Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Buffers, and Chemicals
2.2. Sampling of Life Cycle Stages for CK Profiling
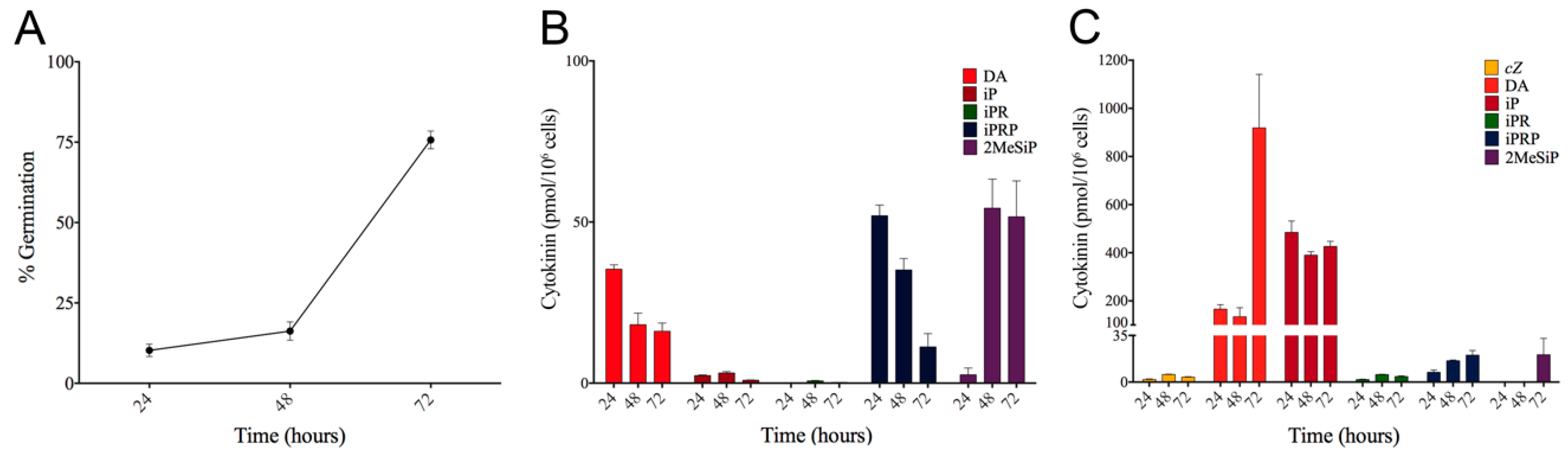
2.3. Spore Germination
2.4. Extraction and Purification of Dictyostelium-Derived CKs
2.5. CK Quantification by UHPLC-(ESI+)-HRMS/MS
2.6. Untargeted Metabolomics
2.7. Growth Assays with Exogenous CK Treatment
3. Results
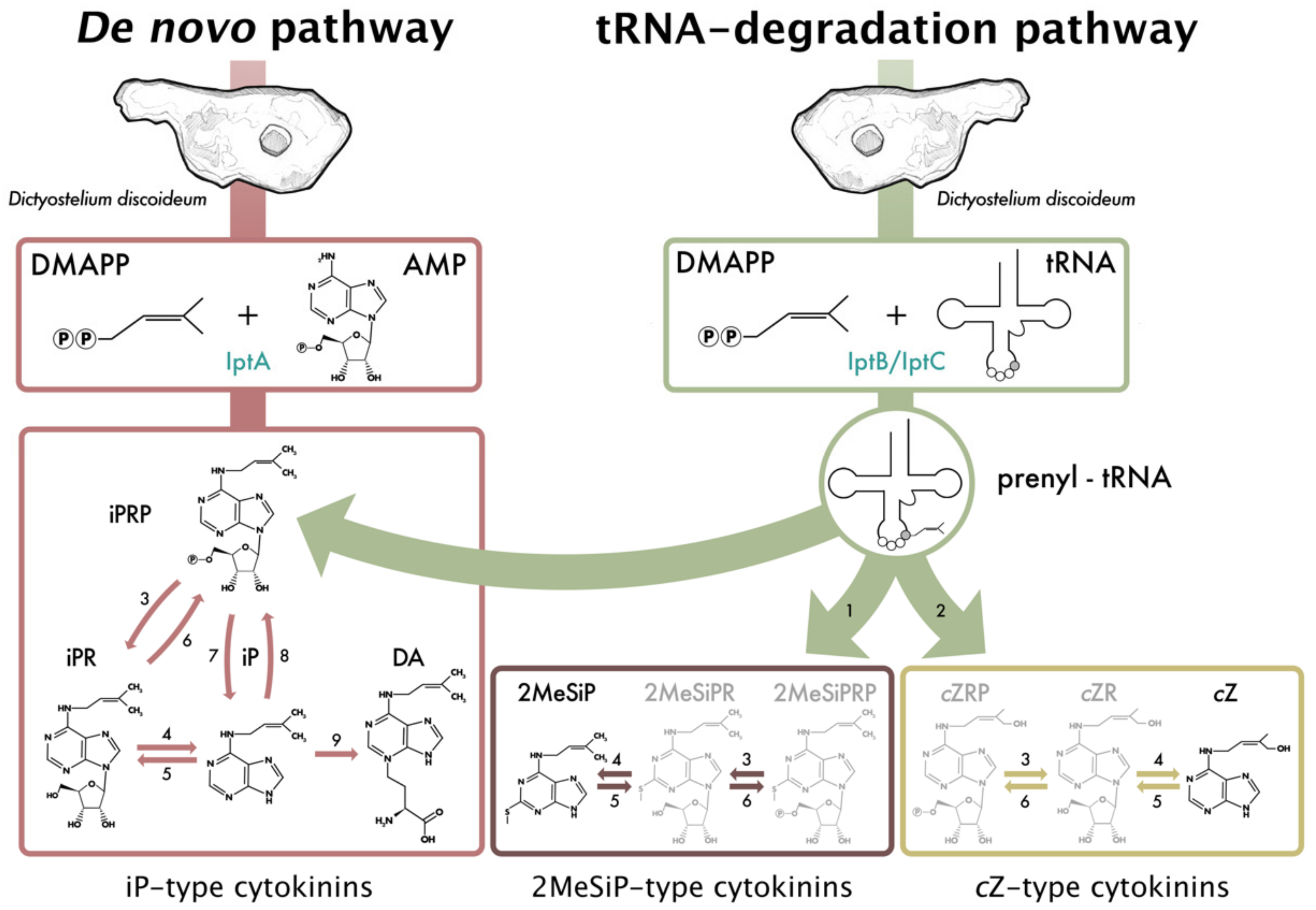
3.1. CKs Are Detected Throughout All Stages of the Dictyostelium Life Cycle
3.2. DA Is One of the Most Up-Regulated Metabolites during Dictyostelium Development
3.3. CK Levels Are Highest During Germination
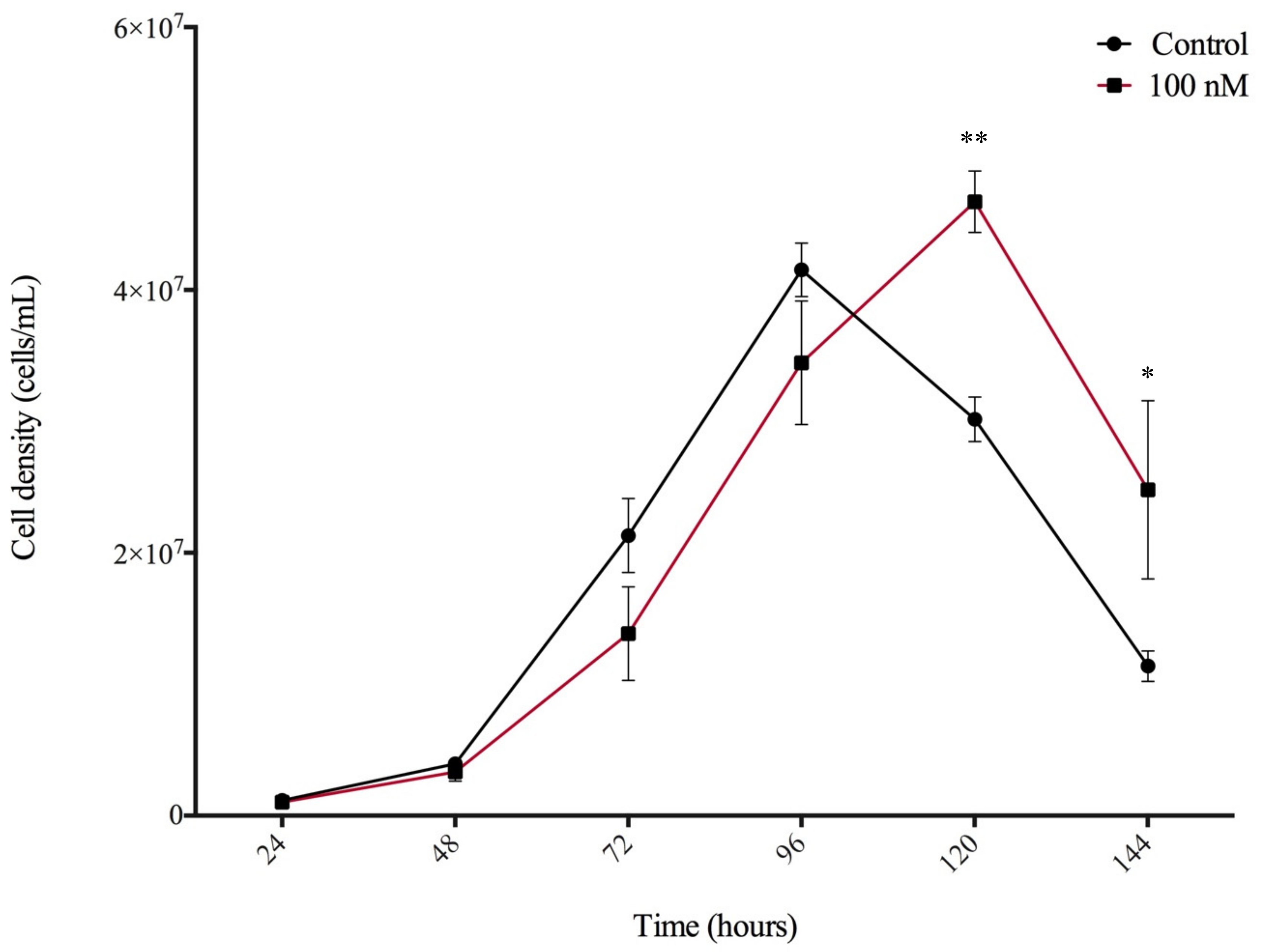
3.4. N6-Isopentenyladenine Prolongs the Stationary Phase of Dictyostelium Growth
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Schilde, C.; Schaap, P. The Amoebozoa; Humana Press: Totowa, NJ, USA, 2013; pp. 1–15. [Google Scholar]
- Loomis, W.F. Cell signaling during development of Dictyostelium. Dev. Biol. 2014, 391, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Konijn, T.M.; Van De Meene, J.G.; Bonner, J.T.; Barkley, D.S. The acrasin activity of adenosine-3’,5’-cyclic phosphate. Proc. Natl. Acad. Sci. USA 1967, 58, 1152–1154. [Google Scholar] [CrossRef] [PubMed]
- Gerisch, G.; Wick, U. Intracellular oscillations and release of cyclic AMP from Dictyostelium cells. Biochem. Biophys. Res. Commun. 1975, 65, 364–370. [Google Scholar] [CrossRef]
- Siegert, F.; Weijer, C.J. Spiral and concentric waves organize multicellular Dictyostelium mounds. Curr. Biol. 1995, 5, 937–943. [Google Scholar] [CrossRef]
- Anjard, C.; Loomis, W.F. Cytokinins induce sporulation in Dictyostelium. Development 2008, 135, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Obata, Y.; Abe, H.; Tanaka, Y.; Yanagisawa, K.; Uchiyama, M. Isolation of a spore germination inhibitor from a cellular slime mold Dictyostelium discoideum. Agric. Biol. Chem. 1973, 37, 1989–1990. [Google Scholar] [CrossRef]
