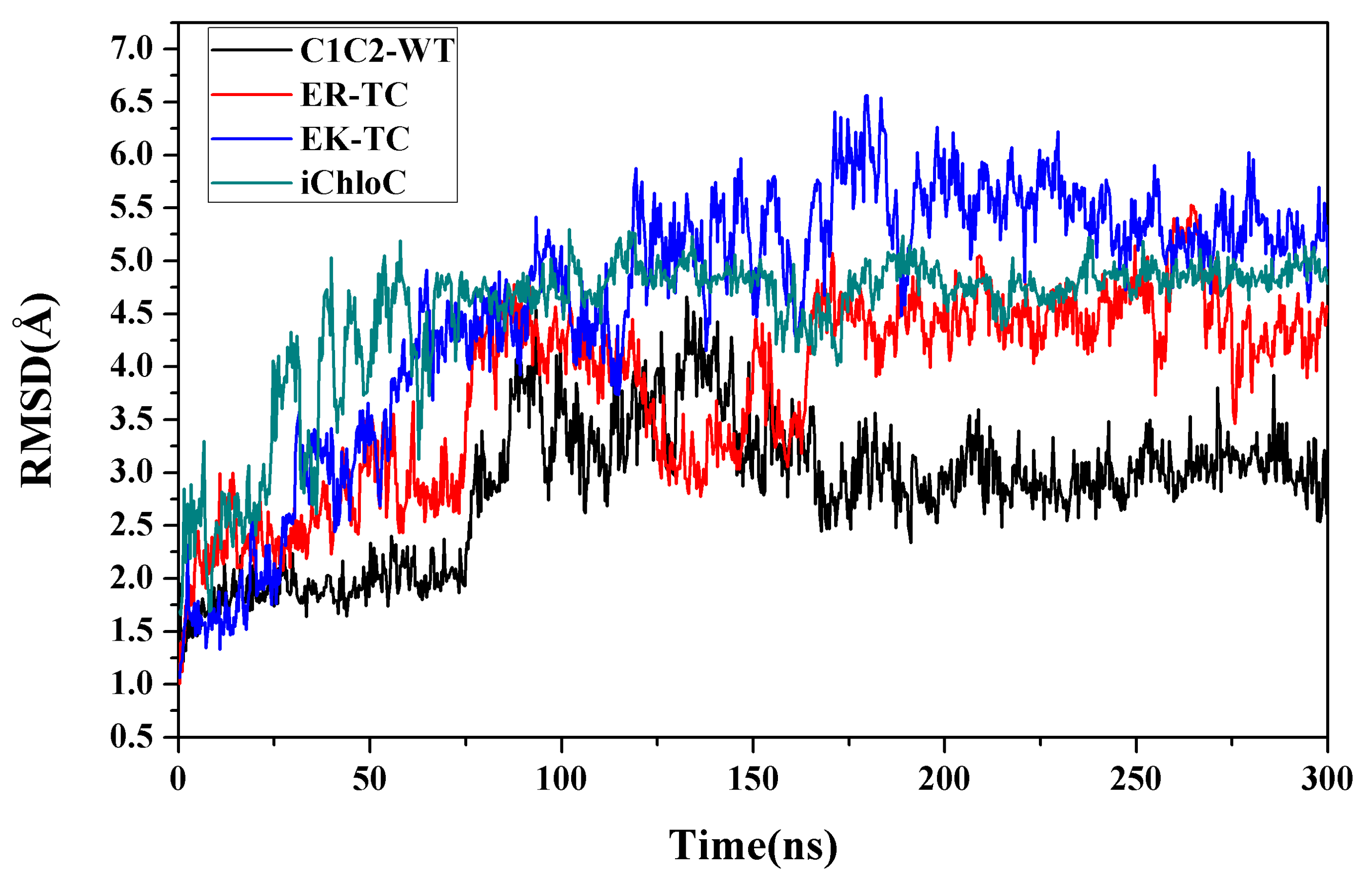
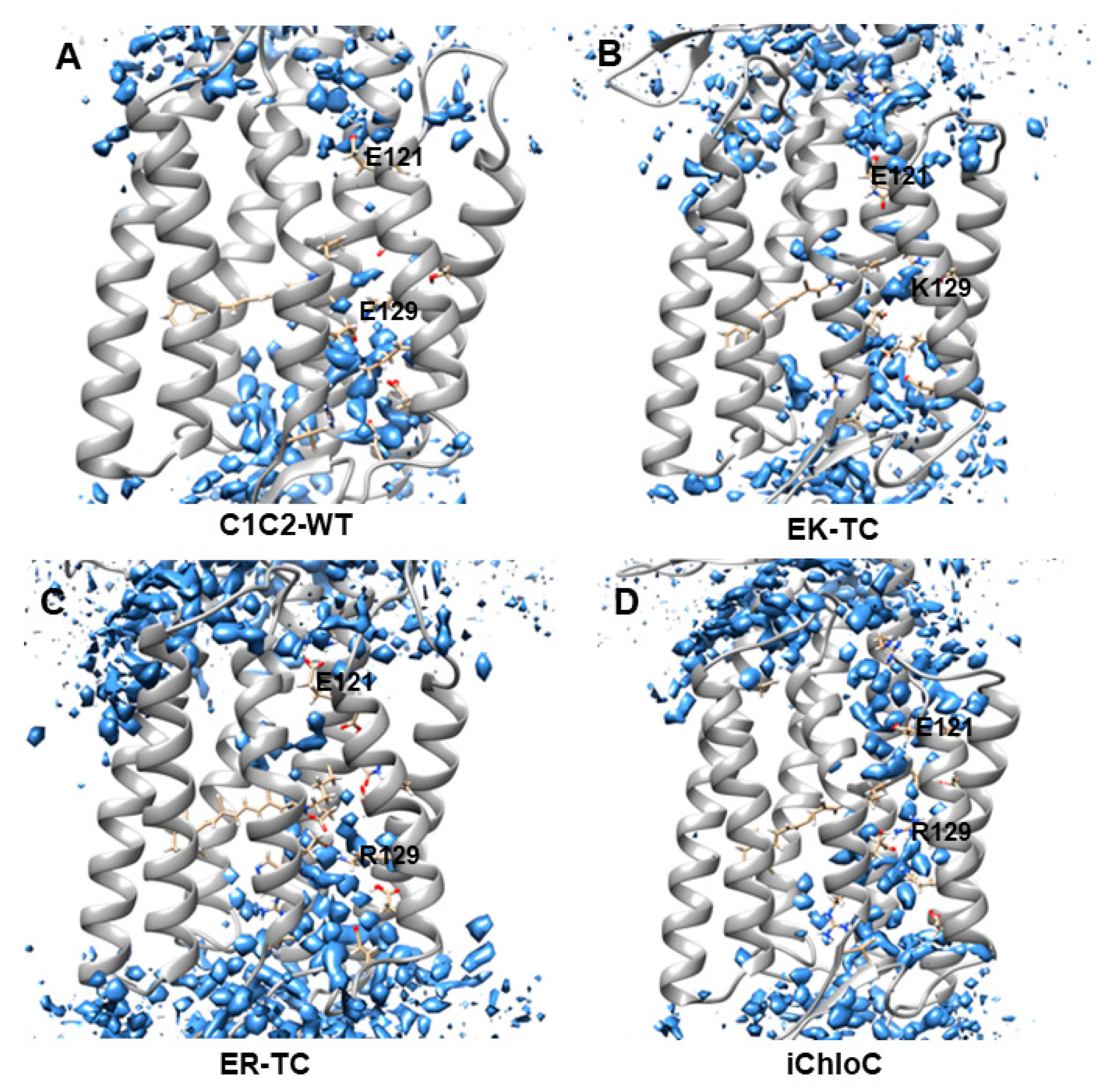
3.1. Water Distributions
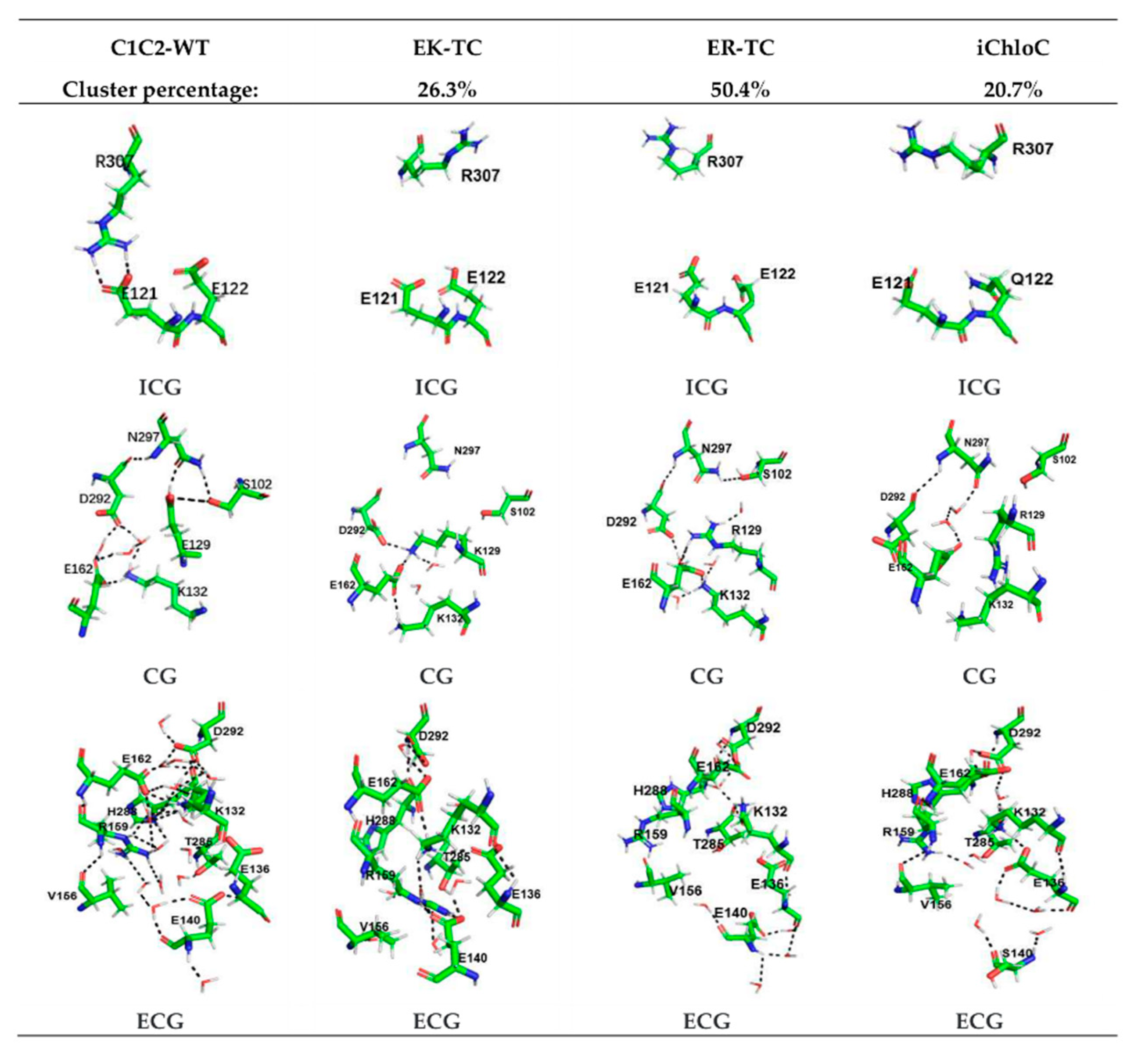
3.2. Structure Changes in Mutants
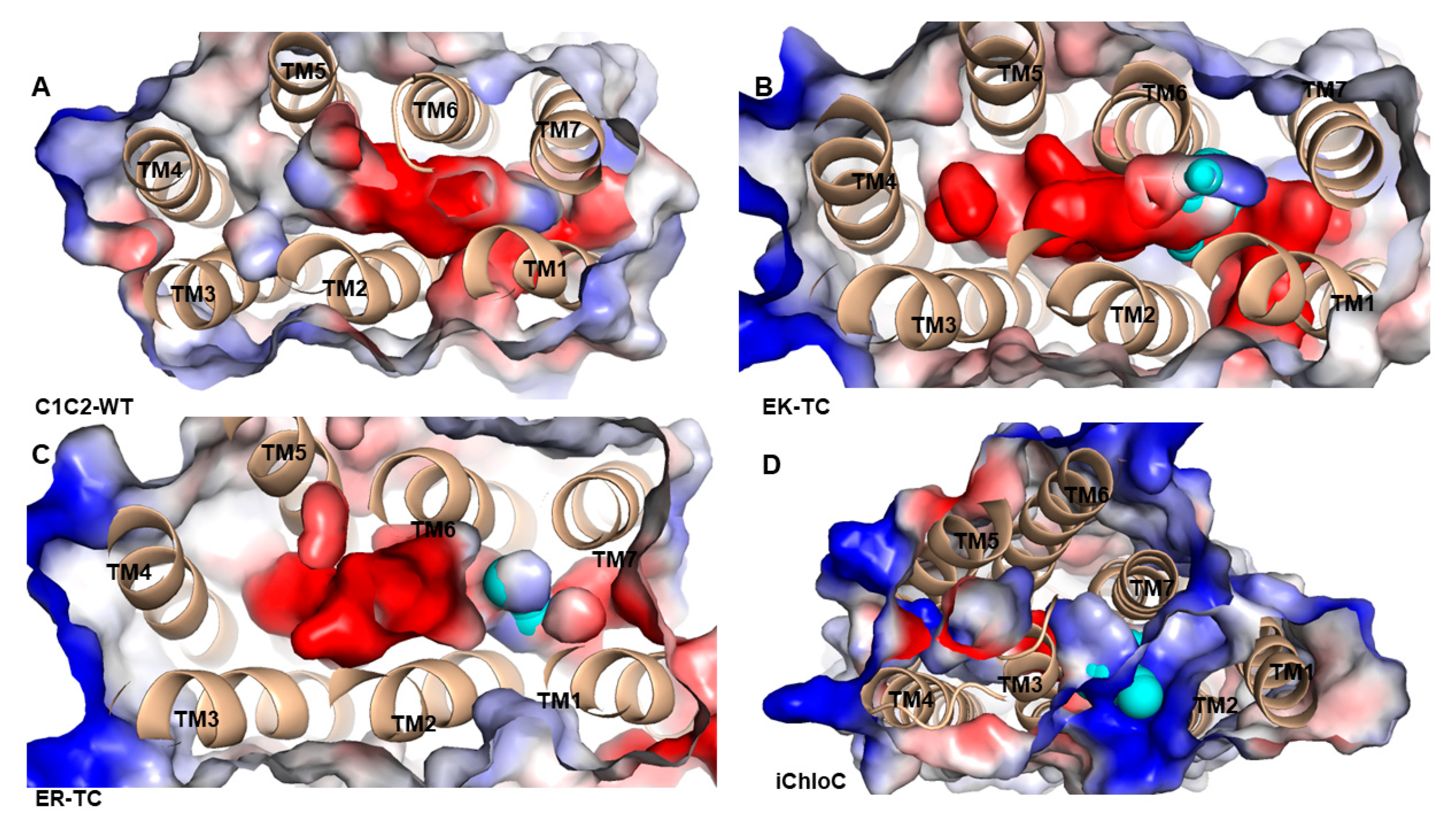
3.3. The Electrostatic Potential Surface of the Channel
3.4. Potential of Mean Force from Umbrella Sampling
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Georg, N.; Doris, O.; Markus, F.; Suneel, K.; Anna Maria, M.; Ernst, B.; Peter, H. Channelrhodopsin-1: A light-gated proton channel in green algae. Science 2002, 296, 2395–2398. [Google Scholar]
- Georg, N.; Tanjef, S.; Wolfram, H.; Suneel, K.; Nona, A.; Peter, B.; Doris, O.; Peter, H.; Ernst, B. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 2003, 100, 13940–13945. [Google Scholar]
- Sineshchekov, O.A.; Jung, K.H.; Spudich, J.L. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 2002, 99, 8689–8694. [Google Scholar] [CrossRef] [PubMed]
- Schneider, F.; Grimm, C.; Hegemann, P. Biophysics of Channelrhodopsin. Annu. Rev. Biophys. 2015, 44, 167–186. [Google Scholar] [CrossRef] [PubMed]
- Saita, M.; Pranga-Sellnau, F.; Resler, T.; Schlesinger, R.; Heberle, J.; Lorenz-Fonfria, V.A. Photoexcitation of the P4(480) State Induces a Secondary Photocycle That Potentially Desensitizes Channelrhodopsin-2. J. Am. Chem. Soc. 2018, 140, 9899–9903. [Google Scholar] [CrossRef] [PubMed]
- Ardevol, A.; Hummer, G. Retinal isomerization and water-pore formation in channelrhodopsin-2. Proc. Natl. Acad. Sci. USA 2018, 115, 3557–3562. [Google Scholar] [CrossRef] [PubMed]
