Knock-Down of a Novel snoRNA in Tetrahymena Reveals a Dual Role in 5.8S rRNA Processing and Generation of a 26S rRNA Fragment
Abstract
1. Introduction
2. Materials and Methods
2.1. Construction of TtnuCD32 Knock-Down Cell Lines
2.2. TpnuCD32 Sequencing
2.3. Tetrahymena Cell Culture
2.4. RNA Extraction and Northern Blot Analysis
2.5. Primer Extension Analysis, Prediction, and Mapping of Methylated Nucleotides
2.6. Cluster Analysis
3. Results
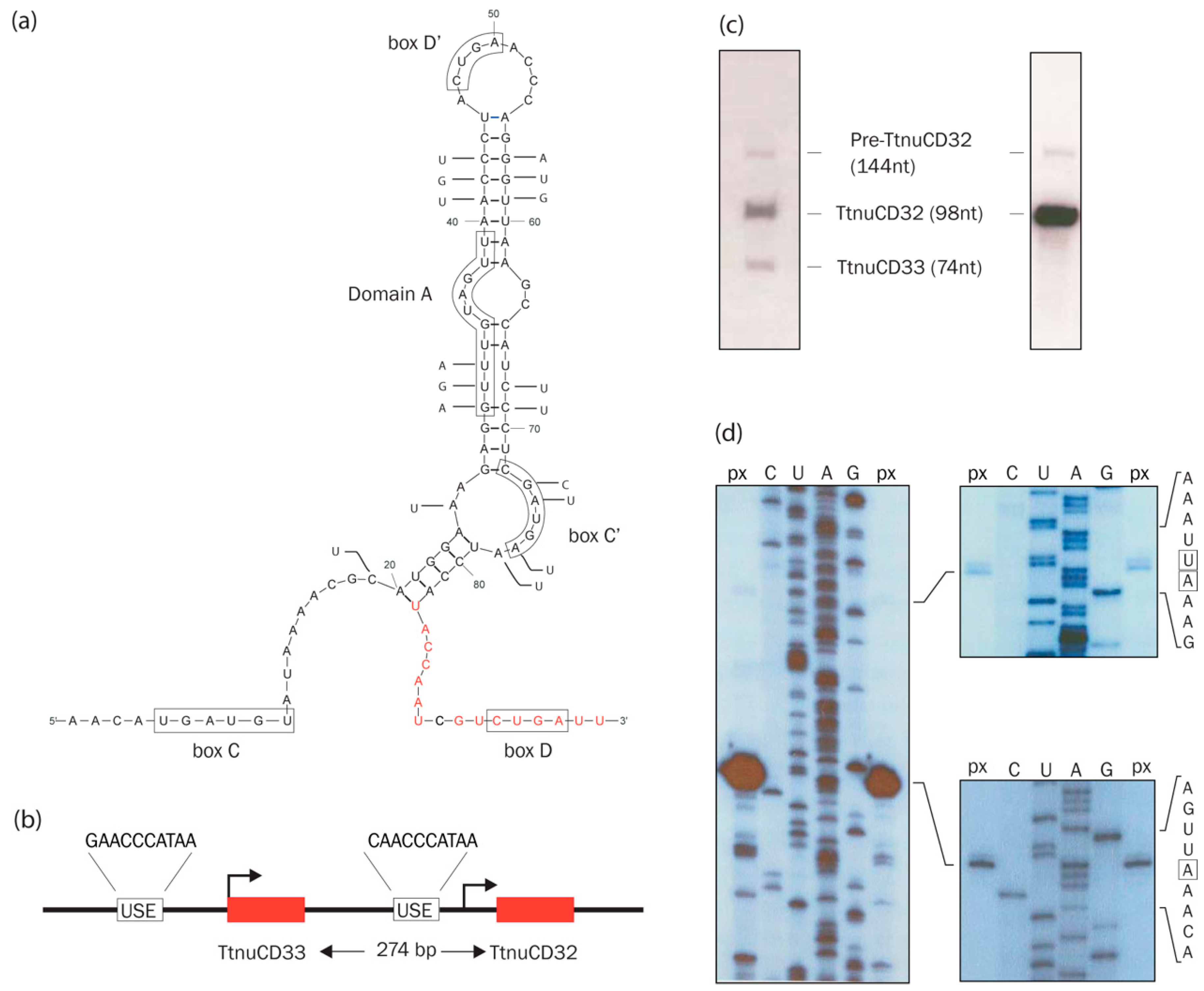
