Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Differential Scanning Calorimetry (DSC) Studies
2.3. Modeling of DSC Curves
2.4. Ultraviolet Absorption Studies
3. Results and Discussion
3.1. The Systems Studied
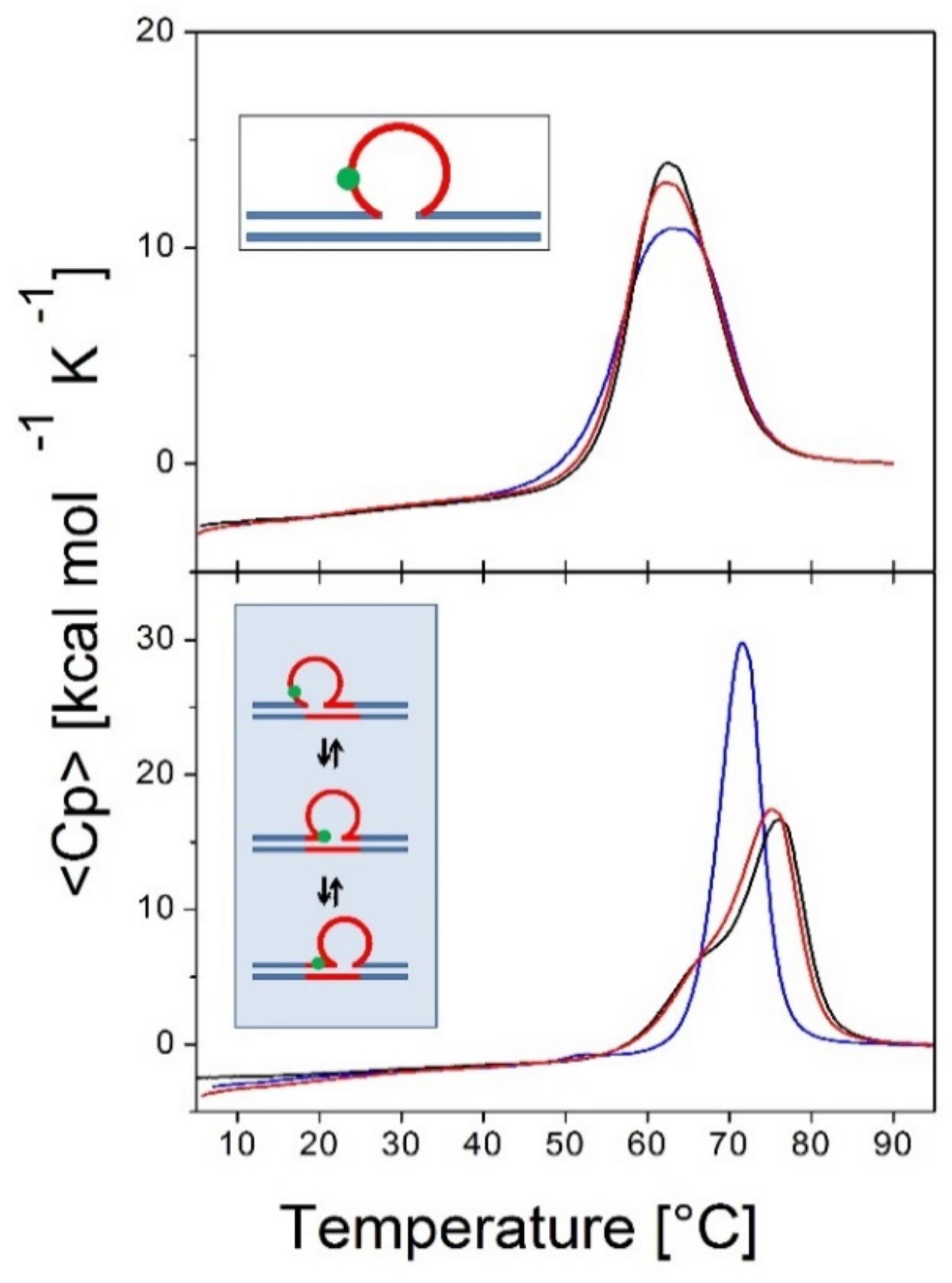
3.1.1. The Shape of the Rollamer Heat Capacity Curve Reflects the Coupling and Merging of Duplex Arm Melting and Loop Migration
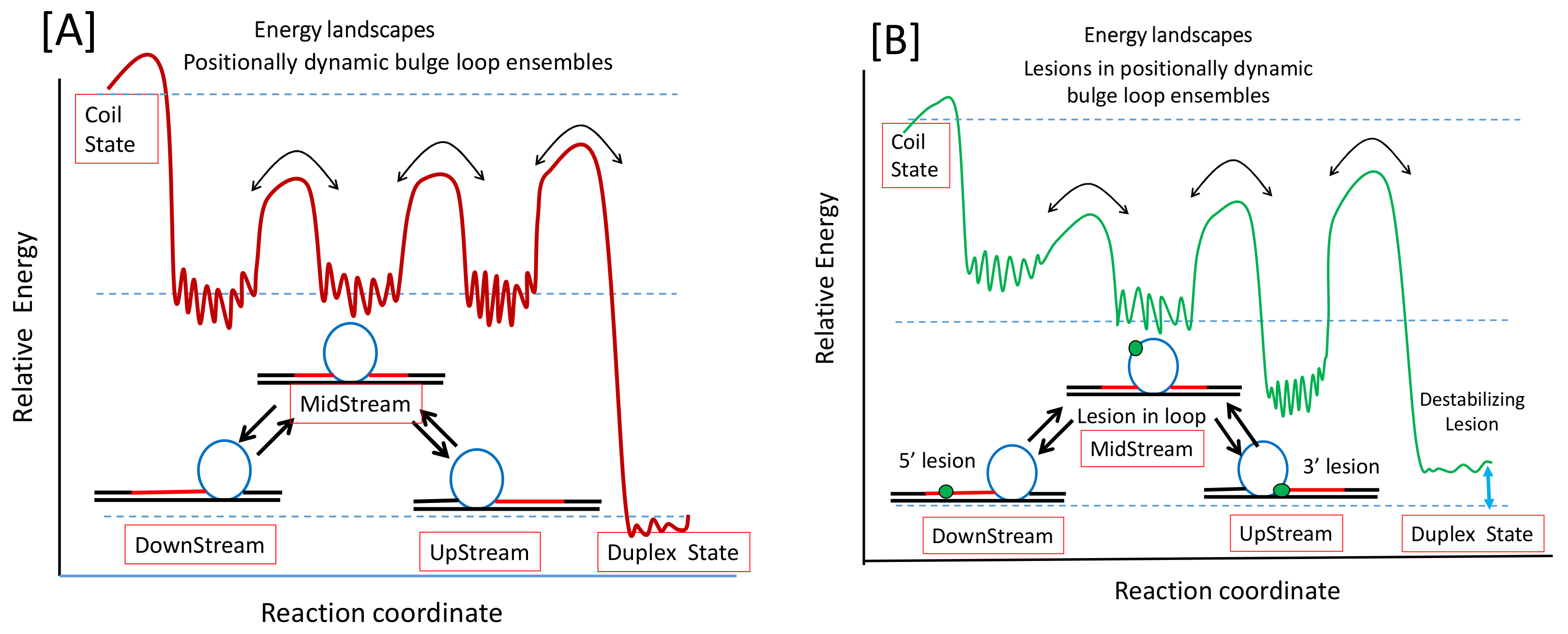
3.1.2. Differential Impact of Lesions on Static Versus Dynamic Loop Landscapes
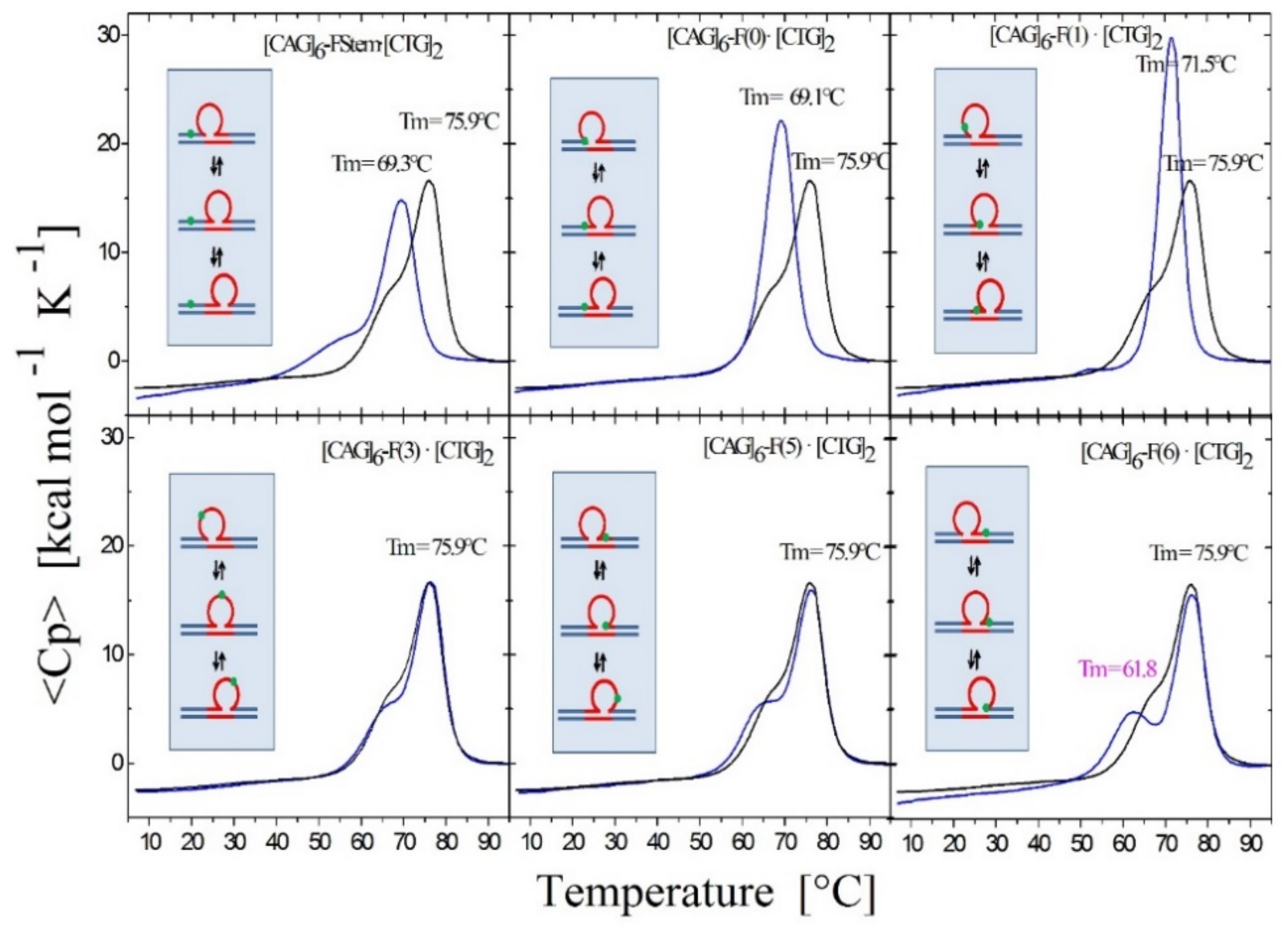
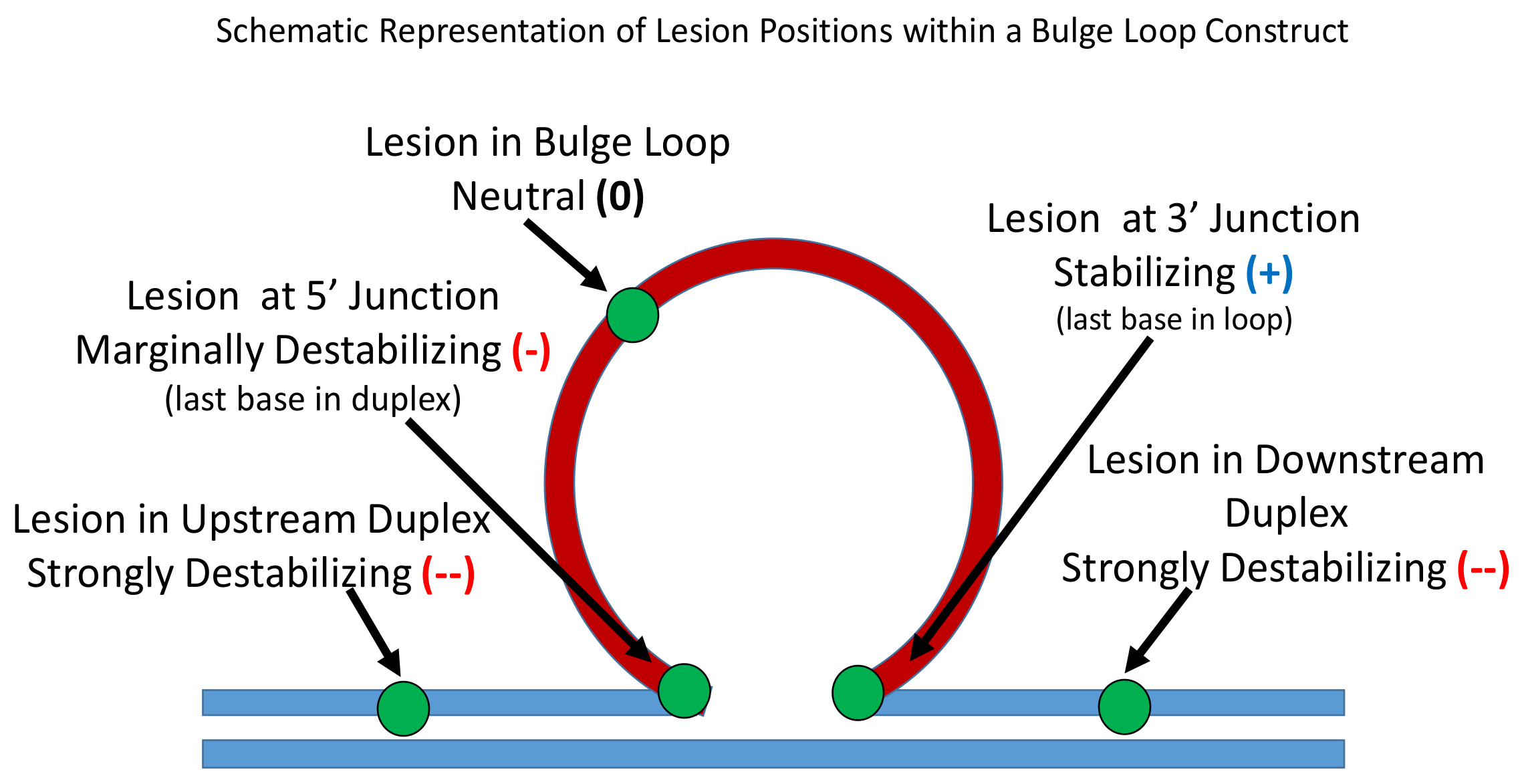
3.1.3. Lesion Impact Is Position Dependent
3.1.4. Position-Dependent Lesion Impact on Rollamer Distribution
3.1.5. Lesions Cause Preferential Occupancy of the Enthalpically Most Stabilizing of the Three Possible Loop Isomer States
3.2. Semi-Empirical Modeling Yields Rollamer Distributions Consistent with Our Experimental Results
3.3. Implications of Dynamic Energy Landscapes on DNA Repair
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bagshaw, A.T.M. Functional Mechanisms of Microsatellite DNA in Eukaryotic Genomes. Genome Biol. Evol. 2017, 9, 2428–2443. [Google Scholar] [CrossRef] [PubMed]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Mishra, R.K.; Singh, L. Genome-Wide analysis of microsatellite repeats in humans: Their abundance and density in specific genomic regions. Genome Biol. 2003, 4, R13. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, G.R.; Richards, R.I. Simple Tandem DNA Repeats and Human Genetic Disease. Proc. Natl. Acad. Sci. USA 1995, 92, 3636–3641. [Google Scholar] [CrossRef] [PubMed]
- Cummings, C.J.; Zoghbi, H.Y. Trinucleotide repeats: Mechanisms and pathophysiology. Annu. Rev. Genom. Hum. Genet. 2000, 1, 281–328. [Google Scholar] [CrossRef] [PubMed]
- Orr, H.T.; Zoghbi, H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007, 30, 575–621. [Google Scholar] [CrossRef] [PubMed]
- Ashley, C.T.; Warren, S.T. Trinucleotide repeat expansion and human disease. Annu. Rev. Genet. 1995, 29, 703–728. [Google Scholar] [CrossRef] [PubMed]
- Bacolla, A.; Wells, R.D. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009, 48, 273–285. [Google Scholar] [CrossRef]
- Wells, R.D.; Dere, R.; Hebert, M.L.; Napierala, M.; Son, L.S. Advances in mechanisms of genetic instability related to hereditary neurological diseases. Nucl. Acids Res. 2005, 33, 3785–3798. [Google Scholar] [CrossRef]