- Kuhne, J.; Eisenhauer, K.; Ritter, E.; Hegemann, P.; Gerwert, K.; Bartl, F. Early formation of the ion-conducting pore in channelrhodopsin-2. Angew. Chem. Int. Ed. Engl. 2015, 54, 4953–4957. [Google Scholar] [CrossRef]
- Flusberg, B.A.; Jung, J.C.; Cocker, E.D.; Anderson, E.P.; Schnitzer, M.J. In vivo brain imaging using a porTable 3.9 gram two-photon fluorescence microendoscope. Optics. Letters 2005, 30, 2272–2274. [Google Scholar] [CrossRef]
- Lin, J.Y.; Lin, M.Z.; Steinbach, P.; Tsien, R.Y. Characterization of Engineered Channelrhodopsin Variants with Improved Properties and Kinetics. Biophys. J. 2009, 96, 1803–1814. [Google Scholar] [CrossRef]
- Berndt, A.; Schoenenberger, P.; Mattis, J.; Tye, K.M.; Deisseroth, K.; Hegemann, P.; Oertner, T.G. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc. Natl. Acad. Sci. USA 2011, 108, 7595–7600. [Google Scholar] [CrossRef]
- Deisseroth, K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat. Neurosci. 2015, 18, 1213–1225. [Google Scholar] [CrossRef] [PubMed]
- Deisseroth, K.; Hegemann, P. The form and function of channelrhodopsin. Science 2017, 357, eaan5544. [Google Scholar] [CrossRef] [PubMed]
- Wiegert, J.S.; Mahn, M.; Prigge, M.; Printz, Y.; Yizhar, O. Silencing Neurons: Tools, Applications, and Experimental Constraints. Neuron 2017, 95, 504–529. [Google Scholar] [CrossRef] [PubMed]
- Berndt, A.; Lee, S.Y.; Ramakrishnan, C.; Deisseroth, K. Structure-Guided Transformation of Channelrhodopsin into a Light-Activated Chloride Channel. Science 2014, 344, 420–424. [Google Scholar] [CrossRef]
