Macromolecule Particle Picking and Segmentation of a KLH Database by Unsupervised Cryo-EM Image Processing
Abstract
1. Introduction
2. Materials and Methods
2.1. Preliminary
2.2. Image Framework and Spectral Properties
2.3. Matrix Decomposition NNMF
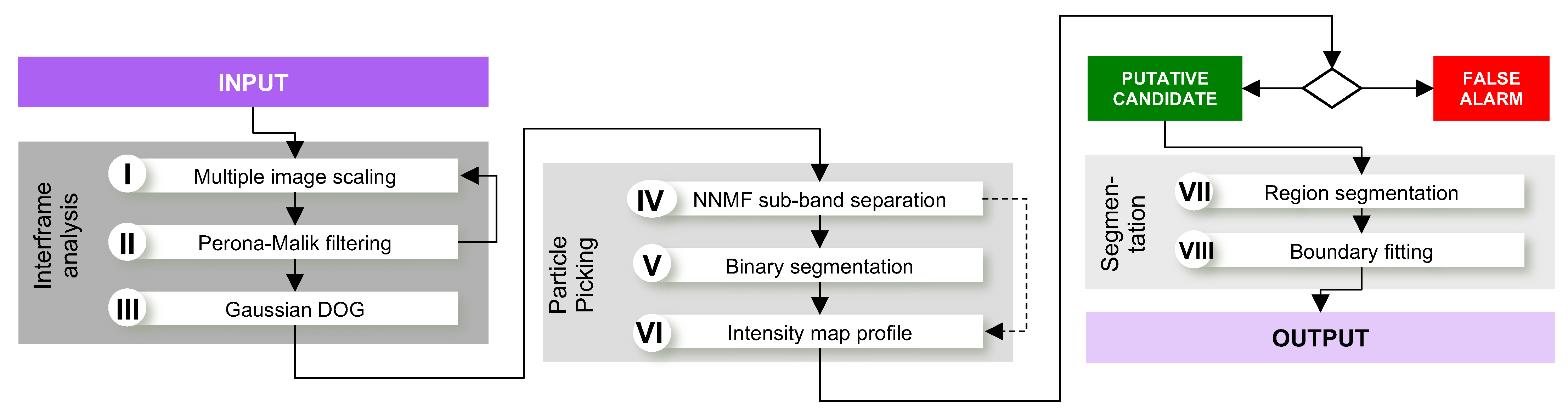
2.4. Proposed Algorithm
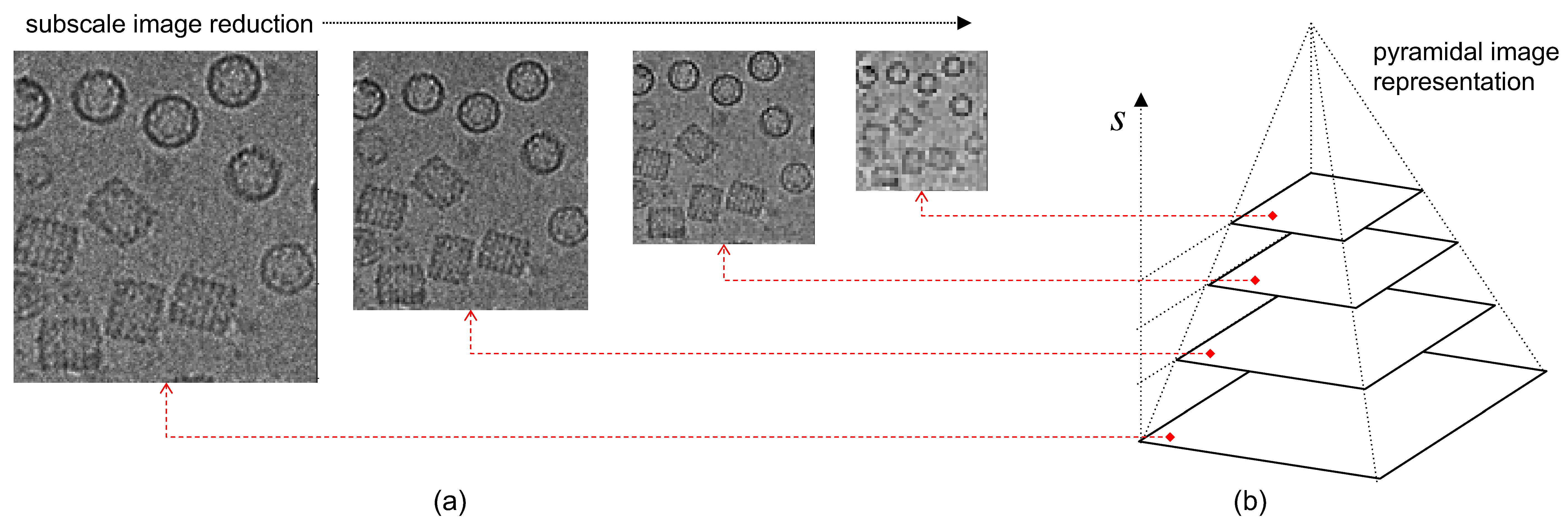
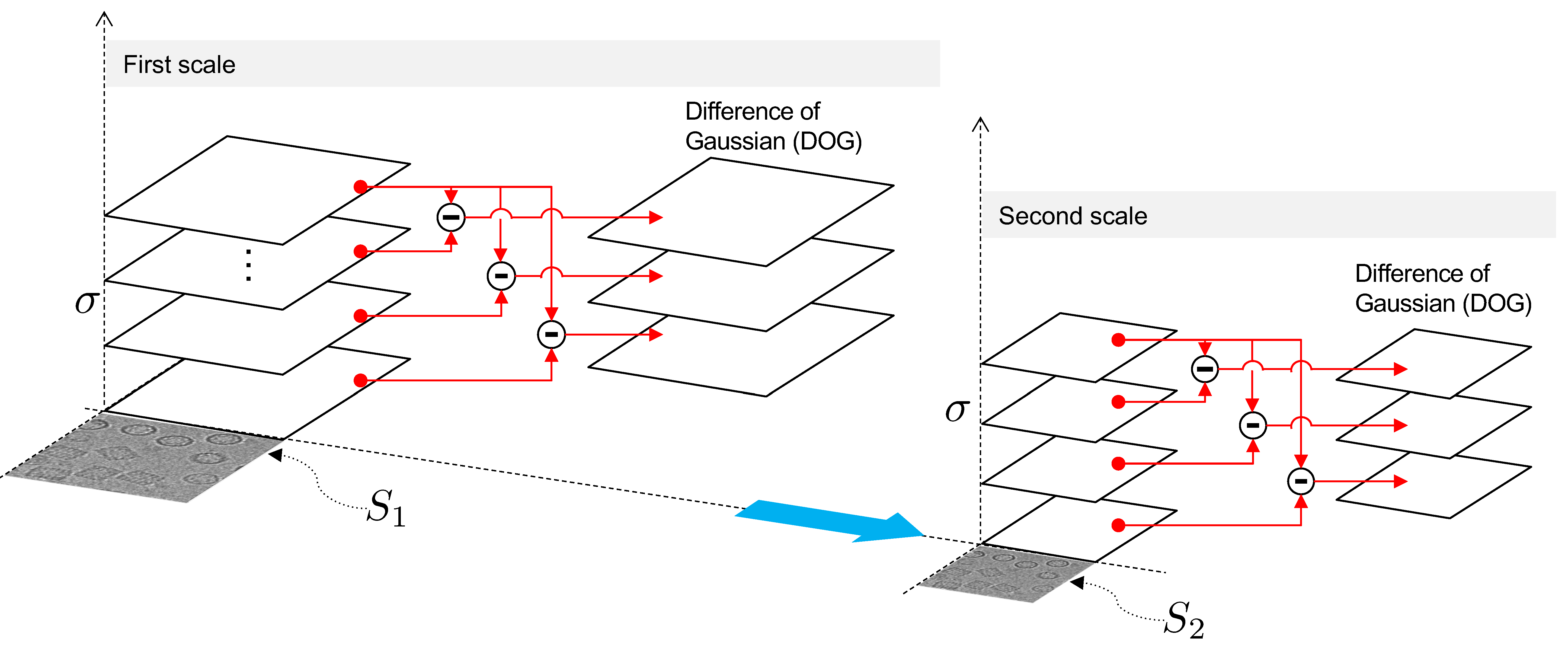
2.5. Phase I: Interframe Analysis
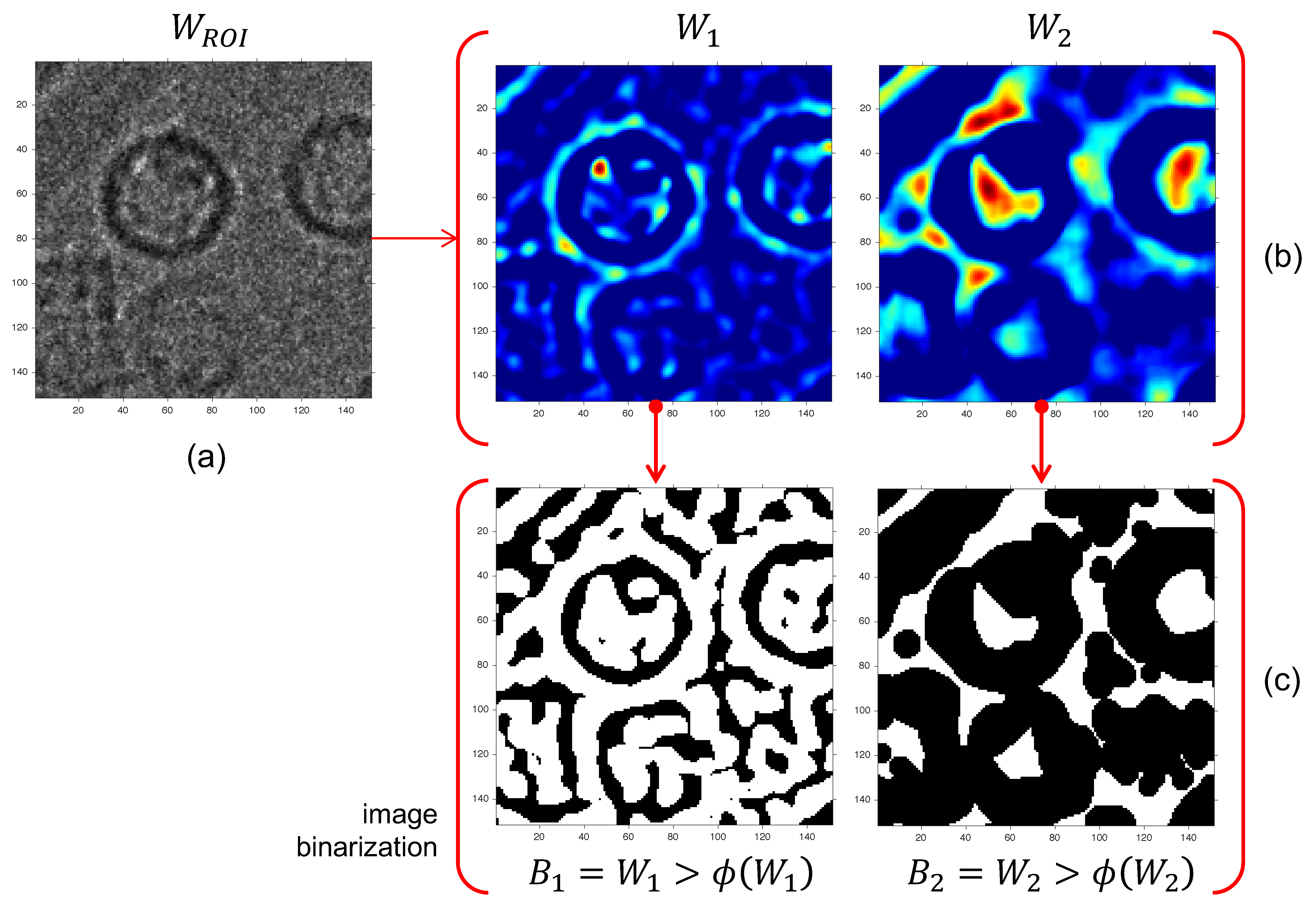
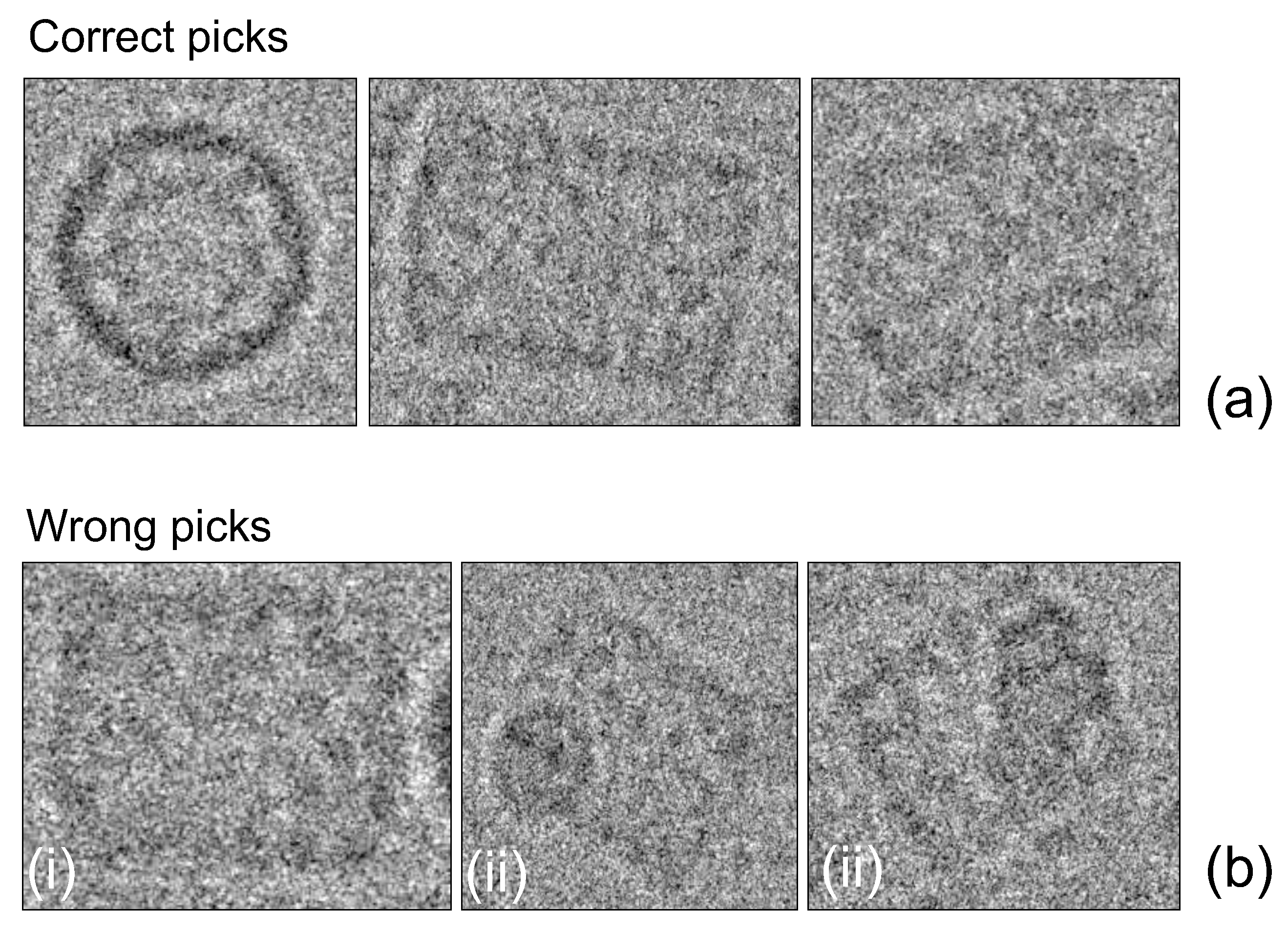
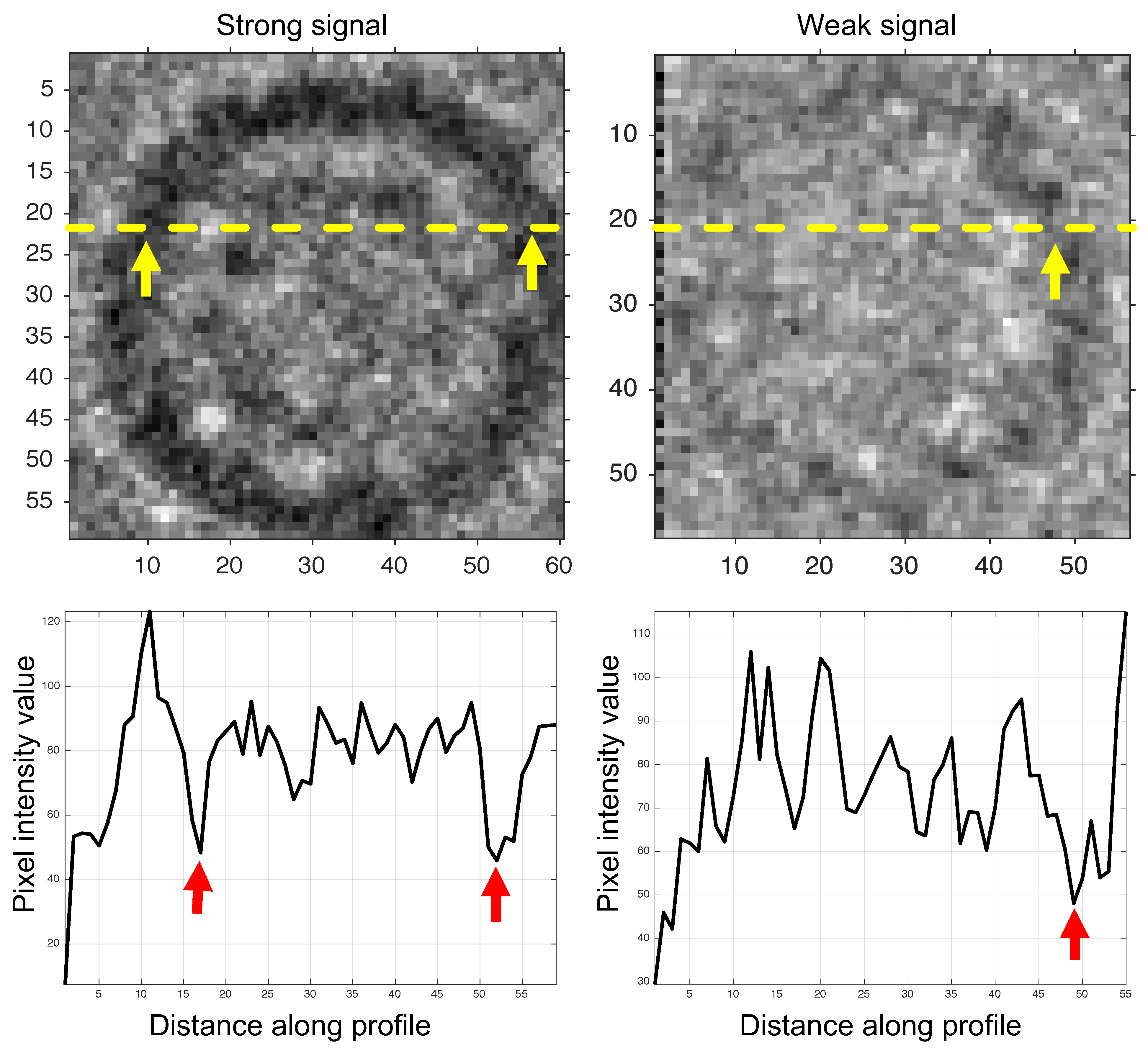
2.6. Phase II: Particle Picking
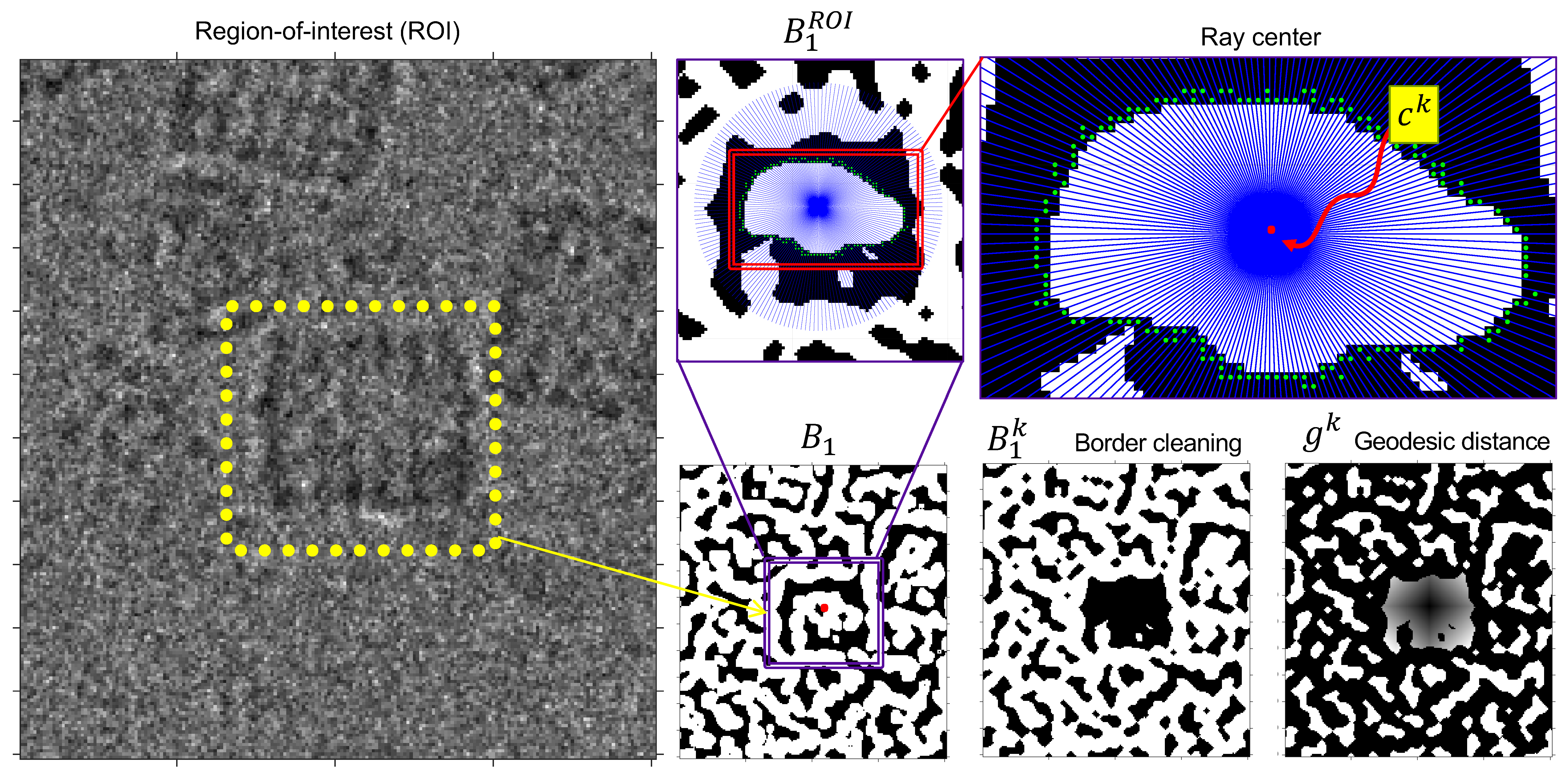
2.7. Phase III: Segmentation
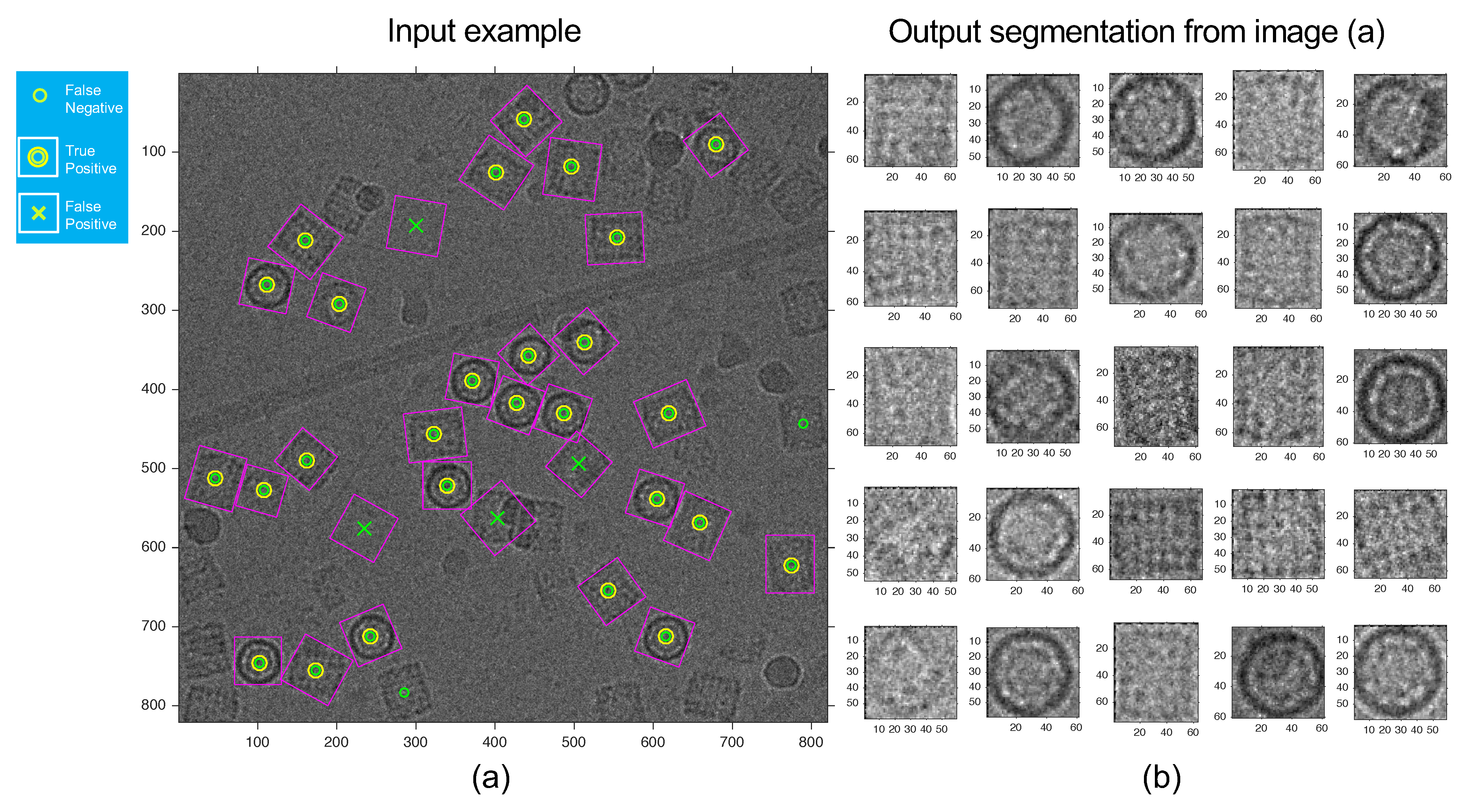
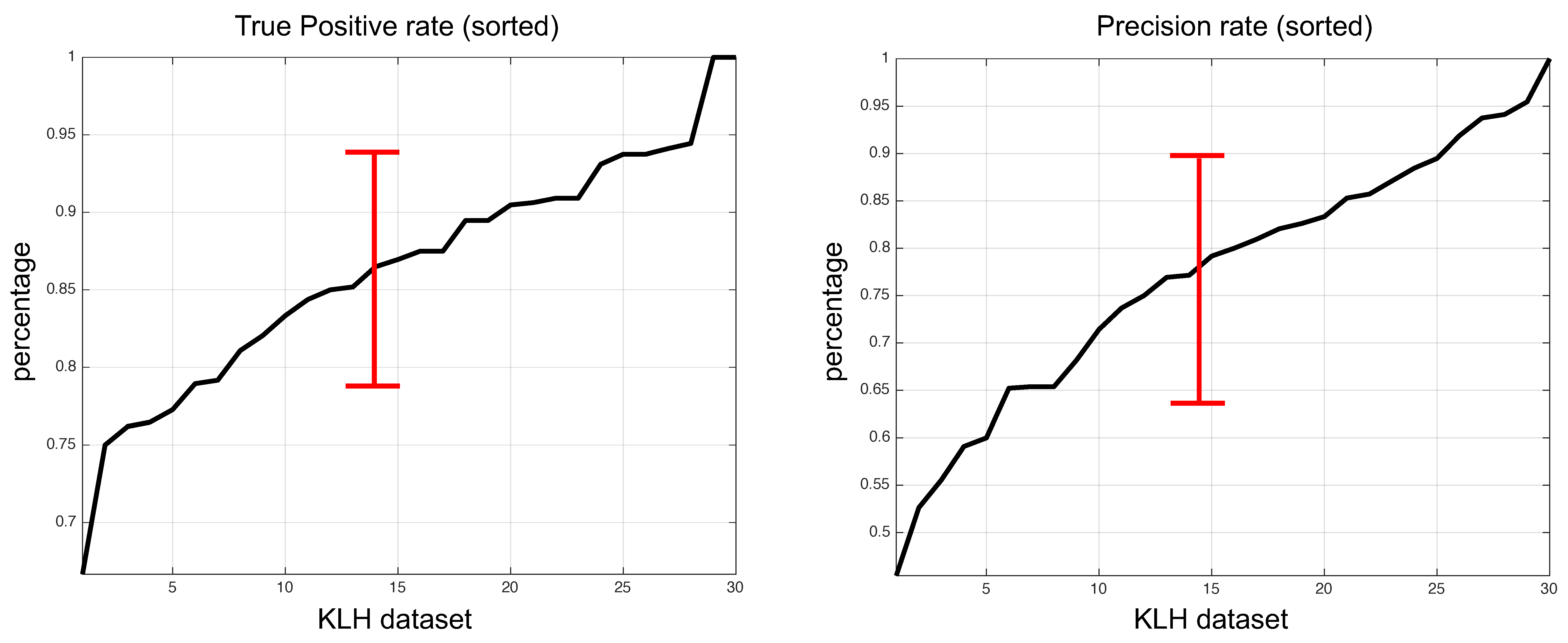
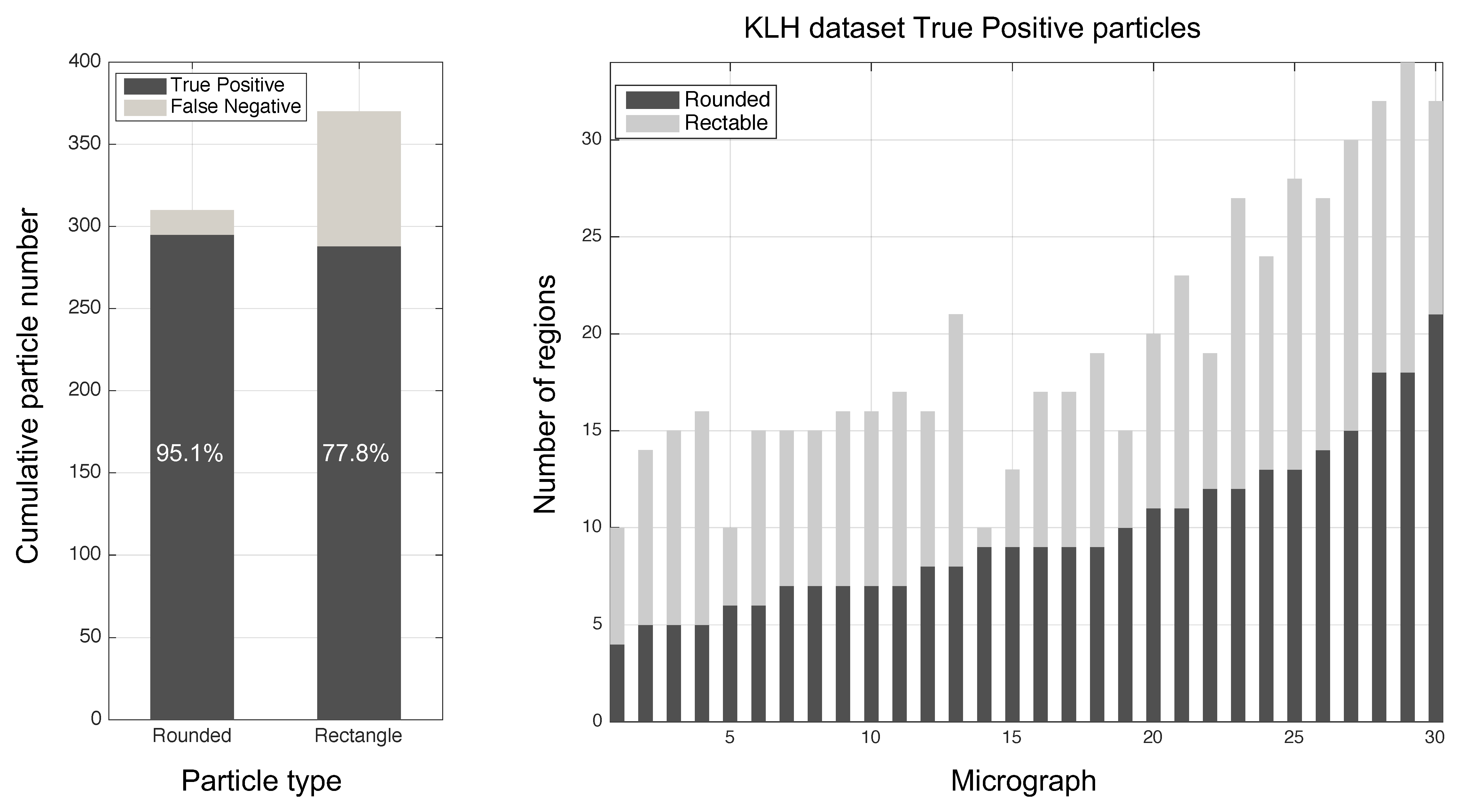
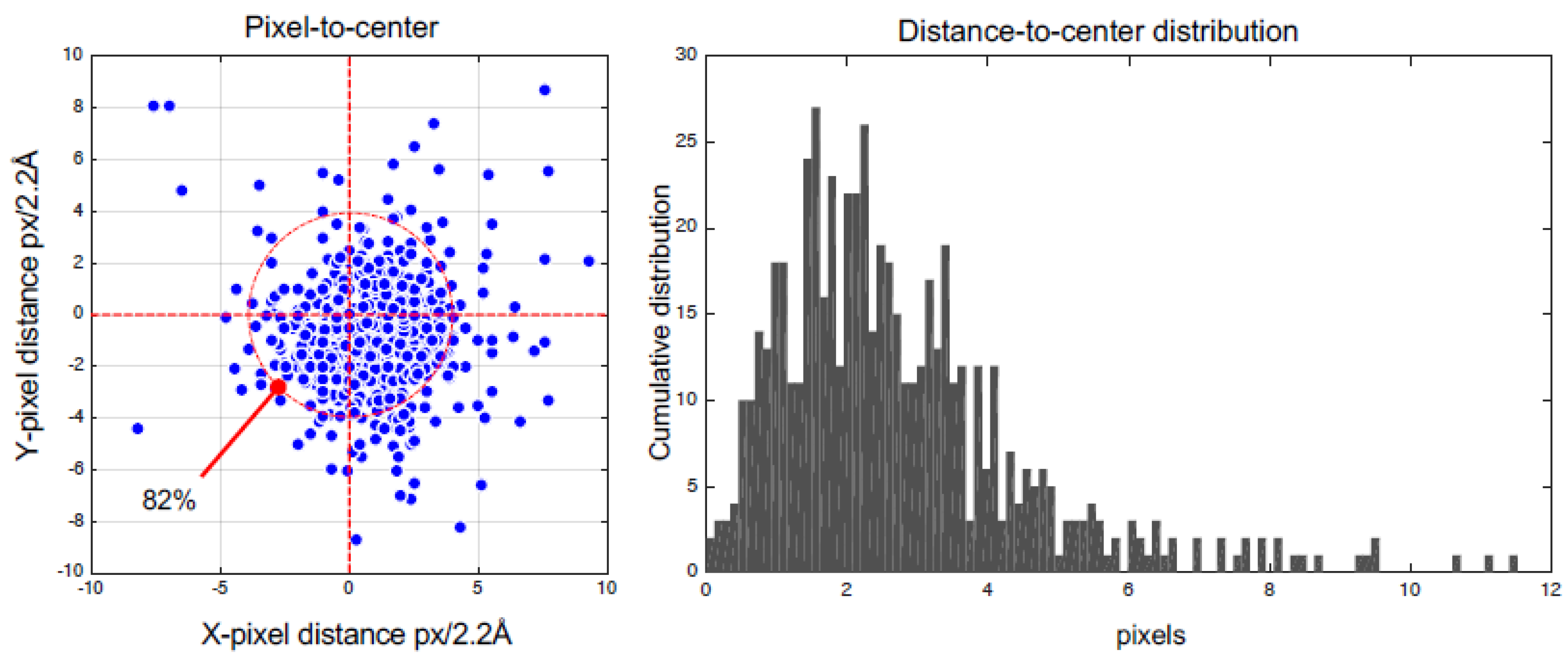
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Henderson, R. Overview and future of single particle electron cryomicroscopy. Arch. Biochem. Biophys. 2015, 581, 19–24. [Google Scholar] [CrossRef]
- Kuhlbrandt, W. The Resolution Revolution. Science 2014, 343, 1443–1444. [Google Scholar] [CrossRef]
- Scheres, S.H.W. Semi-automated selection of cryo-EM particles in RELION-1.3. J. Struct. Biol. 2015, 189, 114–122. [Google Scholar] [CrossRef]
- Egelman, E.H. The Current Revolution in Cryo-EM. Biophys. J. 2016, 110, 1008–1012. [Google Scholar] [CrossRef]