- Tanaka, Y.; Hashimoto, Y.; Yanagisawa, K.; Abe, H.; Uchiyama, M. Partial structure of a spore germination inhibitor from a cellular slime mold, Dictyostelium discoideum. Agric. Biol. Chem. 1975, 39, 1929–1932. [Google Scholar]
- Mok, D.W.; Mok, M.C. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Uchiyama, M.; Tanaka, Y.; Saitô, H. Structure of discadenine, a spore germination inhibitor from the cellular slime mold, Dictyostelium discoideum. Tetrahedron Lett. 1976, 17, 3807–3810. [Google Scholar] [CrossRef]
- Tanaka, Y.; Abe, H.; Uchiyama, M.; Taya, Y.; Nishimura, S. Isopentenyladenine from Dictyostelium discoideum. Phytochemistry 1978, 17, 543–544. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef] [PubMed]
- Spíchal, L. Cytokinins - recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267. [Google Scholar] [CrossRef]
- Taya, Y.; Tanaka, Y.; Nishimura, S. Cell-free biosynthesis of discadenine, a spore germination inhibitor of Dictyostelium discoideum. FEBS Lett. 1978, 89, 326–328. [Google Scholar] [CrossRef]
- Taya, Y.; Yamada, T.; Nishimura, S. Correlation between acrasins and spore germination inhibitors in cellular slime molds. J. Bacteriol. 1980, 143, 715–719. [Google Scholar]
- Abe, H.; Hashimoto, K.; Uchiyama, M. Discadenine distribution in cellular slime molds and its inhibitory activity on spore germination. Agric. Biol. Chem. 1981, 45, 1295–1296. [Google Scholar]
- Schaap, P.; Winckler, T.; Nelson, M.; Alvarez-Curto, E.; Elgie, B.; Hagiwara, H.; Cavender, J.; Milano-Curto, A.; Rozen, D.E.; Dingermann, T.; et al. Molecular phylogeny and evolution of morphology in the social amoebas. Science 2006, 314, 661–663. [Google Scholar] [CrossRef]
- Kamada-Nobusada, T.; Sakakibara, H. Molecular basis for cytokinin biosynthesis. Phytochemistry 2009, 70, 444–449. [Google Scholar] [CrossRef]