- Wietek, J.; Wiegert, J.S.; Adeishvili, N.; Schneider, F.; Watanabe, H.; Tsunoda, S.P.; Vogt, A.; Elstner, M.; Oertner, T.G.; Hegemann, P. Conversion of channelrhodopsin into a light-gated chloride channel. Science 2014, 344, 409–412. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Janz, R.; Liu, X.; Spudich, J.L. Natural light-gated anion channels: A family of microbial rhodopsins for advanced optogenetics. Science 2015, 349, 647–650. [Google Scholar] [CrossRef]
- Berndt, A.; Lee, S.Y.; Wietek, J.; Ramakrishnan, C.; Steinberg, E.E.; Rashid, A.J.; Kim, H.; Park, S.; Santoro, A.; Frankland, P.W. Structural foundations of optogenetics: Determinants of channelrhodopsin ion selectivity. Proc. Natl. Acad. Sci. USA 2015, 113, 822–829. [Google Scholar] [CrossRef]
- Wietek, J.; Beltramo, R.; Scanziani, M.; Hegemann, P.; Oertner, T.G.; Wiegert, J.S. An improved chloride-conducting channelrhodopsin for light-induced inhibition of neuronal activity in vivo. Sci. Rep. 2015, 5, 14807. [Google Scholar] [CrossRef]
- Wietek, J.; Rodriguez-Rozada, S.; Tutas, J.; Tenedini, F.; Grimm, C.; Oertner, T.G.; Soba, P.; Hegemann, P.; Wiegert, J.S. Author Correction: Anion-conducting channelrhodopsins with tuned spectra and modified kinetics engineered for optogenetic manipulation of behavior. Sci. Rep. 2017, 7, 14957. [Google Scholar] [CrossRef]
- Govorunova, E.G.; Sineshchekov, O.A.; Spudich, J.L. Proteomonas sulcata ACR1: A Fast Anion Channelrhodopsin. Photochem. Photobiol. 2016, 92, 257–263. [Google Scholar] [CrossRef]
- Wietek, J.; Broser, M.; Krause, B.S.; Hegemann, P. Identification of a Natural Green Light Absorbing Chloride Conducting Channelrhodopsin from Proteomonas sulcata. J. Biol. Chem. 2016, 291, 4121–4127. [Google Scholar] [CrossRef] [PubMed]