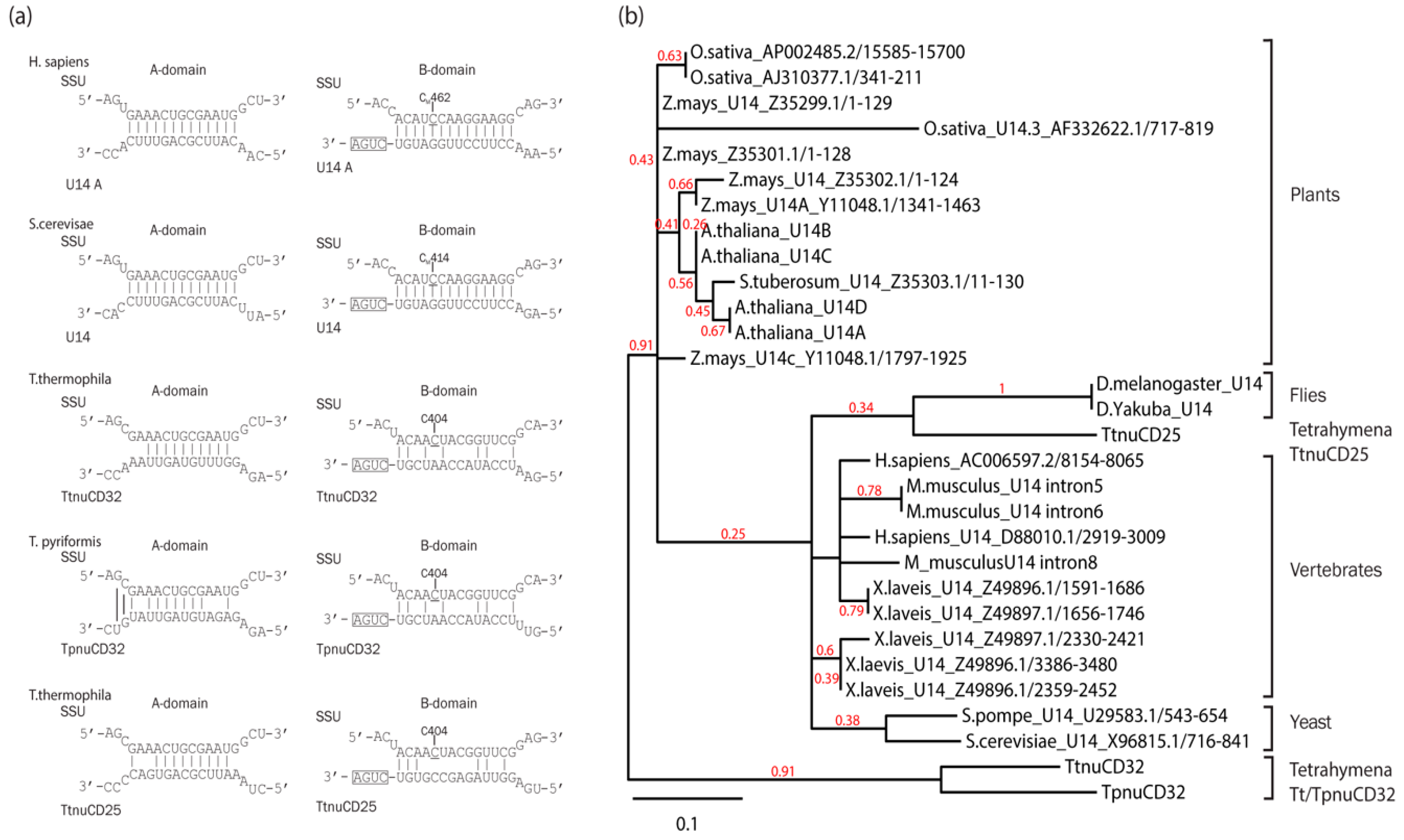
3.1. TtnuCD32 Structure and Conservation
3.2. Genomic Organization, Expression, and Processing of TtnuCD32
3.3. TtnuCD32 is Unlikely to Function as a 2′-O-methylation Guide snoRNA
