Marine Invertebrate Extracts Induce Colon Cancer Cell Death via ROS-Mediated DNA Oxidative Damage and Mitochondrial Impairment
Abstract
1. Introduction
2. Results
2.1. Marine Extracts Derived from Selected Invertebrates Inhibit the Proliferation of Colon Cancer Cells
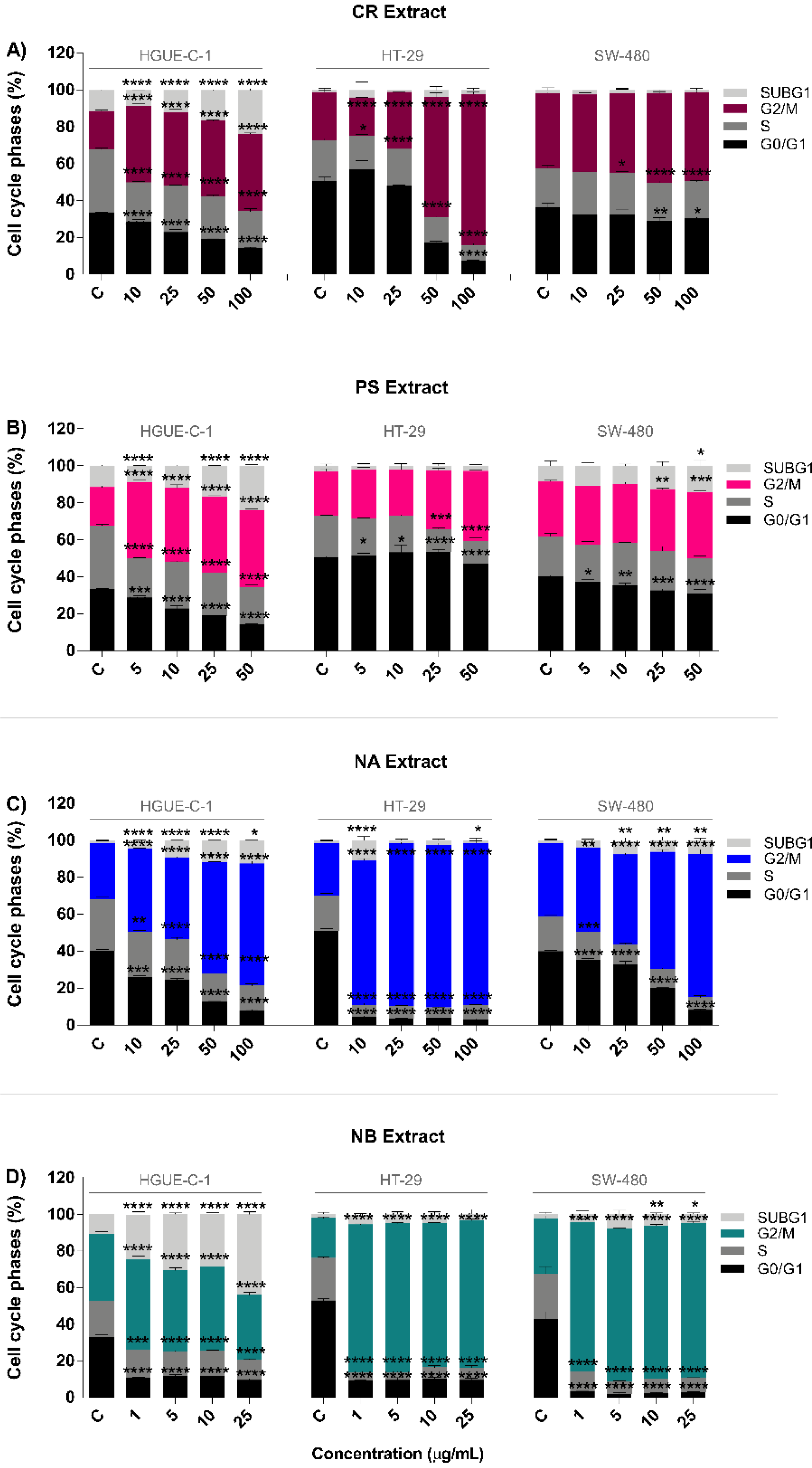
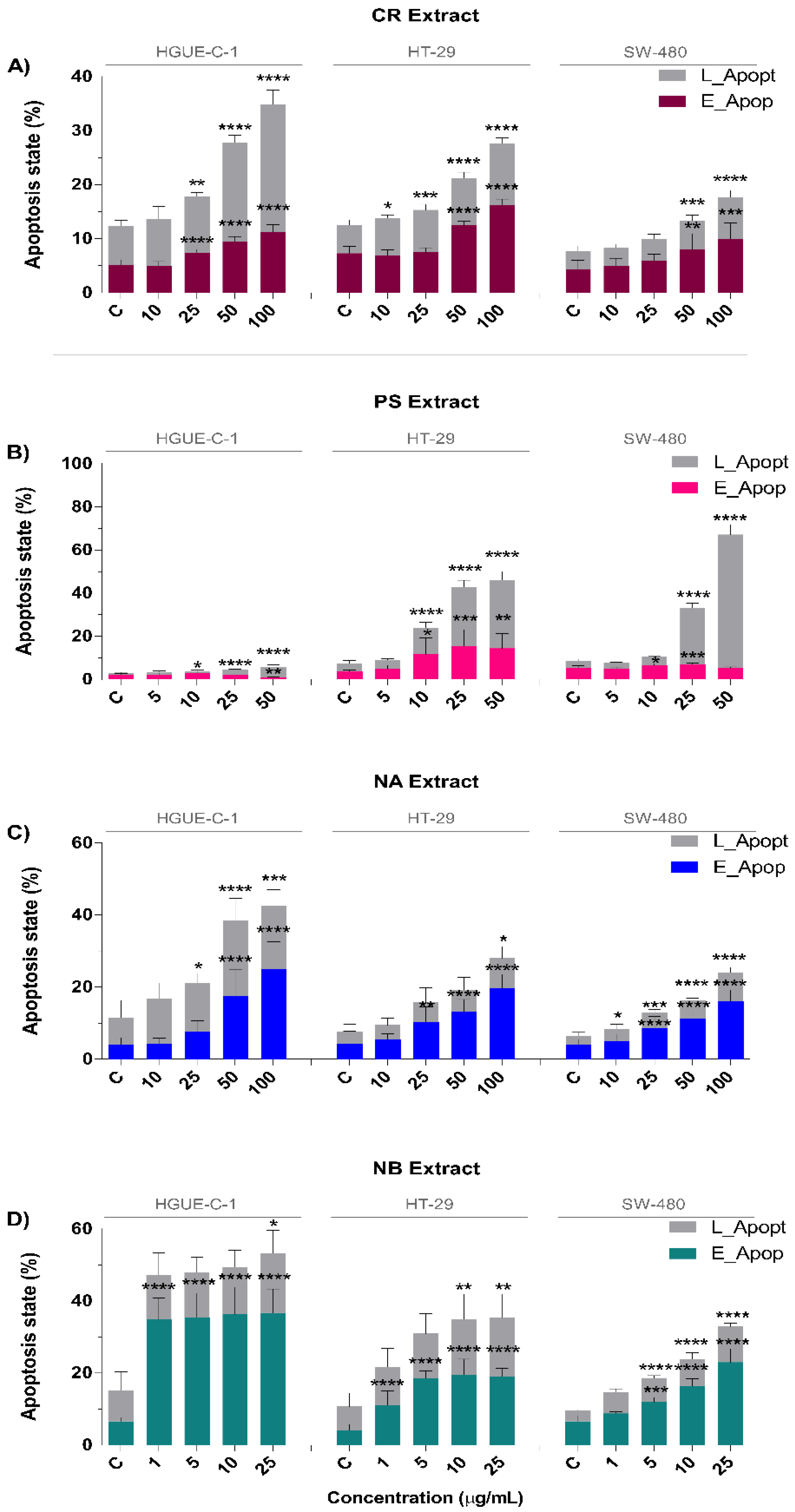
2.2. Invertebrate Marine Extracts Induce Cell Cycle Arrest and Apoptosis
2.3. Effect of Marine Extracts on the Nonapoptotic Cell Death of Colon Cancer Cells
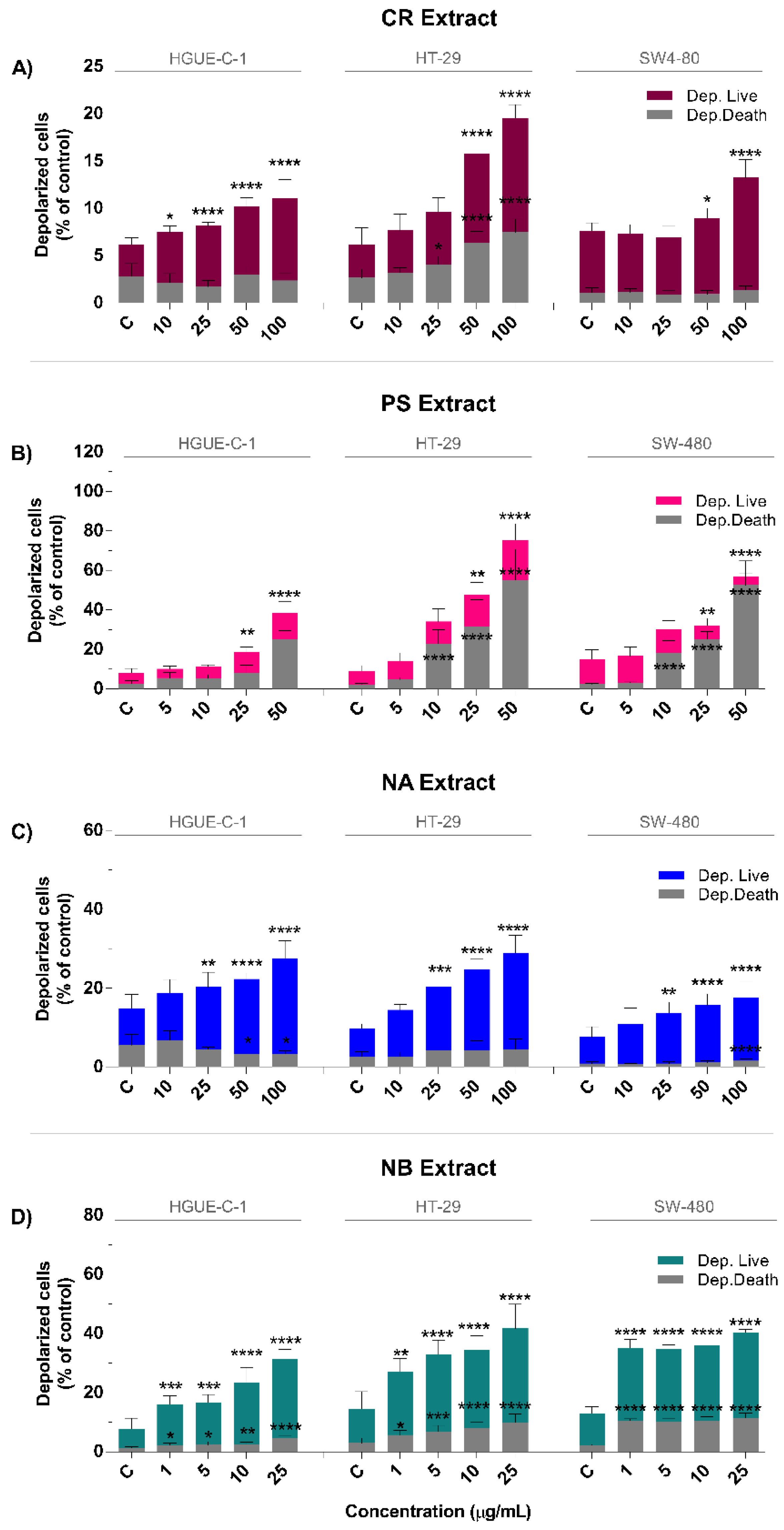
2.4. Marine Extracts Promote Intracellular ROS Generation and Mitochondrial Membrane Depolarization
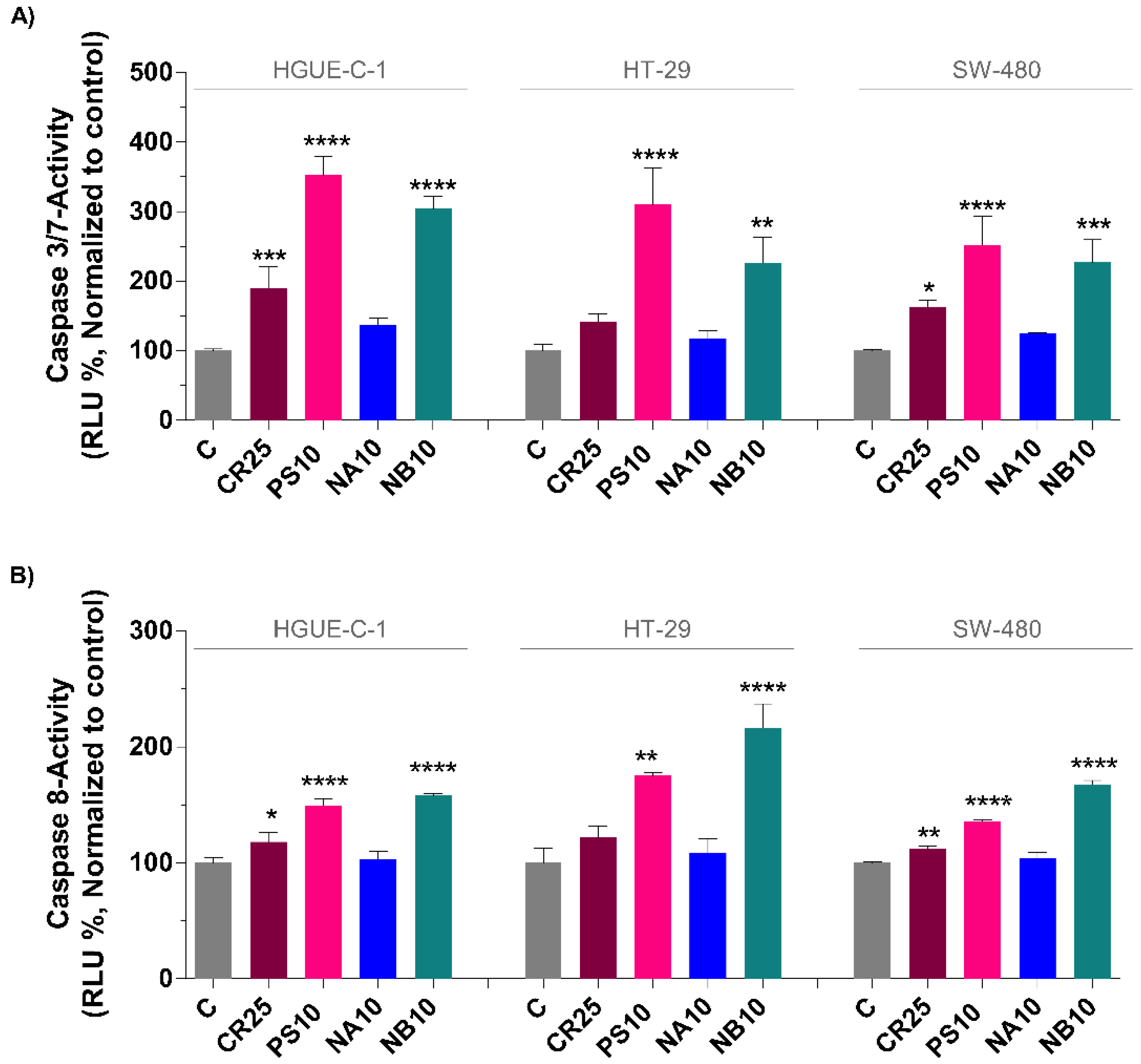
2.5. Marine Extracts Induce Cell Death by Activating Caspases
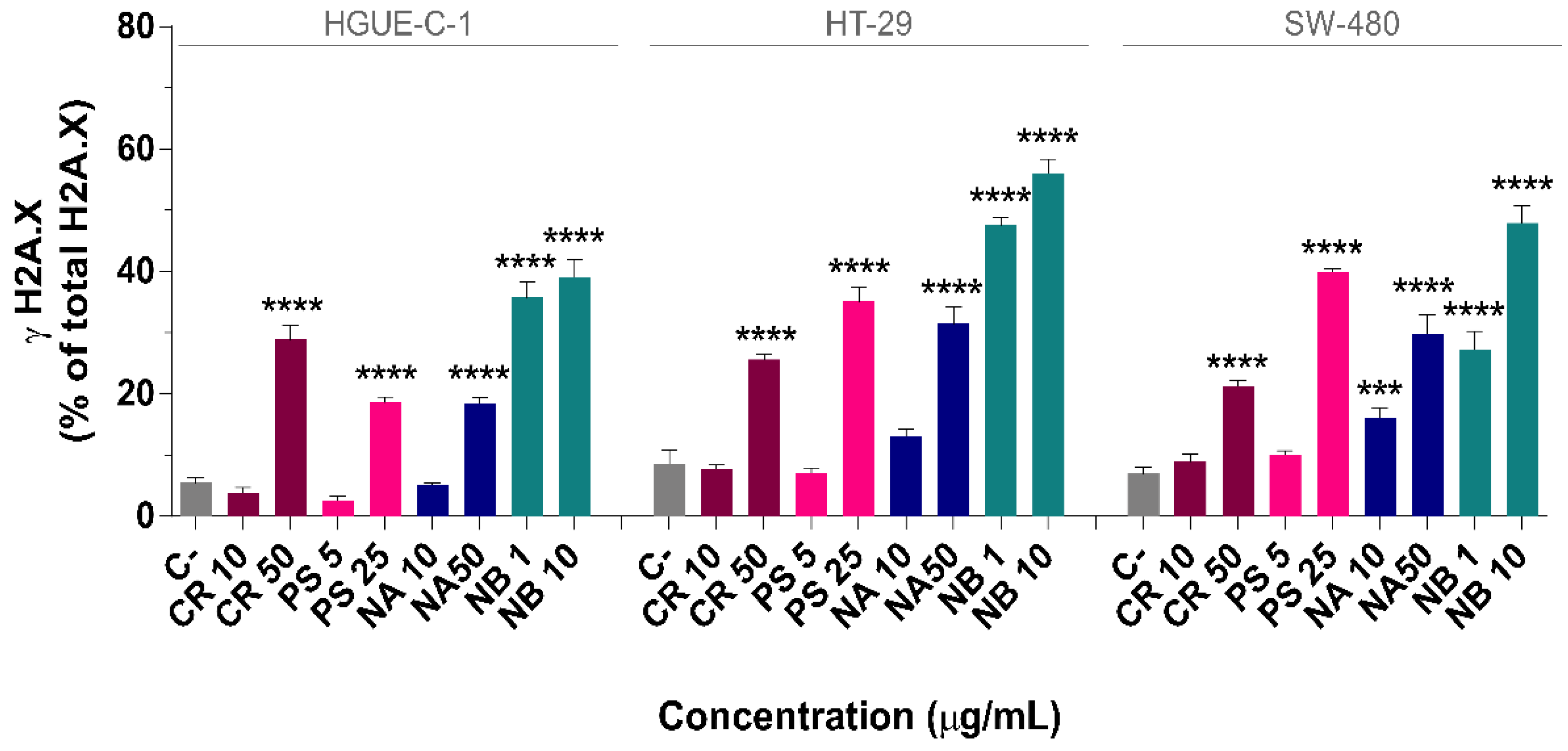
2.6. DNA Damage
2.7. Characterization of Marine Invertebrate Extracts Using HPLC-ESI-TOF-MS
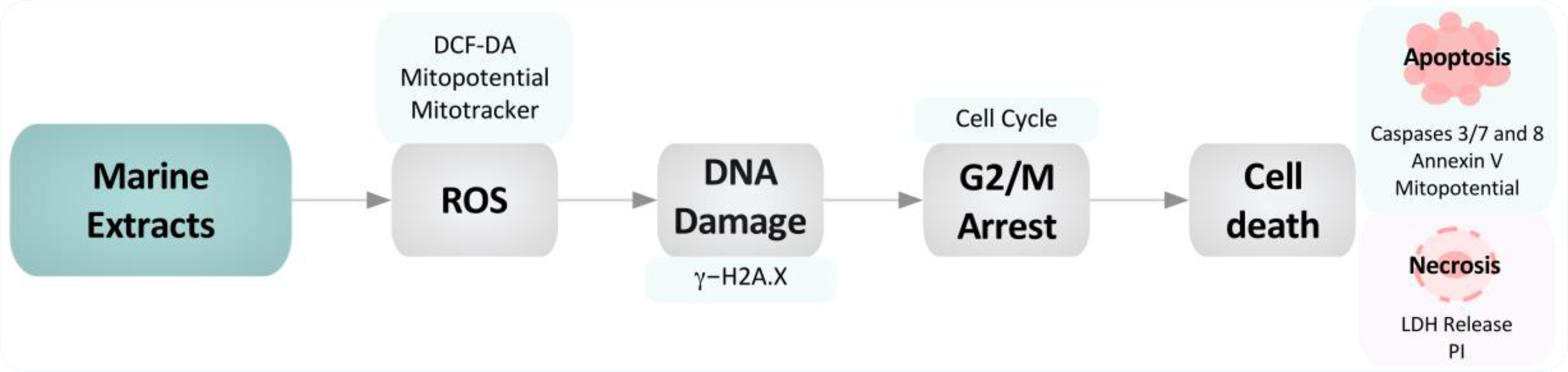
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Marine Invertebrate Material
4.3. Extraction Method and Preparation of Crude Extracts
4.4. Cell Culture
4.5. MTT Cell Viability Assay and IC50 Determination
4.6. RTCA Proliferation Assay
4.7. Clonogenic Assay
4.8. Cell Cycle Analysis
4.9. Measurement of Apoptosis by Annexin V and Mitopotential
4.10. Caspase 3/7 and 8 Activation
4.11. Lactate Dehydrogenase (LDH) Measurement for Necrosis Assay
4.12. Nuclear Staining with Hoechst 33342/Propidium Iodide
4.13. Mitochondrial Membrane Potential (MMP)
4.14. Detection of Reactive Oxygen Species (ROS)
4.15. Detection of Phosphorylated H2A.X
4.16. Determination of Secondary Metabolites by HPLC-ESI-TOF-MS
4.17. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in globocan 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Di Maio, M.; Gallo, C.; Leighl, N.B.; Piccirillo, M.C.; Daniele, G.; Nuzzo, F.; Gridelli, C.; Gebbia, V.; Ciardiello, F.; De Placido, S.; et al. Symptomatic toxicities experienced during anticancer treatment: Agreement between patient and physician reporting in three randomized trials. J. Clin. Oncol. 2015, 33, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Herranz-López, M.; Losada-Echeberría, M.; Barrajón-Catalán, E. The multitarget activity of natural extracts on cancer: Synergy and xenohormesis. Medicines 2018, 6, 6. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.-X.; Jiang, Y.-Y.; Zhang, H.-Y. Marine natural products as sources of novel scaffolds: Achievement and concern. Drug Discov. Today 2010, 15, 884–886. [Google Scholar] [CrossRef]
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. [Google Scholar] [CrossRef] [PubMed]
- Seca, A.M.L.; Pinto, D.C.G.A. Plant secondary metabolites as anticancer agents: Successes in clinical trials and therapeutic application. Int. J. Mol. Sci. 2018, 19, 263. [Google Scholar] [CrossRef]
- Nwodo, J.N.; Ibezim, A.; Simoben, C.V.; Ntie-Kang, F. Exploring cancer therapeutics with natural products from african medicinal plants, part II: Alkaloids, terpenoids and flavonoids. Anticancer Agents Med. Chem. 2016, 16, 108–127. [Google Scholar] [CrossRef]
- Leal, M.C.; Madeira, C.; Brandão, C.A.; Puga, J.; Calado, R. Bioprospecting of marine invertebrates for new natural products—A chemical and zoogeographical perspective. Molecules 2012, 17, 9842–9854. [Google Scholar] [CrossRef]
- Martins, A.; Vieira, H.M.; Gaspar, H.; Santos, S. Marketed marine natural products in the pharmaceutical and cosmeceutical industries: Tips for success. Mar. Drugs 2014, 12, 1066–1101. [Google Scholar] [CrossRef]
- Ruiz-Torres, V.; Encinar, J.A.; Herranz-López, M.; Pérez-Sánchez, A.; Galiano, V.; Barrajón-Catalán, E.; Micol, V. An updated review on marine anticancer compounds: The use of virtual screening for the discovery of small-molecule cancer drugs. Molecules 2017, 22, 1037. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, J.; Hu, G.; Yu, J.; Zhu, X.; Lin, Y.; Chen, S.; Yuan, J. Statistical research on the bioactivity of new marine natural products discovered during the 28 years from 1985 to 2012. Mar. Drugs 2015, 13, 202–221. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, J.R.; Steindler, L.; Henkel, T.P.; Beer, S.; Ilan, M. Chemical warfare on coral reefs: Sponge metabolites differentially affect coral symbiosis in situ. Limnol. Oceanogr. 2007, 52, 907–911. [Google Scholar] [CrossRef]
- Chadwick, N.E.; Morrow, K.M. Competition among sessile organisms on coral reefs. In Coral Reefs: An Ecosystem in Transition; Dubinsky, Z., Stambler, N., Eds.; Springer: New York, NY, USA, 2011; pp. 347–371. [Google Scholar]
- Ribeiro, F.V.; Sá, J.A.; Fistarol, G.O.; Salomon, P.S.; Pereira, R.C.; Souza, M.L.A.; Neves, L.M.; Amado-Filho, G.M.; Francini-Filho, R.B.; Salgado, L.T.; et al. Long-term effects of competition and environmental drivers on the growth of the endangered coral Mussismilia braziliensis (Verril, 1867). PeerJ 2018, 6, e5419. [Google Scholar] [CrossRef] [PubMed]
- Horwitz, R.; Hoogenboom, M.O.; Fine, M. Spatial competition dynamics between reef corals under ocean acidification. Sci. Rep. 2017, 7, 40288. [Google Scholar] [CrossRef] [PubMed]
- Margulis, L.; Chapman, M.J. Chapter three—Animalia. In Kingdoms and Domains; Academic Press: London, UK, 2009; pp. 231–377. [Google Scholar]
- Leal, M.C.; Puga, J.; Serôdio, J.; Gomes, N.C.M.; Calado, R. Trends in the discovery of new marine natural products from invertebrates over the last two decades—Where and what are we bioprospecting? PLoS ONE 2012, 7, e30580. [Google Scholar] [CrossRef] [PubMed]
- Limame, R.; Wouters, A.; Pauwels, B.; Fransen, E.; Peeters, M.; Lardon, F.; De Wever, O.; Pauwels, P. Comparative analysis of dynamic cell viability, migration and invasion assessments by novel real-time technology and classic endpoint assays. PLoS ONE 2012, 7, e46536. [Google Scholar] [CrossRef] [PubMed]
- Roshan, M.M.; Young, A.; Reinheimer, K.; Rayat, J.; Dai, L.J.; Warnock, G.L. Dynamic assessment of cell viability, proliferation and migration using real time cell analyzer system (RTCA). Cytotechnology 2015, 67, 379–386. [Google Scholar] [CrossRef]
- Pérez-Sánchez, A.; Barrajón-Catalán, E.; Ruiz-Torres, V.; Agulló-Chazarra, L.; Herranz-López, M.; Valdés, A.; Cifuentes, A.; Micol, V. Rosemary (Rosmarinus officinalis) extract causes ros-induced necrotic cell death and inhibits tumor growth in vivo. Sci. Rep. 2019, 9. [Google Scholar] [CrossRef]