- Henderson, R.; McMullan, G. Problems in obtaining perfect images by single-particle electron cryomicroscopy of biological structures in amorphous ice. Microscopy 2013, 62, 43–50. [Google Scholar] [CrossRef]
- White, H.E.; Ignatiou, A.; Clare, D.K.; Orlova, E.V. Structural Study of Heterogeneous Biological Samples by Cryoelectron Microscopy and Image Processing. BioMed Res. Int. 2017, 2017, 1–23. [Google Scholar] [CrossRef]
- Yoshioka, C.; Lyumkis, D.; Carragher, B.; Potter, C.S. Maskiton: Interactive, web-based classification of single-particle electron microscopy images. J. Struct. Biol. 2013, 182, 155–163. [Google Scholar] [CrossRef]
- Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016, 193, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Peng, L.; Baldwin, P.R.; Mann, D.S.; Jiang, W.; Rees, I.; Ludtke, S.J. EMAN2: An extensible image processing suite for electron microscopy. J. Struct. Biol. 2007, 157, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Z.; Grigorieff, N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J. Struct. Biol. 2007, 157, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Voss, N.R.; Yoshioka, C.K.; Radermacher, M.; Potter, C.S.; Carragher, B. DoG Picker and TiltPicker: Software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 2009, 166, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Sorzano, C.O.S.; Recarte, E.; Alcorlo, M.; Bilbao-Castro, J.R.; San-Martin, C.; Marabini, R.; Carazo, J.M. Automatic