- Lindner, A.C.; Lang, D.; Seifert, M.; Podlešáková, K.; Novák, O.; Strnad, M.; Reski, R.; Von Schwartzenberg, K. Isopentenyltransferase-1 (IPT1) knockout in Physcomitrella together with phylogenetic analyses of IPTs provide insights into evolution of plant cytokinin biosynthesis. J. Exp. Bot. 2014, 65, 2533–2543. [Google Scholar] [CrossRef]
- Nishii, K.; Wright, F.; Chen, Y.Y.; Möller, M. Tangled history of a multigene family: The evolution of isopentenyltransferase genes. PLoS ONE 2018, 13, 1–23. [Google Scholar] [CrossRef]
- Eichinger, L.; Pachebat, J.A.; Glöckner, G.; Rajandream, M.-A.; Sucgang, R.; Berriman, M.; Song, J.; Olsen, R.; Szafranski, K.; Xu, Q.; et al. The genome of the social amoeba Dictyostelium discoideum. Nature 2005, 435, 43–57. [Google Scholar] [CrossRef]
- Ihara, M.; Taya, Y.; Nishimura, S. Developmental regulation of cytokinin, spore germination inhibitor discadenine and related enzymes in Dictyostelium discoideum. Exp. Cell Res. 1980, 126, 273–278. [Google Scholar] [CrossRef]
- Ihara, M.; Taya, Y.; Nishimura, S.; Tanaka, Y. Purification and some properties of ∆2-isopentenylpyrophosphate:5′AMP ∆2-isopententeyltransferase from the cellular slime mold Dictyostelium discoideum. Arch. Biochem. Biophys. 1984, 230, 652–660. [Google Scholar] [CrossRef]
- Fey, P.; Kowal, A.S.; Gaudet, P.; Pilcher, K.E.; Chisholm, R.L. Protocols for growth and development of Dictyostelium discoideum. Nat. Protoc. 2007, 2, 1307–1316. [Google Scholar] [CrossRef] [PubMed]
- Mik, V.; Mičková, Z.; Doležal, K.; Frébort, I.; Pospíšil, T. Activity of (+)-Discadenine as a plant cytokinin. J. Nat. Prod. 2017, 80, 2136–2140. [Google Scholar] [CrossRef]
- Quesnelle, P.E.; Emery, R.J.N. cis-cytokinins that predominate in Pisum sativum during early embryogenesis will accelerate embryo growth in vitro. Can. J. Bot. 2007, 85, 91–103. [Google Scholar] [CrossRef]
- Farrow, S.C.; Emery, R.J.N. Concurrent profiling of indole-3-acetic acid, abscisic acid, and cytokinins and structurally related purines by high-performance-liquid-chromatography tandem electrospray mass spectrometry. Plant Methods 2012, 8, 42. [Google Scholar] [CrossRef]