- Demchenko, A.P. Beyond annexin v: Fluorescence response of cellular membranes to apoptosis. Cytotechnology 2013, 65, 157–172. [Google Scholar] [CrossRef]
- Hingorani, R.; Deng, J.; Elia, J.; McIntyre, C.; Mittar, D. Detection of Apoptosis Using the bd annexin v fitc Assay on the bd Facsverse™ System. Available online: https: //www.bdbiosciences.com/documents/BD_FACSVerse_Apoptosis_Detection_AppNote.pdf (accessed on 1 July 2019).
- Hearps, A.C.; Burrows, J.; Connor, C.E.; Woods, G.M.; Lowenthal, R.M.; Ragg, S.J. Mitochondrial cytochrome c release precedes transmembrane depolarisation and caspase-3 activation during ceramide-induced apoptosis of Jurkat T cells. Apoptosis 2002, 7, 387–394. [Google Scholar] [CrossRef]
- Lamkanfi, M.; Festjens, N.; Declercq, W.; Vanden Berghe, T.; Vandenabeele, P. Caspases in cell survival, proliferation and differentiation. Cell Death Differ. 2007, 14, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Kuranaga, E.; Miura, M. Nonapoptotic functions of caspases: Caspases as regulatory molecules for immunity and cell-fate determination. Trends Cell Biol. 2007, 17, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Lakhani, S.A.; Masud, A.; Kuida, K.; Porter, G.A.; Booth, C.J.; Mehal, W.Z.; Inayat, I.; Flavell, R.A. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science 2006, 311, 847–851. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S. Mechanisms mediating caspase activation in cell death. Cell Death Differ. 1999, 6, 1060–1066. [Google Scholar] [CrossRef]
- Sedelnikova, O.A.; Pilch, D.R.; Redon, C.; Bonner, W.M. Histone H2AX in DNA damage and repair. Cancer Biol. 2003, 2, 233–235. [Google Scholar]
- Gomes, N.G.M.; Dasari, R.; Chandra, S.; Kiss, R.; Kornienko, A. Marine Invertebrate Metabolites with Anticancer Activities: Solutions to the “Supply Problem”. Mar. Drugs 2016, 14, 98. [Google Scholar] [CrossRef]
- Abou-ElWafa, G.S.E.; Shaaban, M.; Shaaban, K.; El-Naggar, M.E.E.; Laatsch, H. Three new unsaturated fatty acids from the marine green alga Ulva fasciata delile. Zeitschrift für Naturforschung B 2010, 64, 1199–1207. [Google Scholar] [CrossRef]
- Hamdy, A.-H.; Mettwally, W.; Abou El Fotouh, M.; Rodriguez, B.; I El-Dewany, A.; El-Toumy, S.; Hussein, A. Bioactive phenolic compounds from the egyptian red sea seagrass thalassodendron ciliatum. Z. Nat. C 2012, 67, 291–296. [Google Scholar]
- Vidal, N.P.; Manzanos, M.J.; Goicoechea, E.; Guillen, M.D. Farmed and wild sea bass (Dicentrarchus labrax) volatile metabolites: A comparative study by spme-gc/ms. J. Sci. Food Agric. 2016, 96, 1181–1193. [Google Scholar] [CrossRef]
- Lei, L.-F.; Chen, M.-F.; Wang, T.; He, X.-X.; Liu, B.-X.; Deng, Y.; Chen, X.-J.; Li, Y.-T.; Guan, S.-Y.; Yao, J.-H.; et al. Novel cytotoxic nine-membered macrocyclic polysulfur cembranoid lactones from the soft coral sinularia sp. Tetrahedron 2014, 70, 6851–6858. [Google Scholar] [CrossRef]
- Yang, S.-W.; Buevich, A.; Chan, T.-M.; Smith, M.; Lachowicz, J.; Pomponi, S.; Wright, A.; Mierzwa, R.; Patel, M.; Gullo, V.; et al. A new sterol sulfate, sch 572423, from a marine sponge, Topsentia sp. Bioorg. Med. Chem. Lett. 2003, 13, 1791–1794. [Google Scholar] [CrossRef]
- Nam, S.J.; Kauffman, C.A.; Paul, L.A.; Jensen, P.R.; Fenical, W. Actinoranone, a cytotoxic meroterpenoid of unprecedented structure from a marine adapted streptomyces sp. Org. Lett. 2013, 15, 5400–5403. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Shanmughapriya, S.; Gandhimathi, R.; Seghal Kiran, G.; Rajeetha Ravji, T.; Natarajaseenivasan, K.; Hema, T.A. Optimization and production of novel antimicrobial agents from sponge associated marine actinomycetes nocardiopsis dassonvillei mad08. Appl. Microbiol. Biotechnol. 2009, 83, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Elkhayat, E.; Ibrahim, S.; Fouad, M.; Mohamed, G.A. Dendronephthols a–c, new sesquiterpenoids from the red sea soft coral dendronephthya sp. Tetrahedron 2014, 70, 3822–3825. [Google Scholar] [CrossRef]
- Elhady, S.S.; Al-Abd, A.M.; El-Halawany, A.M.; Alahdal, A.M.; Hassanean, H.A.; Ahmed, S.A. Antiproliferative scalarane-based metabolites from the red sea sponge hyrtios erectus. Mar. Drugs 2016, 14, 130. [Google Scholar] [CrossRef] [PubMed]
- Molinski, T.F.; Faulkner, D.J. 6.Alpha.,7.Alpha.,17.Beta.-trihydroxy-15.Beta.,17-oxidospongian-16-one 7-butyrate: A new diterpene lactone from an Australian Aplysilla species. J. Org. Chem. 1986, 51, 1144–1146. [Google Scholar] [CrossRef]
- Karuso, P.; Scheuer, P. Natural products from three nudibranchs: Nembrotha kubaryana, hypselodoris infucata and chromodoris petechialis. Molecules 2002, 7, 1–6. [Google Scholar] [CrossRef]
- Imbs, A.B.; Demina, O.A.; Demidkova, D.A. Lipid class and fatty acid composition of the boreal soft coral gersemia rubiformis. Lipids 2006, 41, 721–725. [Google Scholar] [CrossRef]
- Imbs, A.B.; Demidkova, D.A.; Latypov, Y.Y.; Pham, L.Q. Application of fatty acids for chemotaxonomy of reef-building corals. Lipids 2007, 42, 1035–1046. [Google Scholar] [CrossRef]
- Sudan, S.; Rupasinghe, H.V. Antiproliferative activity of long chain acylated esters of quercetin-3-o-glucoside in hepatocellular carcinoma hepg2 cells. Exp. Biol Med. 2015, 240, 1452–1464. [Google Scholar] [CrossRef]
- Rashid, Z.; Ali, A.; Douzenel, P.; Bourgougnon, N.; Shaari, K.; Andriani, Y.; Tengku Muhammad, T.; Mohamad, H. Phenolics, fatty acids composition and biological activities of various extracts and fractions of malaysian Aaptos aaptos. Asian Pac. J. Trop. Biomed. 2018, 8, 554–564. [Google Scholar]
- Cateni, F.; Zilic, J.; Zacchigna, M.; Procida, G. Cerebrosides with antiproliferative activity from Euphorbia peplis. Fitoterapia 2010, 81, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Taritla, S.; Sharma, A.; Jayabaskaran, C. Antiproliferative and antioxidative bioactive compounds in extracts of marine-derived endophytic fungus talaromyces purpureogenus. Front. Microbiol 2018, 9, 1777. [Google Scholar] [CrossRef] [PubMed]
- Isbilen, O.; Rizaner, N.; Volkan, E. Anti-proliferative and cytotoxic activities of Allium autumnale P.H. Davis (amaryllidaceae) on human breast cancer cell lines MCF-7 and MDA-MB-231. BMC Complement. Altern. Med. 2018, 18, 30. [Google Scholar] [CrossRef] [PubMed]