- Govorunova, E.G.; Sineshchekov, O.A.; Rodarte, E.M.; Janz, R.; Morelle, O.; Melkonian, M.; Wong, G.K.-S.; Spudich, J.L. The Expanding Family of Natural Anion Channelrhodopsins Reveals Large Variations in Kinetics, Conductance, and Spectral Sensitivity. Sci. Rep. 2017, 7, 43358. [Google Scholar] [CrossRef] [PubMed]
- Iyer, S.M.; Vesuna, S.; Ramakrishnan, C.; Huynh, K.; Young, S.; Berndt, A.; Lee, S.Y.; Gorini, C.J.; Deisseroth, K.; Delp, S.L. Optogenetic and chemogenetic strategies for sustained inhibition of pain. Sci. Rep. 2016, 6, 30570. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, L.; Gao, W.; Hu, F.; Zhang, J.; Ren, Y.; Lin, R.; Feng, Q.; Cheng, M.; Ju, D. A Central Catecholaminergic Circuit Controls Blood Glucose Levels during Stress. Neuron 2017, 95, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, F.; Stewart, J.C.; Ott, S.; Chlebikova, K.; Chua, J.Y.; Koh, T.W.; Ho, J.; Claridgechang, A. Optogenetic inhibition of behavior with anion channelrhodopsins. Nature Methods 2016, 14, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Kato, H.E.; Zhang, F.; Yizhar, O.; Ramakrishnan, C.; Nishizawa, T.; Hirata, K.; Ito, J.; Aita, Y.; Tsukazaki, T.; Hayashi, S.; et al. Crystal structure of the channelrhodopsin light-gated cation channel. Nature 2012, 482, 369–374. [Google Scholar] [CrossRef]
- Volkov, O.; Kovalev, K.; Polovinkin, V.; Borshchevskiy, V.; Bamann, C.; Astashkin, R.; Marin, E.; Popov, A.; Balandin, T.; Willbold, D. Structural insights into ion conduction by channelrhodopsin 2. Science 2017, 358, eaan8862. [Google Scholar] [CrossRef]
- Yuka, S.; Hongxia, W.; Takuya, H.; Minami, S.; Jun, K.; Tetsuo, T.; Toru, I.; Hiromu, Y. Photocurrent attenuation by a single polar-to-nonpolar point mutation of channelrhodopsin-2. Photochem. Photobiol. Sci. 2009, 8, 328–336. [Google Scholar]