- Wells, R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007, 32, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Bacolla, A.; Wang, G.; Vasquez, K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2009, 67, 43–62. [Google Scholar] [CrossRef] [PubMed]
- Gadgil, R.; Barthelemy, J.; Lewis, T.; Leffak, M. Replication stalling and DNA microsatellite instability. Biophys. Chem. 2017, 225, 38–48. [Google Scholar] [CrossRef] [PubMed]
- McMurray, C.T. DNA secondary structure: A common and causative factor for expansion in human disease. Proc. Natl. Acad. Sci. USA 1999, 96, 1823–1825. [Google Scholar] [CrossRef]
- Bacolla, A.; Larson, J.E.; Collins, J.R.; Li, J.; Milosavljevic, A.; Stenson, P.D.; Cooper, D.N.; Wells, R.D. Abundance and length of simple repeats in vertebrate genomes are determined by their structural properties. Genome Res. 2008, 18, 1545–1553. [Google Scholar] [CrossRef]
- Wells, R.D. Discovery of the role of non-B DNA structures in mutagenesis and human genomic disorders. J. Biol. Chem. 2009, 284, 8997–9009. [Google Scholar] [CrossRef]
- Bacolla, A.; Wojciechowska, M.; Kosmider, B.; Larson, J.E.; Wells, R.D. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst.) 2006, 5, 1161–1170. [Google Scholar] [CrossRef]
- Liu, G.; Chen, X.; Bissler, J.J.; Sinden, R.R.; Leffak, M. Replication-Dependent instability at (CTG) x (CAG) repeat hairpins in human cells. Nat. Chem. Biol. 2010, 6, 652–659. [Google Scholar] [CrossRef]
- Liu, G.; Chen, X.; Gao, Y.; Lewis, T.; Barthelemy, J.; Leffak, M. Altered replication in human cells promotes DMPK (CTG)(n) (CAG)(n) repeat instability. Mol. Cell. Biol. 2012, 32, 1618–1632. [Google Scholar] [CrossRef]
- Liu, G.; Leffak, M. Instability of (CTG)n*(CAG)n trinucleotide repeats and DNA synthesis. Cell Biosci. 2012, 2, 7. [Google Scholar] [CrossRef]
- Jakupciak, J.P.; Wells, R.D. Gene conversion (recombination) mediates expansions of CTG[middle dot]CAG repeats. J. Biol. Chem. 2000, 275, 40003–40013. [Google Scholar] [CrossRef] [PubMed]
- Jakupciak, J.P.; Wells, R.D. Genetic instabilities of triplet repeat sequences by recombination. IUBMB Life 2000, 50, 355–359. [Google Scholar] [CrossRef] [PubMed]
- Usdin, K.; House, N.C.; Freudenreich, C.H. Repeat instability during DNA repair: Insights from model systems. Crit. Rev. Biochem. Mol. Biol. 2015, 50, 142–167. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, G.B.; Lau, R.; Montgomery, S.E.; Leonard, M.R.; Pearson, C.E. Slipped (CTG)*(CAG) repeats can be correctly repaired, escape repair or undergo error-prone repair. Nat. Struct. Mol. Biol. 2005, 12, 654–662. [Google Scholar] [CrossRef]