- Su, J.H.; Huang, C.Y.; Li, P.J.; Lu, Y.; Wen, Z.H.; Kao, Y.H.; Sheu, J.H. Bioactive cadinane-type compounds from the soft coral sinularia scabra. Arch. Pharmacal Res. 2012, 35, 779–784. [Google Scholar] [CrossRef] [PubMed]
- Ivanisevic, J.; Perez, T.; Ereskovsky, A.V.; Barnathan, G.; Thomas, O.P. Lysophospholipids in the Mediterranean sponge Oscarella tuberculata: Seasonal variability and putative biological role. J. Chem. Ecol. 2011, 37, 537–545. [Google Scholar] [CrossRef] [PubMed]
- da Costa, E.; Melo, T.; Moreira, A.S.; Bernardo, C.; Helguero, L.; Ferreira, I.; Cruz, M.T.; Rego, A.M.; Domingues, P.; Calado, R.; et al. Valorization of lipids from gracilaria sp. Through lipidomics and decoding of antiproliferative and anti-inflammatory activity. Mar. Drugs 2017, 15, 62. [Google Scholar] [CrossRef]
- Alam, N.; Bae, B.H.; Hong, J.; Lee, C.O.; Shin, B.A.; Im, K.S.; Jung, J.H. Additional bioactive lyso-paf congeners from the sponge spirastrella abata. J. Nat. Prod. 2001, 64, 533–535. [Google Scholar] [CrossRef]
- Moritz, M.I.; Marostica, L.L.; Bianco, E.M.; Almeida, M.T.; Carraro, J.L.; Cabrera, G.M.; Palermo, J.A.; Simoes, C.M.; Schenkel, E.P. Polyoxygenated steroids from the octocoral leptogorgia punicea and in vitro evaluation of their cytotoxic activity. Mar. Drugs 2014, 12, 5864–5880. [Google Scholar] [CrossRef]
- Zidar, N.; Montalvao, S.; Hodnik, Z.; Nawrot, D.A.; Zula, A.; Ilas, J.; Kikelj, D.; Tammela, P.; Masic, L.P. Antimicrobial activity of the marine alkaloids, clathrodin and oroidin, and their synthetic analogues. Mar. Drugs 2014, 12, 940–963. [Google Scholar] [CrossRef]
- Ibrahim, S.R.; Mohamed, G.A. Ingenine E, a new cytotoxic beta-carboline alkaloid from the indonesian sponge Acanthostrongylophora ingens. J. Asian Nat. Prod. Res. 2017, 19, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Netz, N.; Opatz, T. Marine indole alkaloids. Mar. Drugs 2015, 13, 4814–4914. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.C.; Ortega, M.J.; Zubia, E.; Carballo, J.L. Cembrane diterpenes from the gorgonian Lophogorgia peruana. J. Nat. Prod. 2006, 69, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.C.; Lin, Y.C.; Ko, C.L.; Wang, L.T. New briaranes from the taiwanese gorgonian Junceella juncea. J. Nat. Prod. 2003, 66, 302–305. [Google Scholar] [CrossRef]
- Li, C.; La, M.P.; Sun, P.; Kurtan, T.; Mandi, A.; Tang, H.; Liu, B.S.; Yi, Y.H.; Li, L.; Zhang, W. Bioactive (3z,5e)-11,20-epoxybriara-3,5-dien-7,18-olide diterpenoids from the south china sea gorgonian Dichotella gemmacea. Mar. Drugs 2011, 9, 1403–1418. [Google Scholar] [CrossRef]
- Suntornchashwej, S.; Chaichit, N.; Isobe, M.; Suwanborirux, K. Hectochlorin and morpholine derivatives from the thai sea hare, Bursatella leachii. J. Nat. Prod. 2005, 68, 951–955. [Google Scholar] [CrossRef]
- Krohn, K.; Sohrab, M.H.; Aust, H.J.; Draeger, S.; Schulz, B. Biologically active metabolites from fungi, 19: New isocoumarins and highly substituted benzoic acids from the endophytic fungus, Scytalidium sp. Nat. Prod. Res. 2004, 18, 277–285. [Google Scholar] [CrossRef]
- Festa, C.; Lauro, G.; De Marino, S.; D’Auria, M.V.; Monti, M.C.; Casapullo, A.; D’Amore, C.; Renga, B.; Mencarelli, A.; Petek, S.; et al. Plakilactones from the marine sponge plakinastrella mamillaris. Discovery of a new class of marine ligands of peroxisome proliferator-activated receptor gamma. J. Med. Chem. 2012, 55, 8303–8317. [Google Scholar] [CrossRef]
- Ueoka, R.; Nakao, Y.; Kawatsu, S.; Yaegashi, J.; Matsumoto, Y.; Matsunaga, S.; Furihata, K.; van Soest, R.W.; Fusetani, N. Gracilioethers a-c, antimalarial metabolites from the marine sponge agelas gracilis. J. Org. Chem. 2009, 74, 4203–4207. [Google Scholar] [CrossRef]
- Festa, C.; D’Amore, C.; Renga, B.; Lauro, G.; De Marino, S.; D’Auria, M.V.; Bifulco, G.; Zampella, A.; Fiorucci, S. Oxygenated polyketides from plakinastrella mamillaris as a new chemotype of PXR agonists. Mar. Drugs 2013, 11, 2314–2327. [Google Scholar] [CrossRef]
- Ortega, M.J.; Zubía, E.; Sánchez, M.C.; Carballo, J.L. Cembrane diterpenes from the gorgonian Leptogorgia laxa. J. Nat. Prod. 2008, 71, 1637–1639. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.J.; Owa, T.; Schreiber, S.L.; Corey, E.J. Phthalascidin, a synthetic antitumor agent with potency and mode of action comparable to ecteinascidin 743. Proc. Natl. Acad. Sci. USA 1999, 96, 3496–3501. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Sun, L.; Yuan, J.; Sun, Y.; Gao, Y.; Zhang, L.; Li, S.; Dai, H.; Hamel, J.-F.; Liu, C.; et al. The sea cucumber genome provides insights into morphological evolution and visceral regeneration. PLoS Biol. 2017, 15, e2003790. [Google Scholar] [CrossRef] [PubMed]
- Castelli, S.; Campagna, A.; Vassallo, O.; Tesauro, C.; Fiorani, P.; Tagliatesta, P.; Oteri, F.; Falconi, M.; Majumder, H.K.; Desideri, A. Conjugated eicosapentaenoic acid inhibits human topoisomerase IB with a mechanism different from camptothecin. Arch. Biochem. Biophys. 2009, 486, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Yonezawa, Y.; Hada, T.; Uryu, K.; Tsuzuki, T.; Eitsuka, T.; Miyazawa, T.; Murakami-Nakai, C.; Yoshida, H.; Mizushina, Y. Inhibitory effect of conjugated eicosapentaenoic acid on mammalian DNA polymerase and topoisomerase activities and human cancer cell proliferation. Biochem. Pharm. 2005, 70, 453–460. [Google Scholar] [CrossRef]
- Berge, J.P.; Debiton, E.; Dumay, J.; Durand, P.; Barthomeuf, C. In vitro anti-inflammatory and anti-proliferative activity of sulfolipids from the red alga Porphyridium cruentum. J. Agric. Food Chem. 2002, 50, 6227–6232. [Google Scholar] [CrossRef]
- Martinez-Luis, S.; Gomez, J.F.; Spadafora, C.; Guzman, H.M.; Gutierrez, M. Antitrypanosomal alkaloids from the marine bacterium bacillus pumilus. Molecules 2012, 17, 11146–11155. [Google Scholar] [CrossRef]
- Bharate, S.B.; Manda, S.; Mupparapu, N.; Battini, N.; Vishwakarma, R.A. Chemistry and biology of fascaplysin, a potent marine-derived CDK-4 inhibitor. Mini Rev. Med. Chem. 2012, 12, 650–664. [Google Scholar] [CrossRef]
- Kimura, K.; Itonori, S.; Kajiwara, C.; Hada, N.; Takeda, T.; Sugita, M. Structural elucidation of the neutral glycosphingolipids, mono-, di-, tri- and tetraglycosylceramides from the marine crab Erimacrus isenbeckii. J. Oleo Sci. 2014, 63, 269–280. [Google Scholar] [CrossRef]