particle selection from electron micrographs using machine learning techniques. J. Struct. Biol. 2009, 167, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Langlois, R.; Pallesen, J.; Ash, J.T.; Ho, D.N.; Rubinstein, J.L.; Frank, J. Automated particle picking for low-contrast macromolecules in cryo-electron microscopy. J. Struct. Biol. 2014, 186, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Roseman, A.M. FindEM—A fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 2004, 145, 91–99. [Google Scholar] [CrossRef]
- Huang, Z.; Penczek, P.A. Application of template matching technique to particle detection in electron micrographs. J. Struct. Biol. 2004, 145, 29–40. [Google Scholar] [CrossRef]
- Tegunov, D.; Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 2019, 16, 1146–1152. [Google Scholar] [CrossRef]
- Adiga, U.; Baxter, W.T.; Hall, R.J.; Rockel, B.; Rath, B.K.; Frank, J.; Glaeser, R. Particle picking by segmentation: A comparative study with SPIDER-based manual particle picking. J. Struct. Biol. 2005, 152, 211–220. [Google Scholar] [CrossRef]
- Woolford, D.; Ericksson, G.; Rothnagel, R.; Muller, D.; Landsberg, M.J.; Pantelic, R.S.; McDowall, A.; Pailthorpe, B.; Young, P.R.; Hankamer, B.; et al. SwarmPS: Rapid, semi-automated single particle selection software. J. Struct. Biol. 2007, 157, 174–188. [Google Scholar] [CrossRef]