- Budworth, H.; McMurray, C.T. Bidirectional transcription of trinucleotide repeats: Roles for excision repair. DNA Repair (Amst.) 2013, 12, 672–684. [Google Scholar] [CrossRef] [PubMed]
- McMurray, C.T. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair (Amst.) 2008, 7, 1121–1134. [Google Scholar] [CrossRef] [PubMed]
- Avila Figueroa, A.; Delaney, S. Mechanistic studies of hairpin to duplex conversion for trinucleotide repeat sequences. J. Biol. Chem. 2010, 285, 14648–14657. [Google Scholar] [CrossRef]
- Volle, C.B.; Jarem, D.A.; Delaney, S. Trinucleotide Repeat DNA Alters Structure To Minimize the Thermodynamic Impact of 8-Oxo-7,8-dihydroguanine. Biochemistry (N. Y.) 2011, 51, 52–62. [Google Scholar] [CrossRef]
- Polyzos, A.A.; McMurray, C.T. Close encounters: Moving along bumps, breaks, and bubbles on expanded trinucleotide tracts. DNA Repair (Amst.) 2017, 56, 144–155. [Google Scholar] [CrossRef]
- Avila-Figueroa, A.; Cattie, D.; Delaney, S. A small unstructured nucleic acid disrupts a trinucleotide repeat hairpin. Biochem. Biophys. Res. Commun. 2011, 413, 532–536. [Google Scholar] [CrossRef][Green Version]
- Figueroa, A.A.; Cattie, D.; Delaney, S. Structure of even/odd trinucleotide repeat sequences modulates persistence of non-B conformations and conversion to duplex. Biochemistry 2011, 50, 4441–4450. [Google Scholar] [CrossRef] [PubMed]
- Völker, J.; Klump, H.H.; Breslauer, K.J. DNA metastability and biological regulation: Conformational dynamics of metastable omega-DNA bulge loops. J. Am. Chem. Soc. 2007, 129, 5272–5280. [Google Scholar] [CrossRef] [PubMed]
- Völker, J.; Plum, G.E.; Gindikin, V.; Klump, H.H.; Breslauer, K.J. Impact of bulge loop size on DNA triplet repeat domains: Implications for DNA repair and expansion. Biopolymers 2014, 101, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Völker, J.; Makube, N.; Plum, G.E.; Klump, H.H.; Breslauer, K.J. Conformational energetics of stable and metastable states formed by DNA triplet repeat oligonucleotides: Implications for triplet expansion diseases. Proc. Natl. Acad. Sci. USA 2002, 99, 14700–14705. [Google Scholar] [CrossRef]
- Degtyareva, N.N.; Reddish, M.J.; Sengupta, B.; Petty, J.T. Structural studies of a trinucleotide repeat sequence using 2-aminopurine. Biochemistry (N. Y.) 2009, 48, 2340–2346. [Google Scholar] [CrossRef]