- Moon, S.H.; Kim, J.Y.; Song, E.H.; Shin, M.K.; Cho, Y.H.; Kim, N.I. Altered levels of sphingosine and sphinganine in psoriatic epidermis. Ann. Derm. 2013, 25, 321–326. [Google Scholar] [CrossRef]
- Bandaranayake, W.M.; Rocher, A.D. Role of secondary metabolites and pigments in the epidermal tissues, ripe ovaries, viscera, gut contents and diet of the sea cucumber Holothuria atra. Mar. Biol. 1999, 133, 163–169. [Google Scholar] [CrossRef]
- Palozza, P.; Torelli, C.; Boninsegna, A.; Simone, R.; Catalano, A.; Mele, M.C.; Picci, N. Growth-inhibitory effects of the astaxanthin-rich alga haematococcus pluvialis in human colon cancer cells. Cancer Lett. 2009, 283, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Galasso, C.; Corinaldesi, C.; Sansone, C. Carotenoids from marine organisms: Biological functions and industrial applications. Antioxidants 2017, 6, 96. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Kong, J.; Li, W.; Molchanova, V.; Chikalovets, I.; Belogortseva, N.; Luk’yanov, P.; Zheng, Y.T. A beta-galactose-specific lectin isolated from the marine worm chaetopterus variopedatus possesses anti-hiv-1 activity. Comp. Biochem. Physiol. C Toxicol. Pharm. 2006, 142, 111–117. [Google Scholar] [CrossRef]
- Watanadilok, R.; Sonchaeng, P.; Kijjoa, A.; Damas, A.M.; Gales, L.; Silva, A.M.; Herz, W. Tetillapyrone and nortetillapyrone, two unusual hydroxypyran-2-ones from the marine sponge tetilla japonica. J. Nat. Prod. 2001, 64, 1056–1058. [Google Scholar] [CrossRef]
- El-Gamal, A.A.; Al-Massarani, S.M.; Shaala, L.A.; Alahdald, A.M.; Al-Said, M.S.; Ashour, A.E.; Kumar, A.; Abdel-Kader, M.S.; Abdel-Mageed, W.M.; Youssef, D.T. Cytotoxic compounds from the Saudi Red Sea sponge Xestospongia testudinaria. Mar. Drugs 2016, 14, 82. [Google Scholar] [CrossRef]
- Manzo, E.; Carbone, M.; Mollo, E.; Irace, C.; Di Pascale, A.; Li, Y.; Ciavatta, M.L.; Cimino, G.; Guo, Y.W.; Gavagnin, M. Structure and synthesis of a unique isonitrile lipid isolated from the marine mollusk actinocyclus papillatus. Org. Lett. 2011, 13, 1897–1899. [Google Scholar] [CrossRef]
- Preethy, C.P.; Padmapriya, R.; Periasamy, V.S.; Riyasdeen, A.; Srinag, S.; Krishnamurthy, H.; Alshatwi, A.A.; Akbarsha, M.A. Antiproliferative property of n-hexane and chloroform extracts of Anisomeles malabarica (L). R. Br. In HPV16-positive human cervical cancer cells. J. Pharmacol. Pharmacother. 2012, 3, 26–34. [Google Scholar]
- Salem, S.M.; Kancharla, P.; Florova, G.; Gupta, S.; Lu, W.; Reynolds, K.A. Elucidation of final steps of the marineosins biosynthetic pathway through identification and characterization of the corresponding gene cluster. J. Am. Chem. Soc. 2014, 136, 4565–4574. [Google Scholar] [CrossRef]
- Blunt, J.W.; Copp, B.R.; Keyzers, R.A.; Munro, M.H.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2016, 33, 382–431. [Google Scholar] [CrossRef]
- Chen, G.; Wang, H.F.; Pei, Y.H. Secondary metabolites from marine-derived microorganisms. J. Asian Nat. Prod. Res. 2014, 16, 105–122. [Google Scholar] [CrossRef] [PubMed]
- Diyabalanage, T.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Palmerolide a, a cytotoxic macrolide from the antarctic tunicate synoicum adareanum. J. Am. Chem. Soc. 2006, 128, 5630–5631. [Google Scholar] [CrossRef] [PubMed]
- Noguez, J.H.; Diyabalanage, T.K.K.; Miyata, Y.; Xie, X.-S.; Valeriote, F.A.; Amsler, C.D.; McClintock, J.B.; Baker, B.J. Palmerolide macrolides from the antarctic tunicate synoicum adareanum. Bioorg. Med. Chem. 2011, 19, 6608–6614. [Google Scholar] [CrossRef] [PubMed]
- Chee, C.F.; Rahman, N.A.; Zain, S.M.; Ng, S.W. Pheophorbide b ethyl ester from a chlorella vulgaris dietary supplement. Acta Cryst. Sect. E Struct. Rep. Online 2008, 64, o1986. [Google Scholar] [CrossRef]
- Cheng, H.H.; Wang, H.K.; Ito, J.; Bastow, K.F.; Tachibana, Y.; Nakanishi, Y.; Xu, Z.; Luo, T.Y.; Lee, K.H. Cytotoxic pheophorbide-related compounds from Clerodendrum calamitosum and C. cyrtophyllum. J. Nat. Prod. 2001, 64, 915–919. [Google Scholar] [CrossRef]
- Gao, S.-S.; Li, X.-M.; Williams, K.; Proksch, P.; Ji, N.-Y.; Wang, B.-G. Rhizovarins a–f, indole-diterpenes from the mangrove-derived endophytic fungus Mucor irregularis QEN-189. J. Nat. Prod. 2016, 79, 2066–2074. [Google Scholar] [CrossRef]
- Zhukova, N.V. Lipids and fatty acids of nudibranch mollusks: Potential sources of bioactive compounds. Mar. Drugs 2014, 12, 4578–4592. [Google Scholar] [CrossRef]
- Kumar, M.S.; Adki, K.M. Marine natural products for multi-targeted cancer treatment: A future insight. Biomed. Pharm. 2018, 105, 233–245. [Google Scholar] [CrossRef]
- Mettu, R.; Narendar Reddy, T.; Oladoye, S.; Vishvakarma, R.; Vaidya, J. Stereoselective synthesis of-triacetyl-D-erythro-sphingosine and N,O,O- triacetyl sphingonine from a common chiral intermediate derived from D-mannitol. Arch. Org. Chem. 2012, vi, 421. [Google Scholar]
- Khalil, Z.G.; Huang, X.C.; Raju, R.; Piggott, A.M.; Capon, R.J. Shornephine A: Structure, chemical stability, and p-glycoprotein inhibitory properties of a rare diketomorpholine from an Australian marine-derived Aspergillus sp. J. Org. Chem. 2014, 79, 8700–8705. [Google Scholar] [CrossRef]
- Ma, Y.-M.; Liang, X.-A.; Kong, Y.; Jia, B. Structural diversity and biological activities of indole diketopiperazine alkaloids from fungi. J. Agric. Food Chem. 2016, 64, 6659–6671. [Google Scholar] [CrossRef] [PubMed]
- Shaala, L.A.; Youssef, D.T.; Badr, J.M.; Harakeh, S.M. Bioactive 2(1h)-pyrazinones and diketopiperazine alkaloids from a tunicate-derived actinomycete Streptomyces sp. Molecules 2016, 21, 1116. [Google Scholar] [CrossRef] [PubMed]
- Mioso, R.; Marante, F.J.; Bezerra, R.S.; Borges, F.V.; Santos, B.V.; Laguna, I.H. Cytotoxic compounds derived from marine sponges. A review (2010–2012). Molecules 2017, 22, 208. [Google Scholar] [CrossRef] [PubMed]
- Casalme, L.O.; Yamauchi, A.; Sato, A.; Petitbois, J.G.; Nogata, Y.; Yoshimura, E.; Okino, T.; Umezawa, T.; Matsuda, F. Total synthesis and biological activity of dolastatin 16. Org. Biomol. Chem. 2017, 15, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Egami, Y.; Kimura, M.; Wakimoto, T.; Abe, I. Metagenomic analysis of the sponge discodermia reveals the production of the cyanobacterial natural product kasumigamide by ‘entotheonella’. PLoS ONE 2016, 11, e0164468. [Google Scholar] [CrossRef] [PubMed]