- Ogura, T.; Sato, C. Auto-accumulation method using simulated annealing enables fully automatic particle pickup completely free from a matching template or learning data. J. Struct. Biol. 2004, 146, 344–358. [Google Scholar] [CrossRef]
- Zhu, Y.; Ouyang, Q.; Mao, Y. A deep convolutional neural network approach to single-particle recognition in cryo-electron microscopy. BMC Bioinform. 2017, 18, 348. [Google Scholar] [CrossRef]
- Bepler, T.; Morin, A.; Rapp, M.; Brasch, J.; Shapiro, L.; Noble, A.J.; Berger, B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 2019, 16, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Wagner, T.; Merino, F.; Stabrin, M.; Moriya, T.; Antoni, C.; Apelbaum, A.; Hagel, P.; Sitsel, O.; Raisch, T.; Prumbaum, D.; et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019, 2, 218. [Google Scholar] [CrossRef] [PubMed]
- Perona, P.; Malik, J. Scale-Space and Edge Detection Using Anisotropic Diffusion. IEEE Trans. Pattern Anal. Mach. Intell. 1990, 12, 629–639. [Google Scholar] [CrossRef]
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Weickert, J. Anisotropic Diffusion in Image Processing; Teubner, B.G., Ed.; European Consortium for Mathematics in Industry: Stuttgart, Germany, 1998. [Google Scholar]
- Buades, A.; Coll, B.; Morel, J.M. A Review of Image Denoising Algorithms, with a New One. Multiscale Model. Simul. 2005, 4, 490–530. [Google Scholar] [CrossRef]