- Saki, T.; Yuka, S.; Tetsuo, T.; Toru, I.; Hiromu, Y. Involvement of glutamate 97 in ion influx through photo-activated channelrhodopsin-2. Neurosci. Res. 2013, 75, 13–22. [Google Scholar]
- Watanabe, H.C.; Welke, K.; Schneider, F.; Tsunoda, S.; Zhang, F.; Deisseroth, K.; Hegemann, P.; Elstner, M. Structural Model of Channelrhodopsin. J. Biol. Chem. 2012, 287, 7456–7466. [Google Scholar] [CrossRef]
- Anna Pia, P.; Nicola, D.F.; Francesca, D.B.; Francesco, Z.; Maria Federica, S.; Francesco, F.; Marco, M. Bioinformatic and mutational analysis of channelrhodopsin-2 protein cation-conducting pathway. J. Biol. Chem. 2012, 287, 4818–4825. [Google Scholar]
- Gradmann, D.; Berndt, A.; Schneider, F.; Hegemann, P. Rectification of the Channelrhodopsin Early Conductance. Biophys. J. 2011, 101, 1057–1068. [Google Scholar] [CrossRef] [PubMed]
- Ruffert, K.; Himmel, B.; Lall, D.; Bamann, C.; Bamberg, E.; Betz, H.; Eulenburg, V. Glutamate residue 90 in the predicted transmembrane domain 2 is crucial for cation flux through channelrhodopsin 2. Biochem. Biophys. Res. Commun. 2011, 410, 737–743. [Google Scholar] [CrossRef] [PubMed]
- Eisenhauer, K.; Kuhne, J.; Ritter, E.; Berndt, A.; Wolf, S.; Freier, E.; Bartl, F.; Hegemann, P.; Gerwert, K. In channelrhodopsin-2 Glu-90 is crucial for ion selectivity and is deprotonated during the photocycle. J. Biol. Chem. 2012, 287, 6904–6911. [Google Scholar] [CrossRef] [PubMed]
- Sybille, U.; Ronnie, G.; Georg, N. Degradation of channelopsin-2 in the absence of retinal and degradation resistance in certain mutants. Biol. Chem. 2013, 394, 271–280. [Google Scholar]