- Gogineni, V.; Hamann, M.T. Marine natural product peptides with therapeutic potential: Chemistry, biosynthesis, and pharmacology. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 81–196. [Google Scholar] [CrossRef]
- Lindel, T.; Hoffmann, H.; Hochgürtel, M.; Pawlik, J.R. Structure–activity relationship of inhibition of fish feeding by sponge-derived and synthetic pyrrole–imidazole alkaloids. J. Chem. Ecol. 2000, 26, 1477–1496. [Google Scholar] [CrossRef]
- Song, X.; Xiong, Y.; Qi, X.; Tang, W.; Dai, J.; Gu, Q.; Li, J. Molecular targets of active anticancer compounds derived from marine sources. Mar. Drugs 2018, 16, 175. [Google Scholar] [CrossRef]
- Pennings, S.C.; Paul, V.J.; Dunbar, D.C.; Hamann, M.T.; Lumbang, W.A.; Novack, B.; Jacobs, R.S. Unpalatable compounds in the marine gastropod dolabella auricularia: Distribution and effect of diet. J. Chem. Ecol. 1999, 25, 735–755. [Google Scholar] [CrossRef]
- Kamio, M.; Nguyen, L.; Yaldiz, S.; Derby, C.D. How to produce a chemical defense: Structural elucidation and anatomical distribution of aplysioviolin and phycoerythrobilin in the sea hare aplysia californica. Chem. Biodivers. 2010, 7, 1183–1197. [Google Scholar] [CrossRef]
- Rudiger, W. On the defensive dyes in aplysia species. I. Aplysioviolin, a new bile pigment. Hoppe Seylers Z. Physiol. Chem. 1967, 348, 129–138. [Google Scholar] [PubMed]
- Buznikov, G.A.; Slotkin, T.A.; Lauder, J.M. Sea urchin embryos and larvae as biosensors for neurotoxicants. Curr. Protoc. Toxicol. 2003, 16, 1–6. [Google Scholar]
- Zhukova, N.V. Lipid classes and fatty acid composition of the tropical nudibranch mollusks chromodoris sp. And Phyllidia coelestis. Lipids 2007, 42, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Eser, P.O.; Vanden Heuvel, J.P.; Araujo, J.; Thompson, J.T. Marine- and plant-derived ω-3 fatty acids differentially regulate prostate cancer cell proliferation. Mol. Clin. Oncol. 2013, 1, 444–452. [Google Scholar] [CrossRef] [PubMed]
- Levina, E.V.; Kalinovsky, A.I.; Andriyashchenko, P.V.; Menzorova, N.I.; Dmitrenok, P.S. Trofosides a and b and other cytostatic steroid-derived compounds from the far east starfish trofodiscus über. Russ. J. Bioorg. Chem. 2007, 33, 334–340. [Google Scholar] [CrossRef]
- Pacheco, B.S.; Dos Santos, M.A.Z.; Schultze, E.; Martins, R.M.; Lund, R.G.; Seixas, F.K.; Colepicolo, P.; Collares, T.; Paula, F.R.; De Pereira, C.M.P. Cytotoxic activity of fatty acids from antarctic macroalgae on the growth of human breast cancer cells. Front. Bioeng. Biotechnol. 2018, 6, 185. [Google Scholar] [CrossRef]
- Fang, E.F.; Zhang, C.Z.Y.; Zhang, L.; Wong, J.H.; Chan, Y.S.; Pan, W.L.; Dan, X.L.; Yin, C.M.; Cho, C.H.; Ng, T.B. Trichosanthin inhibits breast cancer cell proliferation in both cell lines and nude mice by promotion of apoptosis. PLoS ONE 2012, 7, e41592. [Google Scholar] [CrossRef]
- Juin, C.; Bonnet, A.; Nicolau, E.; Bérard, J.-B.; Devillers, R.; Thiéry, V.; Cadoret, J.-P.; Picot, L. Uplc-mse profiling of phytoplankton metabolites: Application to the identification of pigments and structural analysis of metabolites in porphyridium purpureum. Mar. Drugs 2015, 13, 2541–2558. [Google Scholar] [CrossRef]
- Baudelet, P.H.; Gagez, A.L.; Berard, J.B.; Juin, C.; Bridiau, N.; Kaas, R.; Thiery, V.; Cadoret, J.P.; Picot, L. Antiproliferative activity of cyanophora paradoxa pigments in melanoma, breast and lung cancer cells. Mar. Drugs 2013, 11, 4390–4406. [Google Scholar] [CrossRef]
- Molinski, T.F.; Dalisay, D.S.; Lievens, S.L.; Saludes, J.P. Drug development from marine natural products. Nat. Rev. Drug Discov. 2008, 8, 69. [Google Scholar] [CrossRef]
- Dobretsov, S.; Tamimi, Y.; Al-Kindi, M.A.; Burney, I. Screening for anti-cancer compounds in marine organisms in oman. Sultan Qaboos Univ. Med. J. 2016, 16, e168–e174. [Google Scholar] [CrossRef] [PubMed]
- Fernando, I.P.S.; Sanjeewa, K.K.A.; Ann, Y.-S.; Ko, C.-i.; Lee, S.-H.; Lee, W.W.; Jeon, Y.-J. Apoptotic and antiproliferative effects of stigmast-5-en-3-ol from dendronephthya gigantea on human leukemia hl-60 and human breast cancer mcf-7 cells. Toxicol. In Vitro 2018, 52, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Shaltiel, I.A.; Krenning, L.; Bruinsma, W.; Medema, R.H. The same, only different—DNA damage checkpoints and their reversal throughout the cell cycle. J. Cell Sci. 2015, 128, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, E.; Adhami, V.M.; Mukhtar, H. Targeting microtubules by natural agents for cancer therapy. Mol. Cancer 2014, 13, 275–284. [Google Scholar] [CrossRef]
- Brown, J.S.; O’Carrigan, B.; Jackson, S.P.; Yap, T.A. Targeting DNA repair in cancer: Beyond parp inhibitors. Cancer Discov. 2017, 7, 20–37. [Google Scholar] [CrossRef]
- Uckun, F.M.; Mao, C.; Jan, S.T.; Huang, H.; Vassilev, A.O.; Navara, C.S.; Narla, R.K. Spongistatins as tubulin targeting agents. Curr. Pharm. Des. 2001, 7, 1291–1296. [Google Scholar] [CrossRef]
- Natsume, T.; Watanabe, J.; Ogawa, K.; Yasumura, K.; Kobayashi, M. Tumor-specific antivascular effect of tzt-1027 (soblidotin) elucidated by magnetic resonance imaging and confocal laser scanning microscopy. Cancer Sci. 2007, 98, 598–604. [Google Scholar] [CrossRef]
- Ahn, M.Y.; Jung, J.H.; Na, Y.J.; Kim, H.S. A natural histone deacetylase inhibitor, psammaplin a, induces cell cycle arrest and apoptosis in human endometrial cancer cells. Gynecol. Oncol. 2008, 108, 27–33. [Google Scholar] [CrossRef]
- Kaminskyy, V.O.; Zhivotovsky, B. Free radicals in cross talk between autophagy and apoptosis. Antioxid. Redox Signal. 2014, 21, 86–102. [Google Scholar] [CrossRef]
- Houtgraaf, J.H.; Versmissen, J.; van der Giessen, W.J. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc. Revasc. Med. 2006, 7, 165–172. [Google Scholar] [CrossRef]
- Taylor, R.C.; Cullen, S.P.; Martin, S.J. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231. [Google Scholar] [CrossRef] [PubMed]
- Kitanaka, C.; Kuchino, Y. Caspase-independent programmed cell death with necrotic morphology. Cell Death Differ. 1999, 6, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Belinchon, M.; Sanchez-Osuna, M.; Martinez-Escardo, L.; Granados-Colomina, C.; Pascual-Guiral, S.; Iglesias-Guimarais, V.; Casanelles, E.; Ribas, J.; Yuste, V.J. An early and robust activation of caspases heads cells for a regulated form of necrotic-like cell death. J. Biol. Chem. 2015, 290, 20841–20855. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, M.; Honda, T.; Proske, R.J.; Yeh, E.T. Regulation of reactive oxygen species-induced apoptosis and necrosis by caspase 3-like proteases. Oncogene 1998, 17, 2753–2760. [Google Scholar] [CrossRef] [PubMed]