- Parrilli, S.; Poderico, M.; Angelino, C.V.; Verdoliva, L. A Nonlocal SAR Image Denoising Algorithm Based on LLMMSE Wavelet Shrinkage. IEEE Trans. Geosci. Remote Sens. 2012, 50, 606–616. [Google Scholar] [CrossRef]
- Liu, K.; Tan, J.; Ai, L. Hybrid regularizers-based adaptive anisotropic diffusion for image denoising. SpringerPlus 2016, 5, 404. [Google Scholar] [CrossRef][Green Version]
- Barbu, T. Robust Anisotropic Diffusion Scheme for Image Noise Removal. Procedia Comput. Sci. 2014, 35, 522–530. [Google Scholar] [CrossRef]
- Tschumperlé, D.; Deriche, R. Anisotropic Diffusion Partial Differential Equations for Multichannel Image Regularization: Framework and Applications. Adv. Imaging Electron Phys. 2007, 145, 149–209. [Google Scholar]
- Pruessner, G. Self-Organised Criticality: Theory, Models, and Characterisation; Cambridge University Press: Cambridge, UK; New York, NY, USA, 2012. [Google Scholar]
- Févotte, C.; Bertin, N.; Durrieu, J.-L. Nonnegative Matrix Factorization with the Itakura-Saito Divergence: With Application to Music Analysis. Neural Comput. 2009, 21, 793–830. [Google Scholar] [CrossRef]
- Koenderink, J.J.; van Doorn, A.J. The internal representation of solid shape with respect to vision. Biol. Cybern. 1979, 32, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.; Zisserman, A.; Munoz, X. Representing Shape with a Spatial Pyramid Kernel. In Proceedings of the 6th ACM International Conference on Image and Video Retrieval—CIVR ’07, Amsterdam, The Netherlands, 9–11 July 2007; ACM Press: New York, NY, USA, 2007; pp. 401–408. [Google Scholar]