- Degtyareva, N.N.; Barber, C.A.; Sengupta, B.; Petty, J.T. Context dependence of trinucleotide repeat structures. Biochemistry (N. Y.) 2010, 49, 3024–3030. [Google Scholar] [CrossRef]
- Degtyareva, N.N.; Barber, C.A.; Reddish, M.J.; Petty, J.T. Sequence length dictates repeated CAG folding in three-way junctions. Biochemistry (N. Y.) 2011, 50, 458–465. [Google Scholar] [CrossRef]
- Mitchell, M.L.; Leveille, M.P.; Solecki, R.S.; Tran, T.; Cannon, B. Sequence-Dependent Effects of Monovalent Cations on the Structural Dynamics of Trinucleotide-Repeat DNA Hairpins. J. Phys. Chem. B 2018. [Google Scholar] [CrossRef]
- Slean, M.M.; Reddy, K.; Wu, B.; Nichol Edamura, K.; Kekis, M.; Nelissen, F.H.; Aspers, R.L.; Tessari, M.; Scharer, O.D.; Wijmenga, S.S.; et al. Interconverting conformations of slipped-DNA junctions formed by trinucleotide repeats affect repair outcome. Biochemistry 2013, 52, 773–785. [Google Scholar] [CrossRef]
- Gacy, A.M.; McMurray, C.T. Influence of hairpins on template reannealing at trinucleotide repeat duplexes: A model for slipped DNA. Biochemistry 1998, 37, 9426–9434. [Google Scholar] [CrossRef]
- Gacy, A.M.; Goellner, G.; Juranic, N.; Macura, S.; McMurray, C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell 1995, 81, 533–540. [Google Scholar] [CrossRef]
- Lee, D.Y.; McMurray, C.T. Trinucleotide expansion in disease: Why is there a length threshold? Curr. Opin. Genet. Dev. 2014, 26, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S.C. Slipped Structures in DNA Triplet Repeat Sequences: Entropic Contributions to Genetic Instabilities. Biochemistry 1997, 36, 3047–3049. [Google Scholar] [CrossRef] [PubMed]
- Völker, J.; Gindikin, V.; Klump, H.H.; Plum, G.E.; Breslauer, K.J. Energy landscapes of dynamic ensembles of rolling triplet repeat bulge loops: Implications for DNA expansion associated with disease states. J. Am. Chem. Soc. 2012, 134, 6033–6044. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Budworth, H.; Beaver, J.M.; Chan, N.L.; Zhang, Z.; McMurray, C.T.; Liu, Y. Crosstalk between MSH2-MSH3 and polbeta promotes trinucleotide repeat expansion during base excision repair. Nat. Commun. 2016, 7, 12465. [Google Scholar] [CrossRef]
- Liu, Y.; Prasad, R.; Beard, W.A.; Hou, E.W.; Horton, J.K.; McMurray, C.T.; Wilson, S.H. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 2009, 284, 28352–28366. [Google Scholar] [CrossRef]
- Gelfand, C.A.; Plum, G.E.; Grollman, A.P.; Johnson, F.; Breslauer, K.J. The impact of a bistrand abasic lesion on DNA duplex properties. Biopolymers 1996, 38, 439–445. [Google Scholar] [CrossRef]