- Hoyerová, K.; Gaudinová, A.; Malbeck, J.; Dobrev, P.I.; Kocábek, T.; Šolcová, B.; Trávníčková, A.; Kamínek, M. Efficiency of different methods of extraction and purification of cytokinins. Phytochemistry 2006, 67, 1151–1159. [Google Scholar] [CrossRef]
- Emery, R.J.N.; Ma, Q.; Atkins, C.A. The forms and sources of cytokinins in developing white lupine seeds and fruits. Plant Physiol. 2000, 123, 1593–1604. [Google Scholar] [CrossRef]
- Kisiala, A.; Kambhampati, S.; Stock, N.L.; Aoki, M.; Emery, R.J.N. Quantification of cytokinins using high-resolution accurate-mass orbitrap mass spectrometry and parallel reaction monitoring (PRM). Anal. Chem. 2019, 1–8. [Google Scholar] [CrossRef]
- Tautenhahn, R.; Patti, G.J.; Rinehart, D.; Siuzdak, G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012, 84, 5035–5039. [Google Scholar] [CrossRef]
- Gowda, H.; Ivanisevic, J.; Johnson, C.H.; Kurczy, M.E.; Benton, H.P.; Rinehart, D.; Nguyen, T.; Ray, J.; Kuehl, J.; Arevalo, B.; et al. Interactive XCMS Online: Simplifying advanced metabolomic data processing and subsequent statistical analyses. Anal. Chem. 2014, 86, 6931–6939. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, C.; Wang, Y.; Renaud, J.; Tian, G.; Kambhampati, S.; Saatian, B.; Nguyen, V.; Hannoufa, A.; Marsolais, F.; et al. Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat. Plants 2017, 3, 814–824. [Google Scholar] [CrossRef] [PubMed]
- Prince, J.T.; Marcotte, E.M. Chromatographic alignment of ESI-LC-MS proteomics data sets by ordered bijective interpolated warping. Anal. Chem. 2006, 78, 6140–6152. [Google Scholar] [CrossRef]
- Morrison, E.N.; Knowles, S.; Hayward, A.; Thorn, R.G.; Saville, B.J.; Emery, R.J.N. Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia 2015, 107, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Frébort, I.; Kowalska, M.; Hluska, T.; Frébortová, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef]