- Mikolajczyk, K.; Schmid, C. An Affine Invariant Interest Point Detector. In Computer Vision—ECCV 2002, Proceedings of the 7th European Conference on Computer Vision, Copenhagen, Denmark, 28–31 May 2002; Heyden, A., Sparr, G., Nielsen, M., Johansen, P., Eds.; Springer: Berlin/Heidelberg, Germany, 2002; Volume 2350, pp. 128–142. [Google Scholar]
- Bay, H.; Tuytelaars, T.; Van Gool, L. SURF: Speeded Up Robust Features. In Computer Vision—ECCV 2006, Proceedings of the 9th European Conference on Computer Vision, Graz, Austria, 7–13 May 2006; Leonardis, A., Bischof, H., Pinz, A., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; Volume 3951, pp. 404–417. [Google Scholar]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Gonzalez, R.; Woods, R. Digital Image Processing, 3rd ed.; Prentice Hall: Upper Saddle River, NJ, USA, 2008. [Google Scholar]
- Oppenheim, A.V.; Schafer, R.W. From frequency to quefrency: A history of the cepstrum. IEEE Signal Process. Mag. 2004, 21, 95–106. [Google Scholar] [CrossRef]
- Fitzgibbon, A.; Pilu, M.; Fisher, R.B. Direct least square fitting of ellipses. IEEE Trans. Pattern Anal. Mach. Intell. 1999, 21, 476–480. [Google Scholar] [CrossRef]