- Gelfand, C.A.; Plum, G.E.; Grollman, A.P.; Johnson, F.; Breslauer, K.J. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry (N. Y.) 1998, 37, 7321–7327. [Google Scholar] [CrossRef]
- Minetti, C.A.; Remeta, D.P.; Zharkov, D.O.; Plum, G.E.; Johnson, F.; Grollman, A.P.; Breslauer, K.J. Energetics of lesion recognition by a DNA repair protein: Thermodynamic characterization of formamidopyrimidine-glycosylase (Fpg) interactions with damaged DNA duplexes. J. Mol. Biol. 2003, 328, 1047–1060. [Google Scholar] [CrossRef]
- Völker, J.; Plum, G.E.; Klump, H.H.; Breslauer, K.J. DNA repair and DNA triplet repeat expansion: The impact of abasic lesions on triplet repeat DNA energetics. J. Am. Chem. Soc. 2009, 131, 9354–9360. [Google Scholar] [CrossRef][Green Version]
- David, S.S.; O’Shea, V.L.; Kundu, S. Base-Excision repair of oxidative DNA damage. Nature 2007, 447, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.S.; Demple, B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. FEBS J. 2006, 273, 1620–1629. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Wilson, D.M. Overview of base excision repair biochemistry. Curr. Mol. Pharmacol. 2012, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Nyberg, B. Rate of depurination of native deoxyribonucleic acid. Biochemistry 1972, 11, 3610–3618. [Google Scholar] [CrossRef]
- Loeb, L.A.; Preston, B.D. Mutagenesis by apurinic/apyrimidinic sites. Annu. Rev. Genet. 1986, 20, 201–230. [Google Scholar] [CrossRef]
- Jarem, D.A.; Wilson, N.R.; Delaney, S. Structure-Dependent DNA Damage and Repair in a Trinucleotide Repeat Sequence. Biochemistry (N. Y.) 2009, 48, 6655–6663. [Google Scholar] [CrossRef]
- Kovtun, I.V.; McMurray, C.T. Features of trinucleotide repeat instability in vivo. Cell Res. 2008, 18, 198–213. [Google Scholar] [CrossRef]
- Kovtun, I.V.; Liu, Y.; Bjoras, M.; Klungland, A.; Wilson, S.H.; McMurray, C.T. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 2007, 447, 447–452. [Google Scholar] [CrossRef]
- Völker, J.; Klump, H.H.; Breslauer, K.J. DNA energy landscapes via calorimetric detection of microstate ensembles of metastable macrostates and triplet repeat diseases. Proc. Natl. Acad. Sci. USA 2008, 105. [Google Scholar] [CrossRef]
- Völker, J.; Plum, G.E.; Klump, H.H.; Breslauer, K.J. Energetic coupling between clustered lesions modulated by intervening triplet repeat bulge loops: Allosteric implications for DNA repair and triplet repeat expansion. Biopolymers 2010, 93, 355–369. [Google Scholar] [CrossRef][Green Version]