- Taya, Y.; Tanaka, Y.; Nishimura, S. 5′-AMP is a direct precursor of cytokinin in Dictyostelium discoideum. Nature 1978, 271, 545–547. [Google Scholar] [CrossRef] [PubMed]
- Rot, G.; Parikh, A.; Curk, T.; Kuspa, A.; Shaulsky, G.; Zupan, B. dictyExpress: A Dictyostelium discoideum gene expression database with an explorative data analysis web-based interface. BMC Bioinform. 2009, 10, 265. [Google Scholar] [CrossRef]
- LeJohn, H.B.; Stevenson, R.M. Cytokinins and magnesium ions may control the flow of metabolites and calcium ions through fungal cell membranes. Biochem. Biophys. Res. Commun. 1973, 54, 1061–1066. [Google Scholar] [CrossRef]
- Gogala, N. Regulation of mycorrhizal infection by hormonal factors produced by hosts and fungi. Experientia 1991, 47, 331–340. [Google Scholar] [CrossRef]
- Schäfer, M.; Brütting, C.; Meza-Canales, I.D.; Großkinsky, D.K.; Vankova, R.; Baldwin, I.T.; Meldau, S. The role of cis -zeatin-type cytokinins in plant growth regulation and mediating responses to environmental interactions. J. Exp. Bot. 2015, 66, 4873–4884. [Google Scholar] [CrossRef]
- Kasahara, H.; Takei, K.; Ueda, N.; Hishiyama, S.; Yamaya, T.; Kamiya, Y.; Yamaguchi, S.; Sakakibara, H. Distinct isoprenoid origins of cis- and trans-Zeatin biosyntheses in Arabidopsis. J. Biol. Chem. 2004, 279, 14049–14054. [Google Scholar] [CrossRef] [PubMed]
- Burrows, W.J.; Armstrong, D.J.; Kamínek, M.; Skoog, F.; Bock, R.M.; Hecht, S.M.; Dammann, L.G.; Leonard, N.J.; Occolowitz, J. Isolation and identification of four cytokinins from wheat germ transfer ribonucleic acid. Biochemistry 1970, 9, 1867–1872. [Google Scholar] [CrossRef] [PubMed]
- Kisiala, A.; Laffont, C.; Emery, R.J.N.; Frugier, F. Bioactive cytokinins are selectively secreted by Sinorhizobium meliloti nodulating and nonnodulating Strains. Mol. Plant Microbe Interact. 2013, 26, 1225–1231. [Google Scholar] [CrossRef] [PubMed]