- Leys, C.; Ley, C.; Klein, O.; Bernard, P.; Licata, L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. J. Exp. Soc. Psychol. 2013, 49, 764–766. [Google Scholar] [CrossRef]
- Carragher, B.; Kisseberth, N.; Kriegman, D.; Milligan, R.A.; Potter, C.S.; Pulokas, J.; Reilein, A. Leginon: An automated system for acquisition of images from vitreous ice specimens. J. Struct. Biol. 2000, 132, 33–45. [Google Scholar] [CrossRef]
- Zhu, Y.; Carragher, B.; Glaeser, R.M.; Fellmann, D.; Bajaj, C.; Bern, M.; Mouche, F.; de Haas, F.; Hall, R.J.; Kriegman, D.J.; et al. Automatic particle selection: Results of a comparative study. J. Struct. Biol. 2004, 145, 3–14. [Google Scholar] [CrossRef][Green Version]
- Bishop, C.M. Pattern Recognition and Machine Learning; Springer-Verlag: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Yu, Z.; Bajaj, C. Detecting circular and rectangular particles based on geometric feature detection in electron micrographs. J. Struct. Biol. 2004, 145, 168–180. [Google Scholar] [CrossRef]
- Abrishami, V.; Zaldívar-Peraza, A.; de la Rosa-Trevín, J.M.; Vargas, J.; Otón, J.; Marabini, R.; Shkolnisky, Y.; Carazo, J.M.; Sorzano, C.O.S. A pattern matching approach to the automatic selection of particles from low-contrast electron micrographs. Bioinformatics 2013, 29, 2460–2468. [Google Scholar] [CrossRef]




| Authors | Reference | FPR% | TPR% | Approach |
|---|---|---|---|---|