- Snell, F.D.; Snell, C.T. Colorimetric Methods of Analysis, including Some Turbidimetric and Nephelometric Methods; R. E. Krieger Pub. Co.: Malabar, FL, USA, 1972. [Google Scholar]
- Plum, G.E. Optical Methods; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2000; pp. 7.3.1–7.3.17. [Google Scholar]
- Job, P. Job’s method of continuous variation. Ann. Chim. 1928, 9, 113–203. [Google Scholar]
- Völker, J.; Blake, R.; Delcourt, S.G.; Breslauer, K.J. High-Resolution calorimetric and optical melting profiles of DNA plasmids: Resolving contributions from intrinsic melting domains and specifically designed inserts. Biopolymers 1999, 50, 303–318. [Google Scholar] [CrossRef]
- Privalov, G.; Kavina, V.; Freire, E.; Privalov, P.L. Precise scanning calorimeter for studying thermal properties of biological macromolecules in dilute solution. Anal. Biochem. 1995, 232, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Marky, L.A.; Breslauer, K.J. Calculating thermodynamic data for transitions of any molecularity from equilibrium melting curves. Biopolymers 1987, 26, 1601–1620. [Google Scholar] [CrossRef]
- Wyman, J.; Gill, S.J. Binding and Linkage. Functional Chemistry of Biological Macromolecules; University Science Books: Mill Valley, CA, USA, 1990. [Google Scholar]
- Gill, S.J.; Richey, B.; Bishop, G.; Wyman, J. Generalized binding phenomena in an allosteric macromolecule. Biophys. Chem. 1985, 21, 1–14. [Google Scholar] [CrossRef]
- Plum, G.E.; Grollman, A.P.; Johnson, F.; Breslauer, K.J. Influence of the oxidatively damaged adduct 8-oxodeoxyguanosine on the conformation, energetics, and thermodynamic stability of a DNA duplex. Biochemistry (N. Y.) 1995, 34, 16148–16160. [Google Scholar] [CrossRef]
- Plum, G.E.; Grollman, A.P.; Johnson, F.; Breslauer, K.J. Influence of an exocyclic guanine adduct on the thermal stability, conformation, and melting thermodynamics of a DNA duplex. Biochemistry 1992, 31, 12096–12102. [Google Scholar] [CrossRef]
- Crenshaw, C.M.; Wade, J.E.; Arthanari, H.; Frueh, D.; Lane, B.F.; Nunez, M.E. Hidden in plain sight: Subtle effects of the 8-oxoguanine lesion on the structure, dynamics, and thermodynamics of a 15-base pair oligodeoxynucleotide duplex. Biochemistry 2011, 50, 8463–8477. [Google Scholar] [CrossRef]