- Seegobin, M.; Kisiala, A.; Noble, A.; Kaplan, D.; Brunetti, C.; Emery, R.J.N. Canis familiaris tissues are characterized by different profiles of cytokinins typical of the tRNA degradation pathway. FASEB J. 2018, 32, 6575–6581. [Google Scholar] [CrossRef]
- Persson, B.C.; Esberg, B.; Ólafsson, Ó.; Björk, G.R. Synthesis and function of isopentenyl adenosine derivatives in tRNA. Biochimie 1994, 76, 1152–1160. [Google Scholar] [CrossRef]
- Schweizer, U.; Bohleber, S.; Fradejas-Villar, N. The modified base isopentenyladenosine and its derivatives in tRNA. RNA Biol. 2017, 14, 1197–1208. [Google Scholar] [CrossRef]
- Persson, B.C.; Björk, G.R. Isolation of the gene (miaE) encoding the hydroxylase involved in the synthesis of 2-methylthio-cis-ribozeatin in tRNA of Salmonella typhimurium and characterization of mutants. J. Bacteriol. 1993, 175, 7776–7785. [Google Scholar] [CrossRef]
- Reiter, V.; Matschkal, D.M.S.; Wagner, M.; Globisch, D.; Kneuttinger, A.C.; Müller, M.; Carell, T. The CDK5 repressor CDK5RAP1 is a methylthiotransferase acting on nuclear and mitochondrial RNA. Nucleic Acids Res. 2012, 40, 6235–6240. [Google Scholar] [CrossRef]
- Hinsch, J.; Vrabka, J.; Oeser, B.; Novák, O.; Galuszka, P.; Tudzynski, P. De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea. Environ. Microbiol. 2015, 17, 2935–2951. [Google Scholar] [CrossRef]

| Endogenous CK Fractions | 2H-labelled Internal Standard |
|---|---|
| Nucleotides (RP) | |
| trans-zeatin riboside-5′-monophosphate (tZRP) | 2H6[9RMP]DZ |
| cis-zeatin riboside-5′-monophosphate (cZRP) | 2H6[9RMP]DZ |
| Dihydrozeatin riboside-5′-monophosphate (DZRP) | 2H6[9RMP]DZ |
| N6-benzyladenosine-5′monophosphate (BARP) | 2H6[9RMP]DZ |
| N6-isopentyladenosine-5′monophosphate (iPRP) | 2H6[9RMP]iP |
| Ribosides (R) | |
| trans-zeatin riboside (tZR) | 2H5[9R]tZ |
| cis-zeatin riboside (cZR) | 2H5[9R]tZ |
| Dihydrozeatin riboside (DZR) | 2H3[9R]DZ |
| N6-isopentyladenosine (iPR) | 2H6[9R]iP |
| N6-benzyladenosine (BAR) | 2H7[9R]BA |
| Free bases (FB) | |