- Tomita, H.; Sugano, E.; Fukazawa, Y.; Isago, H.; Sugiyama, Y.; Hiroi, T.; Ishizuka, T.; Mushiake, H.; Kato, M.; Hirabayashi, M. Visual properties of transgenic rats harboring the channelrhodopsin-2 gene regulated by the thy-1.2 promoter. PLoS ONE 2009, 4, e7679. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Cheng, X.; Jo, S.; Mackerell, A.D.; Im, W. CHARMM-GUI Input Generator for NAMD, Gromacs, Amber, Openmm, and CHARMM/OpenMM Simulations using the CHARMM36 Additive Force Field. J. Chem. Theory Comput. 2016, 12, 405–413. [Google Scholar] [CrossRef]
- Wietek, J.; Wiegert, J.S.; Adeishvili, N.; Schneider, F.; Watanabe, H.; Tsunoda, S.P.; Vogt, A.; Elstner, M.; Oertner, T.G.; Hegemann, P. Conversion of channelrhodopsin into a light-gated chloride channel. Science 2014, 344, 409–412. [Google Scholar] [CrossRef]
- Phillips, J.C.; Rosemary, B.; Wei, W.; James, G.; Emad, T.; Elizabeth, V.; Christophe, C.; Skeel, R.D.; Laxmikant, K.; Klaus, S. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2010, 26, 1781–1802. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hammonds, K.D.; Ryckaert, J.P. On the convergence of the SHAKE algorithm. Comput. Phys. Commun. 1991, 62, 336–351. [Google Scholar] [CrossRef]
- Darve, E. Numerical Methods for Calculating the Potential of Mean Force. Lect. Notes Comput. Sci. Eng. 2006, 49, 213–249. [Google Scholar]
- Patel, J.S.; Berteotti, A.; Ronsisvalle, S.; Rocchia, W.; Cavalli, A. Steered molecular dynamics simulations for studying protein-ligand interaction in cyclin-dependent kinase 5. J. Chem. Inf. Model. 2014, 54, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Jalily Hasani, H.; Ganesan, A.; Ahmed, M.; Barakat, K.H. Effects of protein-protein interactions and ligand binding on the ion permeation in KCNQ1 potassium channel. PLoS ONE 2018, 13, e0191905. [Google Scholar] [CrossRef]