- Steenken, S.; Jovanovic, S.V. How Easily Oxidizable Is DNA? One-Electron Reduction Potentials of Adenosine and Guanosine Radicals in Aqueous Solution. J. Am. Chem. Soc. 1997, 119, 617. [Google Scholar] [CrossRef]
- Yennie, C.J.; Delaney, S. Thermodynamic consequences of the hyperoxidized guanine lesion guanidinohydantoin in duplex DNA. Chem. Res. Toxicol. 2012, 25, 1732–1739. [Google Scholar] [CrossRef]
- Wilson, D.M., III; Bohr, V.A. The mechanics of base excision repair, and its relationship to aging and disease. DNA Repair (Amst.) 2007, 6, 544–559. [Google Scholar] [CrossRef] [PubMed]
- Zharkov, D.O. Base excision DNA repair. Cell. Mol. Life Sci. 2008, 65, 1544–1565. [Google Scholar] [CrossRef] [PubMed]
- Völker, J.; Plum, G.E.; Klump, H.H.; Breslauer, K.J. Energy crosstalk between DNA lesions: Implications for allosteric coupling of DNA repair and triplet repeat expansion pathways. J. Am. Chem. Soc. 2010, 132, 4095–4097. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McMurray, C.T. Mechanisms of trinucleotide repeat instability during human development. Nat. Rev. Genet. 2010, 11, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Lenzmeier, B.A.; Freudenreich, C.H. Trinucleotide repeat instability: A hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet. Genome Res. 2003, 100, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Pearson, C.E.; Tam, M.; Wang, Y.H.; Montgomery, S.E.; Dar, A.C.; Cleary, J.D.; Nichol, K. Slipped-Strand DNAs formed by long (CAG)*(CTG) repeats: Slipped-Out repeats and slip-out junctions. Nucleic Acids Res. 2002, 30, 4534–4547. [Google Scholar] [CrossRef]
- Li, M.; Völker, J.; Breslauer, K.J.; Wilson, D.M. APE1 incision activity at abasic sites in tandem repeat sequences. J. Mol. Biol. 2014, 426, 2183–2198. [Google Scholar] [CrossRef]
- Wilson, S.H.; Kunkel, T.A. Passing the baton in base excision repair. Nat. Struct. Biol. 2000, 7, 176–178. [Google Scholar] [CrossRef]
- Goula, A.V.; Pearson, C.E.; Della Maria, J.; Trottier, Y.; Tomkinson, A.E.; Wilson, D.M.; Merienne, K. The nucleotide sequence, DNA damage location, and protein stoichiometry influence the base excision repair outcome at CAG/CTG repeats. Biochemistry 2012, 51, 3919–3932. [Google Scholar] [CrossRef]
- Prasad, R.; Batra, V.K.; Yang, X.; Krahn, J.M.; Pedersen, L.C.; Beard, W.A.; Wilson, S.H. Structural insight into the DNA polymerase beta deoxyribose phosphate lyase mechanism. DNA Repair 2005, 4, 1347–1357. [Google Scholar] [CrossRef]


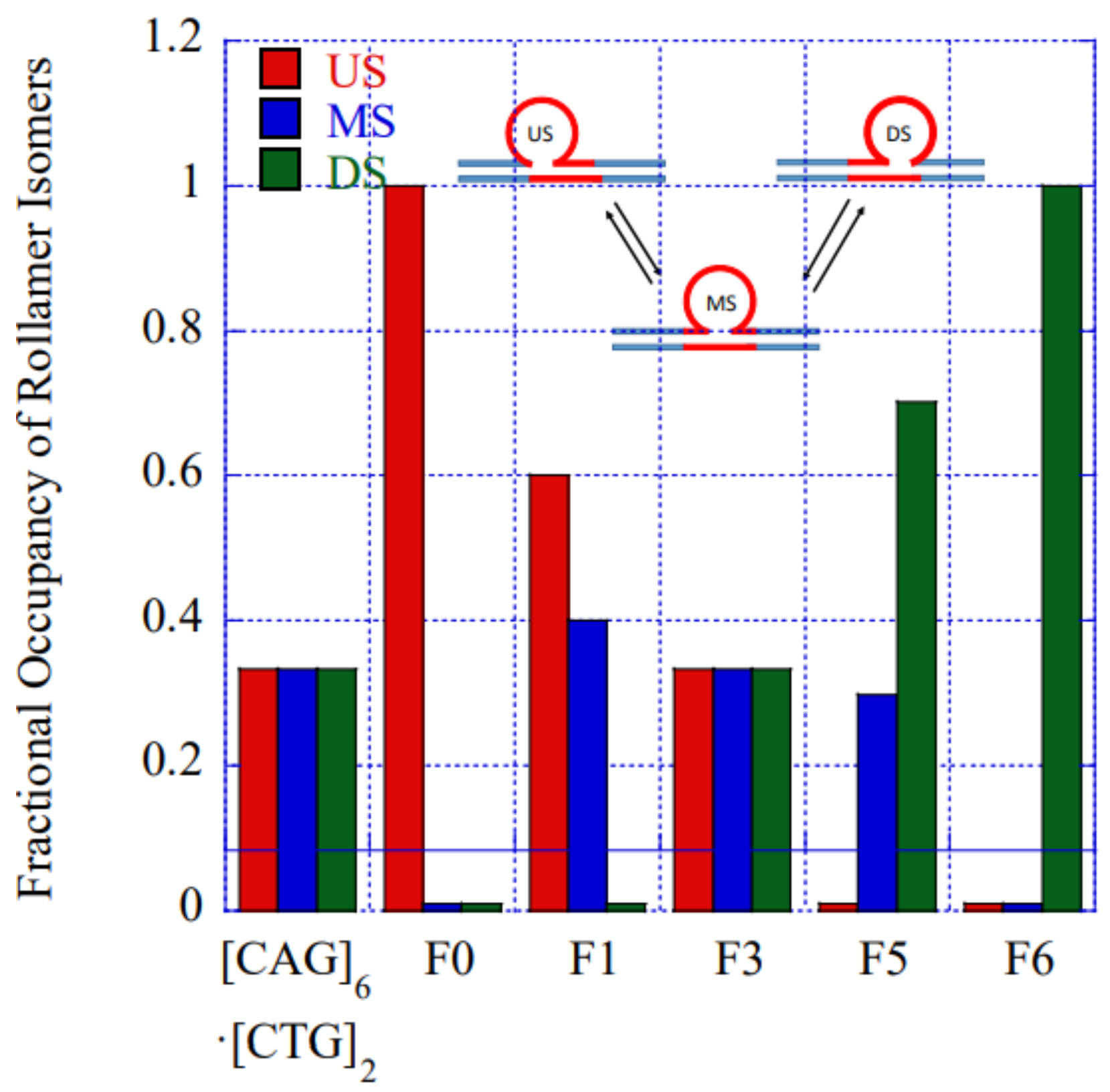