| Zhu et al. (2004) | [43] | 13.7 | 90.3 | Unsupervised |
| Yu and Bajaj (2004) | [45] | 24.7 | 91.7 | Unsupervised |
| Sorzano et al. (2009) | [12] | 9.3 | 69.1 | Supervised (Ensemble) |
| Abrishami et al. (2013) | [46] | 16.2 | 93.3 | Supervised (SVM) |
| Scheres (2015) | [3] | 12 | 90 | Semi-supervised |
| Zhu et al. (2017) | [20] | 10 | 90 | Supervised (Deep Learning) |
| Proposed (2019) | 14.3 | 95.1 | Unsupervised |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrasco, M.; Toledo, P.; Tischler, N.D. Macromolecule Particle Picking and Segmentation of a KLH Database by Unsupervised Cryo-EM Image Processing. Biomolecules 2019, 9, 809. https://doi.org/10.3390/biom9120809
Carrasco M, Toledo P, Tischler ND. Macromolecule Particle Picking and Segmentation of a KLH Database by Unsupervised Cryo-EM Image Processing. Biomolecules. 2019; 9(12):809. https://doi.org/10.3390/biom9120809
Chicago/Turabian StyleCarrasco, Miguel, Patricio Toledo, and Nicole D. Tischler. 2019. "Macromolecule Particle Picking and Segmentation of a KLH Database by Unsupervised Cryo-EM Image Processing" Biomolecules 9, no. 12: 809. https://doi.org/10.3390/biom9120809
APA StyleCarrasco, M., Toledo, P., & Tischler, N. D. (2019). Macromolecule Particle Picking and Segmentation of a KLH Database by Unsupervised Cryo-EM Image Processing. Biomolecules, 9(12), 809. https://doi.org/10.3390/biom9120809