- Mahassni, S.H.; Al-Reemi, R.M. Apoptosis and necrosis of human breast cancer cells by an aqueous extract of garden cress (Lepidium sativum) seeds. Saudi J. Biol. Sci. 2013, 20, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Nikoletopoulou, V.; Markaki, M.; Palikaras, K.; Tavernarakis, N. Crosstalk between apoptosis, necrosis and autophagy. Biochim. Biophys. Acta 2013, 1833, 3448–3459. [Google Scholar] [CrossRef]
- Arakawa, S.; Nakanomyo, I.; Kudo-Sakamoto, Y.; Akazawa, H.; Komuro, I.; Shimizu, S. Identification of a novel compound that inhibits both mitochondria-mediated necrosis and apoptosis. Biochem. Biophys. Res. Commun. 2015, 467, 1006–1011. [Google Scholar] [CrossRef]
- Kepp, O.; Galluzzi, L.; Lipinski, M.; Yuan, J.; Kroemer, G. Cell death assays for drug discovery. Nat. Rev. Drug Discov. 2011, 10, 221–237. [Google Scholar] [CrossRef]
- Qu, X.; Zou, Z.; Sun, Q.; Luby-Phelps, K.; Cheng, P.; Hogan, R.N.; Gilpin, C.; Levine, B. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 2007, 128, 931–946. [Google Scholar] [CrossRef]
- Shimizu, S.; Kanaseki, T.; Mizushima, N.; Mizuta, T.; Arakawa-Kobayashi, S.; Thompson, C.B.; Tsujimoto, Y. Role of BCL-2 family proteins in a non-apoptotic programmed cell death dependent on autophagy genes. Nat. Cell Biol. 2004, 6, 1221–1228. [Google Scholar] [CrossRef]
- Vitale, I.; Galluzzi, L.; Castedo, M.; Kroemer, G. Mitotic catastrophe: A mechanism for avoiding genomic instability. Nat. Rev. Mol. Cell Biol. 2011, 12, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Ma, L.; Merlin, D. Nanoparticle-mediated co-delivery of chemotherapeutic agent and sirna for combination cancer therapy. Expert Opin. Drug Deliv. 2017, 14, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O. The biochemistry of apoptosis. Nature 2000, 407, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Kocarnik, J.M.; Shiovitz, S.; Phipps, A.I. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol. Rep. 2015, 3, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Grasso, S.; Martínez-Lacaci, I.; Barberá, V.M.; Castillejo, A.; Soto, J.L.; Gallego-Plazas, J.; López-Riquelme, N.; García-Morales, P.; Mata-Balaguer, T.; Ferragut, J.A.; et al. Hgue-C-1 is an atypical and novel colon carcinoma cell line. BMC Cancer 2015, 15, 240. [Google Scholar] [CrossRef] [PubMed]
- Roma, C.; Rachiglio, A.M.; Pasquale, R.; Fenizia, F.; Iannaccone, A.; Tatangelo, F.; Antinolfi, G.; Parrella, P.; Graziano, P.; Sabatino, L.; et al. Braf v600e mutation in metastatic colorectal cancer: Methods of detection and correlation with clinical and pathologic features. Cancer Biol. Ther. 2016, 17, 840–848. [Google Scholar] [CrossRef]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknæs, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Chan, F.K.-M.; Moriwaki, K.; De Rosa, M.J. Detection of necrosis by release of lactate dehydrogenase (ldh) activity. Methods Mol. Biol. 2013, 979, 65–70. [Google Scholar]




| Class | Scientific Name | Code | Photo | Class | Scientific Name | Code | Photo |
|---|---|---|---|---|---|---|---|
| Soft Coral | Parazoanthus sp. | P |  | Soft Coral | Euphyllia ancora | Eu |  |
| Discosoma sp. | D | Carotalcyon sp. | CR | ||||
| Lemmalia sp. | L | Anemone | Aiptasia sp. | A | |||
| Capnella sp. | C | Hard Coral | Wellsophyllia sp. | W | |||
| Nepthea sp. | N | Echynophyllia sp. | E | ||||
| Sarcophyton sp. | SII | Fungia sp. | F | ||||
| Sinularia sp. | Si | Duncanopsamia sp. | Du | ||||
| Cataphyllia sp. | Cy | Nudibranch | Phyllidia varicosa | NA | |||
| Xenia sp. | X | Dolabella auricularia | NB | ||||
| Palythoa sp. | Py | Holothurian | Pseudocolochirus violaceus | PS |
| HGUE-C-1 | HT-29 | SW-480 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Code | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h |
| CR | 250.9 ± 92.1 | 82.0 ± 5.9 | 58.1 ± 3.4 | 15.0 ± 4.4 | 9.4 ± 1.4 | 10.6 ± 1.0 | 105.0 ± 10.9 | 27.6 ± 2.8 | 14.8 ± 1.6 |
| PS | 37.5 ± 3. 0 | 37.4 ± 1.3 | 48.0 ± 1.8 | 3.3 ± 1.1 | 0.7 ± 0.4 | 2.1 ± 0.7 | 24.3 ± 2.0 | 18.6 ± 1.2 | 16.9 ± 0.6 |
| NA | 146.0 ± 29.0 | 61.8 ± 2.9 | 78.8 ± 3.4 | 13.0 ± 2.7 | 10.0 ± 0.7 | 9.3 ± 1.0 | 57.2 ± 6.9 | 13.6 ± 1.5 | 13.0 ± 2.0 |
| NB | 11.4 ± 3.4 | 0.3 ± 0.3 | 0.1 ± 0.2 | 5.0 ± 3.6 | 0.1 ± 0.1 | 0.1 ± 0.1 | 54.3 ± 24.2 | 0.6 ± 0.4 | 0.2 ± 0.1 |
| Peak | RT a | m/z Experimental | Molecular Formula (M-H) | m/z Calculated | Error (ppm) | mSigma | Identified Compound | Area b | Identification References | Antiproliferative Activity |
| 1 | 17.1 | 171.1017 | C9H15O3 | 171.1027 | 5.4 | 29.2 | Octenoic acid hydroxy methyl ester isomer 1 * | 0.16 | [30] | |
| 2 | 19.12 | 171.1017 | C9H15O3 | 171.1027 | 5.4 | 25.5 | Octenoic acid hydroxy methyl ester isomer 2 * | 0.08 | [30] | |
| 3 | 25.43 | 449.1448 | C22H25O10 | 449.1453 | 1.3 | 32.9 | Asebotin isomer 1 * | 0.11 | [31] | [31] |
| 4 | 25.66 | 153.1277 | C10H17O | 153.1285 | 4.9 | 62.5 | Terpineol * | 0.12 | [32] | [31] |
| 5 | 26.13 | 449.1457 | C22H25O10 | 449.1453 | −0.8 | 36.9 | Asebotin isomer 2 * | 0.19 | [31] | [31] |
| 6 | 26.65 | 353.2311 | C20H33O5 | 353.2333 | 6.3 | 29.3 | Sinulariaoid D | 0.05 | [33] | [33] |
| 7 | 26.7 | 363.2502 | C18H31N6O2 | 363.2514 | 3.4 | 64.3 | Sch 575948 * | 0.04 | [34] | |
| 8 | 28.36 | 439.3304 | C32H45O 4 | 439.3323 | 3.8 | 39.9 | Actinoranone * | 0.36 | [35] | [35] |
| 9 | 29.61 | 255.1588 | C14H23O 4 | 255.1602 | 5.4 | 87.2 | Oxalic acid, allyl nonyl ester * | 0.77 | [36] | [36] |
| 10 | 29.66 | 265.1461 | C15H21O4 | 265.1445 | −5.7 | 24.8 | Dendronephthol C | 1.73 | [37] | [37] |
| 11 | 33.43 | 429.2977 | C27H41O4 | 429.3010 | 7.7 | 6.5 | Deoxoscalarin * | 1.65 | [37] | [38] |
| 12 | 36.18 | 303.2354 | C20H31O2 | 303.2330 | −7.9 | 35.8 | Spongian-16-one * | 15.00 | [39] | [40] |
| 13 | 37.02 | 283.2620 | C18H35O2 | 283.2643 | 8.5 | 11.7 | Stearic acid | 3.45 | [41,42] | [43] |
| 14 | 37.18 | 267.2312 | C17H31 O 2 | 267.2330 | 6.7 | 3.1 | Heptadecenoic acid | 6.46 | [41,42] | [44] |
| 15 | 37.7 | 327.2897 | C20H39O3 | 327.2905 | 2.4 | 13 | 2-Hydroxyeicosanoic acid | 4.59 | [41,42] | [45] |
| 16 | 37.84 | 255.2317 | C16H 31O2 | 255.2330 | 5.3 | 11.7 | Hexadecanoic acid | 5.62 | [41,42] | [46] |