| Rollamer Constructs | Lesion (x) Position in Sequence | Lesion Positions in Each of the Three Rollamers | Predicted Impacts from Static Loop Studies | Observed Impact on Rollamer Loop Distributions Assessed Calorimetrically and Computationally | ||
|---|---|---|---|---|---|---|
| Upstream | Midstream | Downstream | ||||
| [CAG]6·[CTG]2 Lesion-free control | N/A | N/A |  |  |  | N/A |
| [CAG]6-X(Stem) ·[CTG]2 | Upstream Stem | 5 b. upstream 8 b. upstream 11 b. upstream | s. destabilizing - | s. destabilizing – | s. destabilizing -  | No impact on loop distribution |
| [CAG]6-F(0) ·[CTG]2 | Last base before 1st repeat | 5′ junction 3b. upstream 6 b. upstream | m. Destabilizing - | s. Destabilizing -  | s. Destabilizing -  | Populates rollamer with lesion at 5′ duplex/loop junction |
| [CAG]6-X(1) ·[CTG]2 | G in 1st repeat | 1 repeat loop 5′ junction 3 b. upstream | Neutral 0  | m. Destabilizing -  | s. Destabilizing --  | Populates rollamer with lesion within loop |
| [CAG]6-X(3) ·[CTG]2 | G in 3rd repeat | 5′ of loop apex Loop apex 3′ of loop apex | Neutral 0  | m. Stabilizing +  | neutral 0  | Populates rollamer with lesion within loop |
| [CAG]6-X(5) ·[CTG]2 | G in 5th repeat | 3 b. downstream 3′ junction 3 b. into loop | Destabilizing -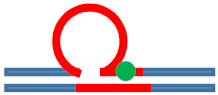 | s. Stabilizing ++ | Neutral 0 | Populates rollamer with lesion at 3/loop/duplex junction |
| [CAG]6-F(6) ·[CTG]2 | G in 6th repeat | 6 b. downstream 3 b. downstream 3′ junction | s. Destabilizing - | s. Destabilizing - | s. Stabilizing ++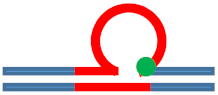 | Populates rollamer with lesion at 3/loop/duplex junction |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Völker, J.; Plum, G.E.; Gindikin, V.; Breslauer, K.J. Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair. Biomolecules 2019, 9, 709. https://doi.org/10.3390/biom9110709
Völker J, Plum GE, Gindikin V, Breslauer KJ. Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair. Biomolecules. 2019; 9(11):709. https://doi.org/10.3390/biom9110709
Chicago/Turabian StyleVölker, Jens, G. Eric Plum, Vera Gindikin, and Kenneth J. Breslauer. 2019. "Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair" Biomolecules 9, no. 11: 709. https://doi.org/10.3390/biom9110709
APA StyleVölker, J., Plum, G. E., Gindikin, V., & Breslauer, K. J. (2019). Dynamic DNA Energy Landscapes and Substrate Complexity in Triplet Repeat Expansion and DNA Repair. Biomolecules, 9(11), 709. https://doi.org/10.3390/biom9110709