- Mücksch, C.; Urbassek, H.M. Accelerating Steered Molecular Dynamics: Toward Smaller Velocities in Forced Unfolding Simulations. J. Chem. Theory Comput. 2016, 12, 1380–1384. [Google Scholar] [CrossRef]
- Kingsley, L.J.; Esquivel-Rodríguez, J.; Yang, Y.; Kihara, D.; Lill, M.A. Ranking protein-protein docking results using steered molecular dynamics and potential of mean force calculations. J. Comput. Chem. 2016, 37, 1861–1865. [Google Scholar] [CrossRef]
- Bonomi, M.; Branduardi, D.; Bussi, G.; Camilloni, C.; Provasi, D.; Raiteri, P.; Donadio, D.; Marinelli, F.; Pietrucci, F.; Broglia, R.A. PLUMED: A portable plugin for free-energy calculations with molecular dynamics. Comput. Phys. Commun. 2009, 180, 1961–1972. [Google Scholar] [CrossRef]
- Isralewitz, B.; Gao, M.; Schulten, K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001, 11, 224–230. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, W.Y.; Cheng, J.; Nie, Y.H.; Xin, Q.; Yuan, S.; Dou, Y.S. Formation Mechanism of Ion Channel in Channelrhodopsin-2: Molecular Dynamics Simulation and Steering Molecular Dynamics Simulations. Int. J. Mol. Sci. 2019, 20. [Google Scholar] [CrossRef]
- Kästner, J. Umbrella sampling. Wires. Comput. Mol. Sci. 2011, 1, 932–942. [Google Scholar] [CrossRef]