| trans-zeatin (tZ) | 2H3DZ |
| cis-zeatin (cZ) | 2H3DZ |
| Discadenine (DA) | 2H3DZ |
| Dihydrozeatin (DZ) | 2H3DZ |
| N6-isopentyladenine (iP) | 2H6iP |
| N6-benzyladenine (BA) | 2H7BA |
| Glucosides (GLUC) | |
| trans-Zeatin-O-glucoside (tZOG) | 2H5tZOG |
| cis-Zeatin-O-glucoside (cZOG) | 2H5tZOG |
| Dihydrozeatin-O-glucoside (DZOG) | 2H7DZOG |
| trans-Zeatin-O-glucoside riboside (tZROG) | 2H5tZROG |
| cis-Zeatin-O-glucoside riboside (cZROG) | 2H5tZROG |
| Dihydrozeatin-O-glucoside riboside (DZROG) | 2H7DZROG |
| trans-Zeatin-7-glucoside (tZ7G) | 2H5tZ7G |
| trans-Zeatin-9-glucoside (tZ9G) | 2H5tZ9G |
| cis-Zeatin-9-glucoside (cZ9G) | 2H5tZ9G |
| Dihydrozeatin-9-glucoside (DZ9G) | 2H3DZ9G |
| Methylthiols (2MeS) | |
| 2-Methylthio-zeatin (2MeSZ) | 2H52MeStZ |
| 2-Methylthio-zeatin riboside (2MeSZR) | 2H52MeStZR |
| 2-Methylthio-N6-isopentenyladenine (2MeSiP) | 2H62MeSiP |
| 2-Methylthio-N6-isopentenyladenosine (2MeSiPR) | 2H62MeSiPR |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aoki, M.M.; Kisiala, A.B.; Li, S.; Stock, N.L.; Brunetti, C.R.; Huber, R.J.; Emery, R.J.N. Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules 2019, 9, 702. https://doi.org/10.3390/biom9110702
Aoki MM, Kisiala AB, Li S, Stock NL, Brunetti CR, Huber RJ, Emery RJN. Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules. 2019; 9(11):702. https://doi.org/10.3390/biom9110702
Chicago/Turabian StyleAoki, Megan M., Anna B. Kisiala, Shaojun Li, Naomi L. Stock, Craig R. Brunetti, Robert J. Huber, and R. J. Neil Emery. 2019. "Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination" Biomolecules 9, no. 11: 702. https://doi.org/10.3390/biom9110702
APA StyleAoki, M. M., Kisiala, A. B., Li, S., Stock, N. L., Brunetti, C. R., Huber, R. J., & Emery, R. J. N. (2019). Cytokinin Detection during the Dictyostelium discoideum Life Cycle: Profiles Are Dynamic and Affect Cell Growth and Spore Germination. Biomolecules, 9(11), 702. https://doi.org/10.3390/biom9110702