| 17 | 38.05 | 281.2462 | C18H33O2 | 281.2486 | 8.5 | 30.8 | 9-Octadecenoic acid | 2.75 | [41,42] | [46,47] |
| 18 | 38.42 | 357.2772 | C24H37O2 | 357.2799 | 7.6 | 8.3 | Tetracosapentaenoic acid | 6.54 | [41,42] | |
| Peak | RT a | m/z experimental | Molecular formula (M+H) | m/z calculated | error (ppm) | mSigma | Identified compound (positive mode) | Area b | Identification references | Antiproliferative activity |
| 1 | 3.6 | 259.1768 | C15H24NaO2 | 259.1669 | −38.5 | 17.8 | Scabralin A | 0.50 | [48] | [48] |
| 2 | 8.70 | 482.3610 | C24H53NO6P | 482.3605 | −1.1 | 8.1 | 1-O-hexadecyl-sn-glycero-3-phosphocholine (lyso-PAF) * | 27.74 | [49] | [50,51] |
| 3 | 11.43 | 462.3596 | C28H48NO4 | 462.3578 | −3.9 | 23.4 | Punicinol D * | 2.35 | [52] | [52] |
| Peak | RT a | m/z Experimental | Molecular Formula (M-H) | m/z Calculated | Error (ppm) | mSigma | Identified Compound | Area b | Identification References | Antiproliferative Activity |
| 1 | 9.81 | 280.1221 | C15H14N5O | 280.1204 | 6.1 | 42.9 | N-[(2E)-3-(2-Amino-1H-imidazol-5-yl)-2-propen-1-yl]-1H-indole-2-carboxamide * | 0.11 | [53] | [53] |
| 2 | 14.36 | 262.1113 | C15H12N5 | 262.1098 | −5.8 | 37.5 | Acanthomine A isomer 1 * | 0.56 | [54] | [55] |
| 3 | 16.1 | 262.1105 | C15H12N5 | 262.1098 | −2.5 | 187.6 | Acanthomine A isomer 2 * | 0.15 | [54] | [55] |
| 4 | 22.36 | 651.2298 | C31H39O15 | 651.2294 | 0.3 | 19 | Juncenolide D * | 0.61 | [56,57] | [58] |
| 5 | 23.09 | 290.1416 | C16H20 NO4 | 290.1398 | −6.3 | 19.8 | Isopropyl-6-(4-methoxybenzyl)-4-methylmorpholine-2,5-dione * | 0.13 | [59] | |
| 6 | 23.43 | 383.2072 | C20H31O7 | 383.2075 | 0.8 | 24.4 | Acetyl-methoxydeacetyldihydrobotrydial isomer 1 * | 0.04 | [60] | |
| 7 | 23.84 | 383.2076 | C20H31O7 | 383.2075 | −0.1 | 20.2 | Acetyl-methoxydeacetyldihydrobotrydial isomer 2 * | 0.04 | [60] | |
| 8 | 23.84 | 353.1972 | C19H29O6 | 353.1970 | −0.7 | 25.5 | Gracilioether A * | 0.04 | [61,62] | [62,63] |
| 9 | 24.14 | 383.2067 | C20H31O7 | 383.2075 | 2.2 | 35.7 | Acetyl-methoxydeacetyldihydrobotrydial isomer 3 * | 0.05 | [60] | |
| 10 | 25.4 | 455.1522 | C26H23N4O2S | 445.1547 | 5.6 | 50.1 | Unknown | 14.16 | ||
| 11 | 25.72 | 445.1491 | C23H25O9 | 445.1504 | 3 | 24 | Lopholide * | 8.67 | [56,57] | [56,64] |
| 12 | 26.19 | 621.2349 | C35H33N4O7 | 621.2355 | 1 | 54.3 | Eictenascidin analog * | 5.59 | [65] | [65] |
| 13 | 26.4 | 643.2497 | C25H39N8O10S | 643.2515 | 2.8 | 27.1 | Unknown | 26.67 | ||
| 14 | 34.93 | 301.2162 | C20H29O2 | 301.2173 | 3.7 | 1.1 | Eicosapentaenoic acid | 0.76 | [66] | [67,68] |
| 15 | 36.2 | 303.2328 | C20H31O 2 | 303.2330 | 0.4 | 6.6 | Arachidonic acid | 1.13 | [66] | [69] |
| 16 | 37.04 | 283.2625 | C18H35O2 | 283.2643 | 6.2 | 20.6 | Stearic acid | 0.82 | [66] | [43] |
| 17 | 37.82 | 255.2319 | C16H31O2 | 255.2330 | 4.1 | 21.8 | Hexadecanoic acid * | 1.18 | [66] | [46] |
| Peak | RT a | m/z experimental | Molecular formula (M+H) | m/z calculated | error (ppm) | mSigma | Identified compound | Area b | Identification references | Antiproliferative activity |
| 1 | 3.96 | 146.0598 | C9H8NO | 146.0600 | 1.7 | 15.3 | 3-Formylindole * | 1.61 | [70] | [71] |
| 2 | 6.61 | 274.2730 | C16H36NO2 | 274.2741 | 3.7 | 8.4 | 2-Amino-1,3-hexadecanediol * | 4.87 | [72] | |
| 3 | 7.20 | 302.3039 | C18H40NO2 | 302.3054 | 4.9 | 11.2 | Sphinganine 1 * | 3 | [72] | [73] |
| 4 | 14.84 | 597.3906 | C40H53O4 | 597.3938 | 5.4 | 29.7 | Astaxanthin isomer 1 * | 0.5 | [74] | [75] |
| 5 | 16.68 | 597.3905 | C40H53O4 | 597.3938 | 5.6 | 27.1 | Astaxanthin isomer 2 * | 0.43 | [74] | [75] |
| 6 | 18.64 | 565.4002 | C40H53O2 | 565.404 | 6.7 | 36.7 | Canthaxanthin isomer 1 * | 0.76 | [74] | [76] |
| 7 | 19.48 | 565.4040 | C40H53O2 | 565.3999 | 7.3 | 12.9 | Canthaxanthin isomer 2 * | 0.42 | [74] | [76] |
| Peak | RT a | m/z Experimental | Molecular Formula (M-H) | m/z Calculated | Error (ppm) | mSigma | Identified Compound | Area b | Identification References | Antiproliferative Activity |
| 1 | 8.03 | 255.0881 | C12H15O6 | 255.0874 | −2.8 | 22.8 | Phenyl β-D-galactopyranoside * | 0.21 | [77] | [77] |
| 2 | 12.68 | 241.0710 | C11H13O6 | 241.0718 | 3.1 | 14.3 | Tetillapyrone * | 0.11 | [78] | [79] |
| 3 | 17.12 | 167.1067 | C10H15O2 | 167.1078 | 0.5 | 17.6 | Geranic acid * | 0.24 | [80] | [81] |
| 4 | 18.04 | 420.2317 | C25H30N3O3 | 420.2293 | −5.9 | 16.7 | Ketopremarineosin A isomer 1 * | 1.92 | [82] | [83] |
| 5 | 20.43 | 420.2299 | C25H30N3O3 | 420.2293 | −1.5 | 31.4 | Ketopremarineosin A isomer 2 * | 1.44 | [82] | [83] |
| 6 | 27.19 | 353.2312 | C20H33O5 | 353.2333 | 6.2 | 24 | 2-Furantridecanoic acid, 2,5-dihydro-2-hydroxy-3,4-dimethyl-5-oxo-, methyl ester * | 0.22 | [84] | |
| 7 | 29.51 | 540.3280 | C32H46NO6 | 540.3331 | 5.1 | 37.7 | Palmerolide A derivative isomer 1 * | 1.87 | [85,86] | [85,86] |
| 8 | 29.75 | 619.2882 | C37H39N4O5 | 619.2926 | 7.1 | 44 | Ethyl pheophorbide A * | 5.87 | [87] | [46,88] |
| 9 | 29.93 | 540.3295 | C 32H 46NO 6 | 540.3331 | 6.6 | 15.3 | Palmerolide A derivative isomer 2 * | 3.93 | [85,86] | [85,86] |
| 10 | 30.22 | 602.3456 | C32H48N3O8 | 602.3487 | 4.5 | 37.2 | Rhizovarin D * | 2.26 | [89] | [89] |
| 11 | 34.95 | 301.2153 | C20H29O2 | 301.2173 | 6.6 | 19.1 | Eicosapentanoic acid isomer 1 | 0.46 | [90] | [67,68] |
| 12 | 35.13 | 301.2162 | C20H29O 2 | 301.2173 | 3.5 | 10.5 | Eicosapentanoic acid isomer 2 | 0.47 | [90] | [67,68] |
| 13 | 35.6 | 227.2013 | C14H27O2 | 227.2017 | 1.5 | 3.8 | Tetradecanoic acid | 1.88 | [90] | |
| 14 | 36.02 | 253.2166 | C16H29O2 | 253.2173 | 2.6 | 33.4 | 9-Hexadecenoic acid | 0.75 | [90] | |
| 15 | 36.22 | 303.2329 | C20H31O2 | 303.2330 | 0.1 | 14.4 | Spongian-16-one | 5.00 | [40] | [40] |
| 16 | 36.56 | 279.2323 | C18H31O2 | 279.2330 | 2.4 | 9.1 | 9,12-Octadecadienoic acid | 3.84 | [90] | [91] |
| 17 | 36.74 | 241.2178 | C15 H29O2 | 241.2173 | −2.1 | 4.2 | Pentadecanoic acid | 5.24 | [90] | [47] |
| 18 | 37.04 | 283.2637 | C18H35O2 | 283.26.43 | 2 | 9.6 | Stearic acid | 6.66 | [90] | [43] |
| 19 | 37.54 | 255.2319 | C16H31O2 | 255.2330 | 2.7 | 1.5 | Hexadecanoic acid isomer 1 | 0.86 | [90] | [46] |
| 20 | 37.64 | 331.2640 | C 22H35O2 | 331.2643 | 0.9 | 11.8 | Docosatetraenoic acid isomer 1 | 3.60 | [90] | |
| 21 | 37.73 | 331.2646 | C22H35O2 | 331.2643 | −1.1 | 5.1 | Docosatetraenoic acid isomer 2 | 2.76 | [90] | |