- Akhshi, P.; Wu, G. Umbrella sampling molecular dynamics simulations reveal concerted ion movement through G-quadruplex DNA channels. Phys. Chem. Chem. Phys. 2017, 19, 11017–11025. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rosenberg, J.M.; Bouzida, D.; Swendsen, R.H.; Kollman, P.A. The Weighted Histogram Analysis Method for Free-Energy Calculations on Biomolecules. I. The Method. J. Comput. Chem. 2010, 13, 1011–1021. [Google Scholar] [CrossRef]
- Lórenz-Fonfría, V.A.; Tom, R.; Nils, K.; Melanie, N.; Michael, G.; Gabriele, F.V.M.; Christian, B.; Ernst, B.; Ramona, S.; Joachim, H. Transient protonation changes in channelrhodopsin-2 and their relevance to channel gating. Proc. Natl. Acad. Sci. USA 2013, 110, E1273–E1281. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Yang, T.; Zhou, S.; Cheng, J.; Yuan, S.; Lo, G.V.; Dou, Y. Molecular Dynamics Simulation of Transmembrane Transport of Chloride Ions in Mutants of Channelrhodopsin. Biomolecules 2019, 9, 852. https://doi.org/10.3390/biom9120852
Zhang W, Yang T, Zhou S, Cheng J, Yuan S, Lo GV, Dou Y. Molecular Dynamics Simulation of Transmembrane Transport of Chloride Ions in Mutants of Channelrhodopsin. Biomolecules. 2019; 9(12):852. https://doi.org/10.3390/biom9120852
Chicago/Turabian StyleZhang, Wenying, Ting Yang, Shuangyan Zhou, Jie Cheng, Shuai Yuan, Glenn V. Lo, and Yusheng Dou. 2019. "Molecular Dynamics Simulation of Transmembrane Transport of Chloride Ions in Mutants of Channelrhodopsin" Biomolecules 9, no. 12: 852. https://doi.org/10.3390/biom9120852
APA StyleZhang, W., Yang, T., Zhou, S., Cheng, J., Yuan, S., Lo, G. V., & Dou, Y. (2019). Molecular Dynamics Simulation of Transmembrane Transport of Chloride Ions in Mutants of Channelrhodopsin. Biomolecules, 9(12), 852. https://doi.org/10.3390/biom9120852