| 22 | 37.83 | 255.2346 | C16H31O2 | 255.2330 | −6.6 | 34 | Hexadecanoic acid isomer 2 | 10.37 | [90] | [46] |
| 23 | 38.16 | 281.2483 | C18H33O2 | 281.2486 | 1.2 | 19.5 | 9-Octadecenoic acid | 2.83 | [90] | [46,47] |
| Peak | RT a | m/z experimental | Molecular formula (M+H) | m/z calculated | error (ppm) | mSigma | Identified compound | Area b | Identification references | Antiproliferative activity |
| 1 | 7.21 | 302.3049 | C18H40NO2 | 302.3054 | 1.5 | 3.2 | Sphinganine * | 1.53 | [92] | [73] |
| 2 | 8.72 | 482.3604 | C24H53NO6P | 482.3602 | −0.4 | 31.4 | 1-O-hexadecyl-sn-glycero-3-phosphocholine (lyso-PAF) * | 9.31 | [49] | [50,51] |
| Peak | RT a | m/z Experimental | Molecular Formula (M-H) | m/z Calculated | Error (ppm) | mSigma | Identified Compound | Area b | Identification References | Antiproliferative Activity |
| 1 | 6.09 | 218.0820 | C12H12NO3 | 218.0823 | 1.2 | 8.5 | 2,5-Morpholinedione, 3-methyl-6-(phenylmethyl)- * | 0.48 | [93] | |
| 2 | 6.81 | 275.1408 | C15H19N2O3 | 275.1401 | −2.4 | 15.8 | 2,5-Piperazinedione, 3-[(4-hydroxyphenyl)methyl]-6-(2-methylpropyl)- * | 0.32 | [94] | [95] |
| 3 | 8.01 | 255.0894 | C12H15O6 | 255.0874 | −7.9 | 13.3 | Phenyl-β-D-galactopyranoside * | 0.10 | [77] | |
| 4 | 13.88 | 454.1905 | C22H32NO7S | 454.1905 | −0.1 | 23.5 | Latrunculol A isomer 1 * | 0.55 | [96] | [96] |
| 5 | 14.87 | 438.1954 | C26H24N5O2 | 438.1935 | −4.1 | 18.9 | 1H-Indole, 3-[[3-[5-[4-(4-methyl-1-piperazinyl)phenyl]-2-furanyl]-1,2,4-oxadiazol-5-yl]methyl]- isomer 1 | 0.58 | [97] | |
| 6 | 15.64 | 454.1910 | C22H32NO7S | 454.1905 | 3.4 | 37.7 | Latrunculol A isomer 2 * | 0.15 | [96] | [96] |
| 7 | 15.94 | 438.1963 | C26H24N5O2 | 438.1935 | −6.2 | 37 | 1H-Indole, 3-[[3-[5-[4-(4-methyl-1-piperazinyl)phenyl]-2-furanyl]-1,2,4-oxadiazol-5-yl]methyl]- isomer 2 * | 0.24 | [97] | |
| 8 | 17.73 | 785.3622 | C40H49N8O9 | 785.3622 | 0.8 | 73.7 | Kasumigamide | 0.20 | [98] | [99] |
| 9 | 20.15 | 352.9135 | C9H11Br2N2O3 | 352.9142 | 2 | 359.9 | 4,5-Dibromo-N-(2,2-dimethoxy-ethyl)-1H-pyrrole-2-carboxamide * | 8.06 | [100] | [100] |
| 10 | 21.64 | 601.3731 | C35H53O8 | 601.3746 | 2.6 | 12.8 | Agosterol E3 * | 0.16 | [101] | [101] |
| 11 | 23.8 | 417.1562 | C22H25O8 | 417.1555 | −1.8 | 2.4 | Unknown | 0.11 | ||
| 12 | 24.45 | 480.2132 | C23H34N3O6S | 480.2174 | 8.8 | 18.1 | Carbamic acid, [(1R)-1-[[[(acetylamino)methyl]thio]methyl]-2-[[(1S)-1-[(acetyloxy)methyl]-2-phenylethyl]amino]-2-oxoethyl]methyl-, 1,1-dimethyl-ethyl ester * | 0.37 | [102] | |
| 13 | 26.07 | 599.2904 | C34H39N4O6 | 599.2862 | 4.9 | 5.9 | Aplysioviolin * | 1.01 | [102] | [103,104] |
| 14 | 29.92 | 540.3353 | C32H46NO6 | 540.3331 | −2.1 | 30.3 | Palmerolide A derivative * | 0.70 | [85,86] | [85,86] |
| 15 | 34.08 | 464.3186 | C30H42NO3 | 464.3170 | −3.4 | 27.9 | (7Z,10Z,13Z,16Z,19Z)-N-[2-(3,4-Dihydroxyphenyl)ethyl]-7,10,13,16,19-docosapentaenamide * | 4.79 | [105] | |
| 16 | 34.95 | 301.2195 | C20H29O2 | 301.2173 | −7.4 | 10.8 | Eicosapentanoic acid | 2.59 | [90] | [67,68] |
| 17 | 35.19 | 277.2195 | C18H29O2 | 277.2173 | −8.1 | 7 | Linolenic acid | 1.72 | [90,106] | [107] |
| 18 | 35.32 | 597.4049 | C33H57O9 | 597.4008 | −6.9 | 23.1 | Trofoside A | 1.89 | [108] | [108] |
| 19 | 35.6 | 227.2039 | C14H27O2 | 227.2017 | −9.8 | 22.5 | Tetradecanoic acid | 0.87 | [90,106] | |
| 20 | 35.82 | 327.2340 | C22H31O2 | 327.2330 | −3.1 | 31.3 | Docosahexaenoic acid | 0.43 | [90,106] | [107] |
| 21 | 35.85 | 303.2342 | C20H31O2 | 303.2330 | −4.1 | 24.8 | Spongian-16-one isomer 1 | 0.43 | [40] | [40] |
| 22 | 35.97 | 253.2189 | C16H29O2 | 253.2173 | −6.4 | 12.2 | Palmitoleic acid | 1.21 | [90,106] | |
| 23 | 36.22 | 303.2373 | C20H31O2 | 303.2350 | −4.5 | 19.3 | Spongian-16-one isomer 2 | 9.03 | [40] | [40] |
| 24 | 36.56 | 279.2353 | C18H31O2 | 279.2330 | −8.3 | 1.1 | 9, 12-Octadecadienoic acid | 7.03 | [90] | [91] |
| 25 | 37.06 | 283.2663 | C18H35O2 | 283.2643 | −7.3 | 3.7 | Stearic acid | 2.73 | [90] | [43] |
| 26 | 37.68 | 331.2670 | C22H35O2 | 331.2643 | −8.2 | 3.6 | Docosatetraenoic acid | 5.69 | [90] | |
| 27 | 37.94 | 255.2350 | C16H31O2 | 255.2330 | −8.2 | 16.5 | Hexadecanoic acid | 5.04 | [90] | [46] |
| 28 | 38.14 | 281.2508 | C18H33O2 | 281.2486 | −8 | 9.1 | 9-Octadecenoic acid | 4.77 | [90] | [46,47] |
| 29 | 38.91 | 307.2650 | C20H35O2 | 307.2643 | −2.3 | 6.9 | Eicosadienoic acid | 0.86 | [90,106] | [109] |
| Peak | RT a | m/z experimental | Molecular formula (M+H) | m/z calculated | error (ppm) | mSigma | Identified compound | Area b | Identification references | Antiproliferative activity |
| 1 | 6.89 | 436.2688 | C24H38NO6 | 436.2694 | 1.2 | 4.2 | Purpurogemutantin * | 3.84 | [110] | [110] |
| 2 | 14.98 | 535.2694 | C33H35N4O3 | 535.2704 | 1.5 | 2.4 | Pyropheophorbide A * | 8.11 | [111] | [112] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruiz-Torres, V.; Rodríguez-Pérez, C.; Herranz-López, M.; Martín-García, B.; Gómez-Caravaca, A.-M.; Arráez-Román, D.; Segura-Carretero, A.; Barrajón-Catalán, E.; Micol, V. Marine Invertebrate Extracts Induce Colon Cancer Cell Death via ROS-Mediated DNA Oxidative Damage and Mitochondrial Impairment. Biomolecules 2019, 9, 771. https://doi.org/10.3390/biom9120771
Ruiz-Torres V, Rodríguez-Pérez C, Herranz-López M, Martín-García B, Gómez-Caravaca A-M, Arráez-Román D, Segura-Carretero A, Barrajón-Catalán E, Micol V. Marine Invertebrate Extracts Induce Colon Cancer Cell Death via ROS-Mediated DNA Oxidative Damage and Mitochondrial Impairment. Biomolecules. 2019; 9(12):771. https://doi.org/10.3390/biom9120771
Chicago/Turabian StyleRuiz-Torres, Verónica, Celia Rodríguez-Pérez, María Herranz-López, Beatriz Martín-García, Ana-María Gómez-Caravaca, David Arráez-Román, Antonio Segura-Carretero, Enrique Barrajón-Catalán, and Vicente Micol. 2019. "Marine Invertebrate Extracts Induce Colon Cancer Cell Death via ROS-Mediated DNA Oxidative Damage and Mitochondrial Impairment" Biomolecules 9, no. 12: 771. https://doi.org/10.3390/biom9120771
APA StyleRuiz-Torres, V., Rodríguez-Pérez, C., Herranz-López, M., Martín-García, B., Gómez-Caravaca, A.-M., Arráez-Román, D., Segura-Carretero, A., Barrajón-Catalán, E., & Micol, V. (2019). Marine Invertebrate Extracts Induce Colon Cancer Cell Death via ROS-Mediated DNA Oxidative Damage and Mitochondrial Impairment. Biomolecules, 9(12), 771. https://doi.org/10.3390/biom9120771