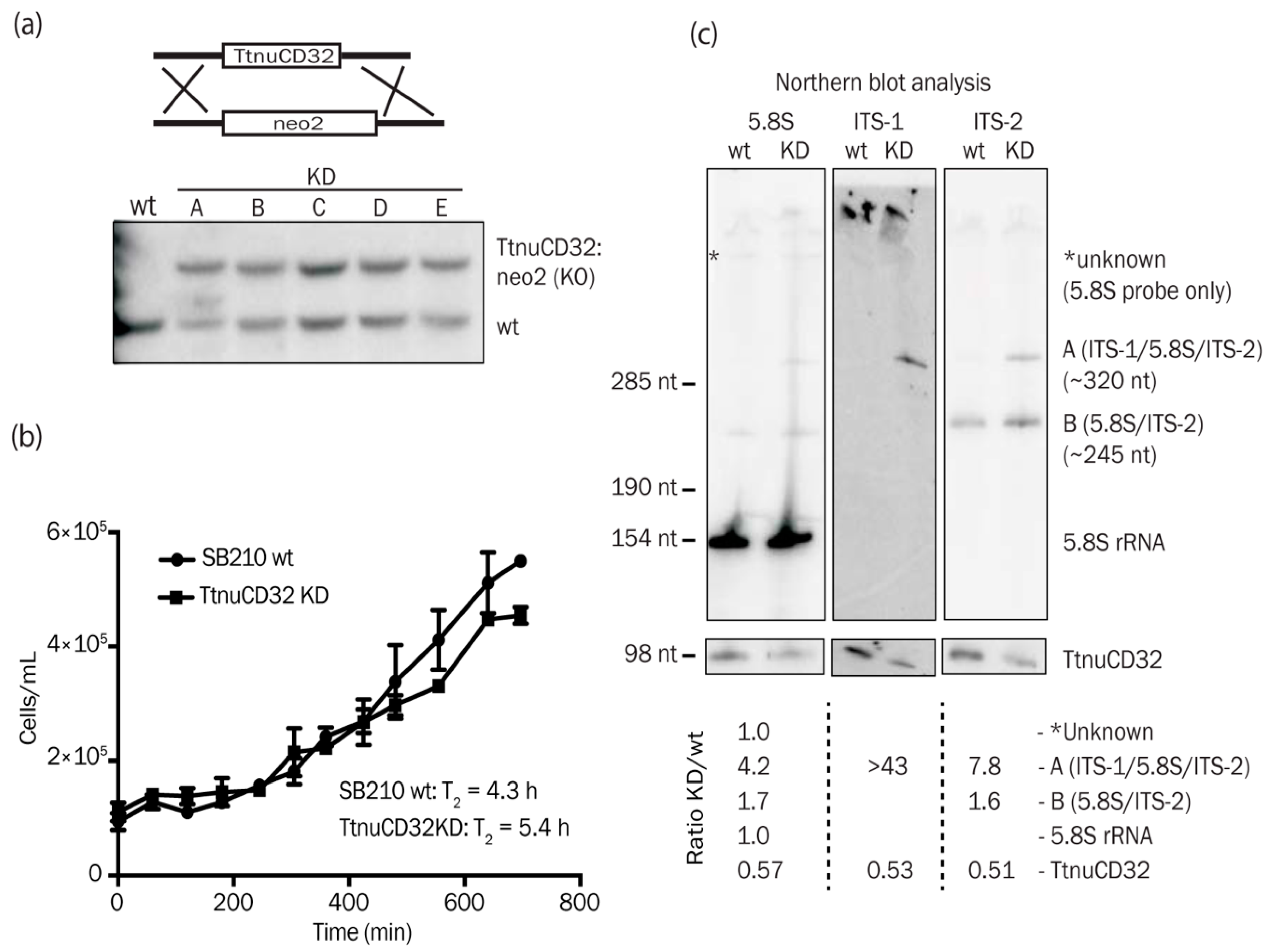
3.4. TtnuCD32 is Critical for Cell Survival
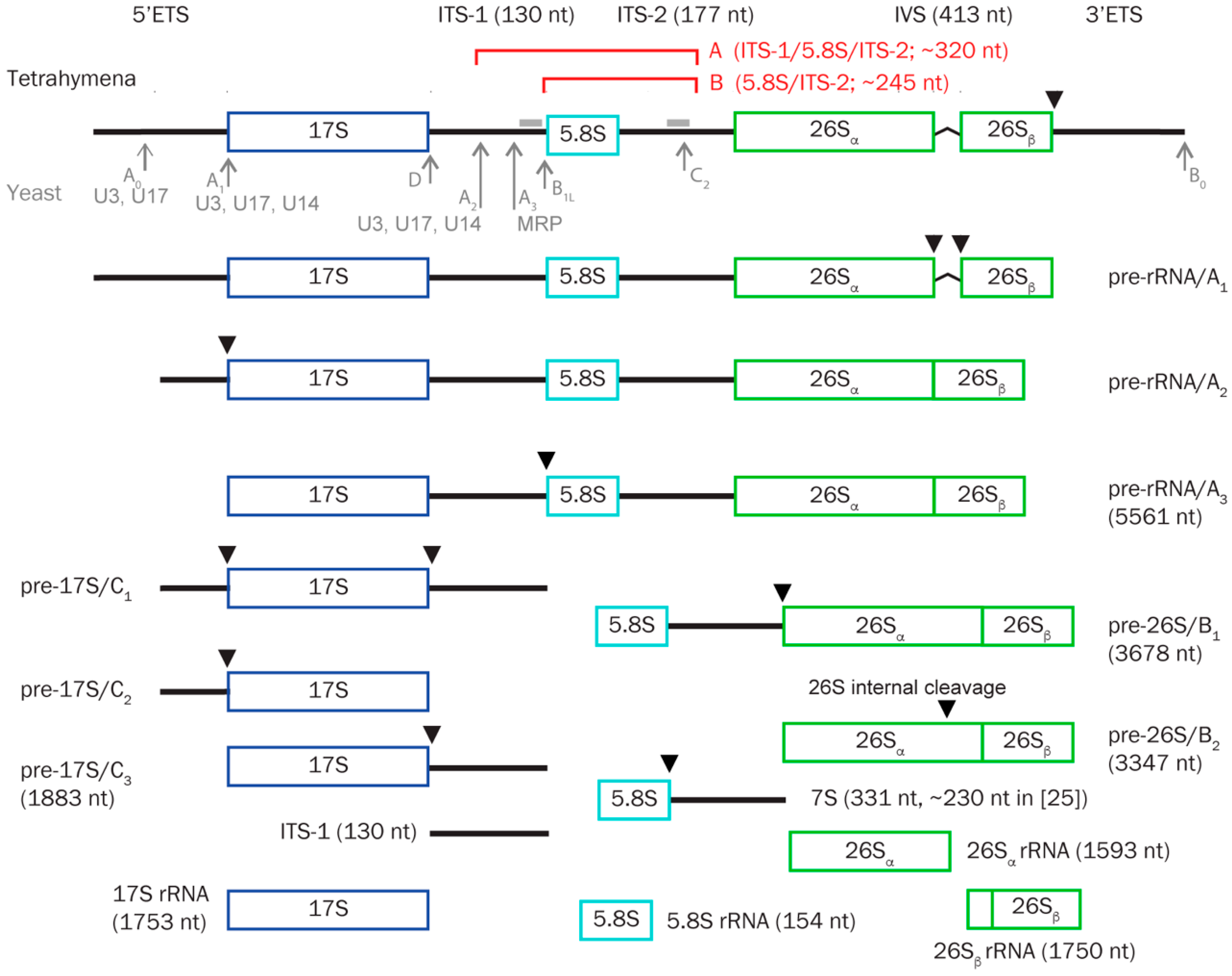
3.5. TtnuCD32 Is Involved in Pre-rRNA Cleavages Adjacent to 5.8S rRNA
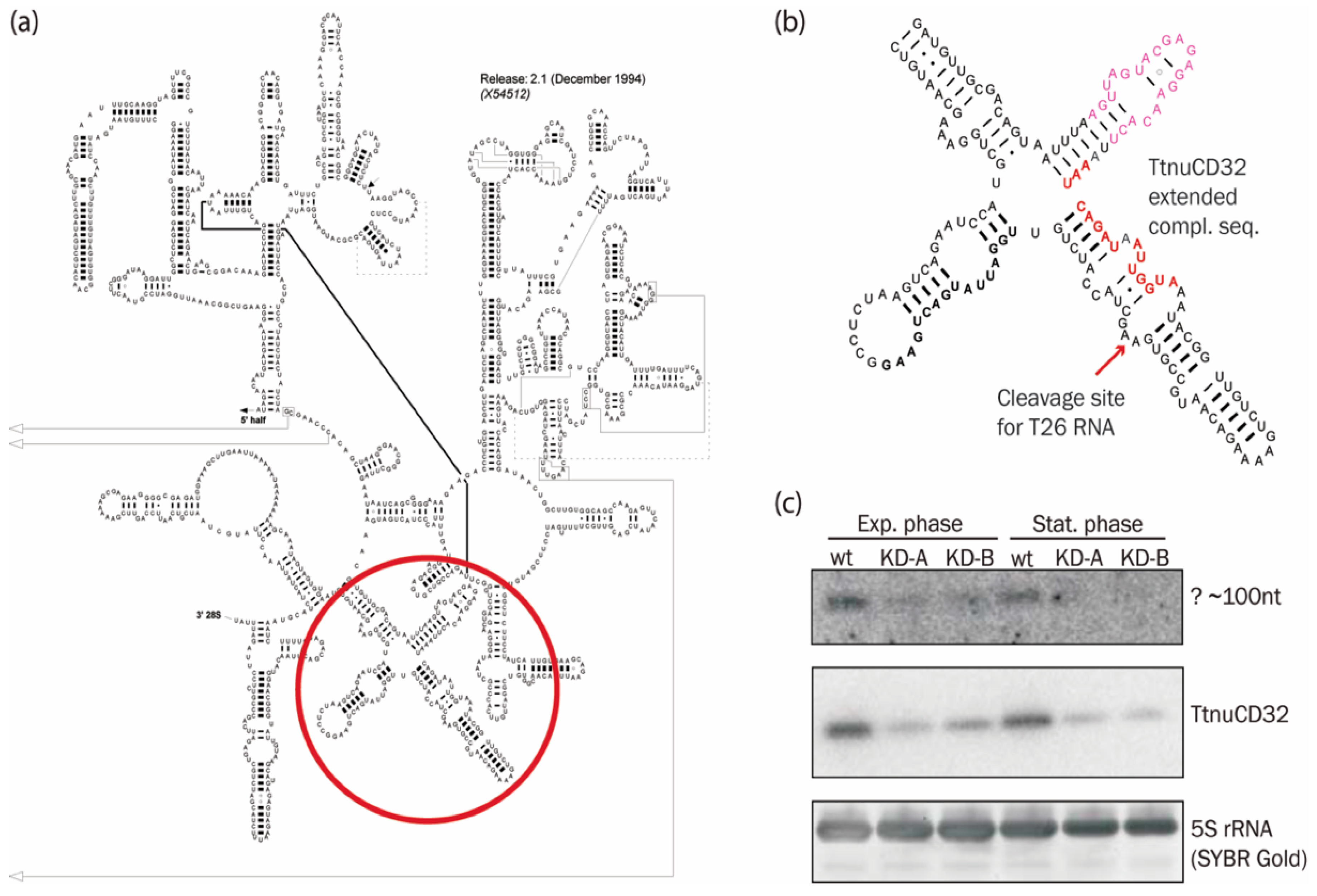
3.6. Abundance of a Small rRNA Fragment Correlates with TtnuCD32 Levels
3.7. TtnuCD32 Is a U14-Like snoRNA without the Cannonical Domain B
4. Discussion
4.1. TtnuCD32 Is Involved in pre-rRNA Processing and Maturation of 5.8S rRNA
4.2. An Additional TtnuCD32 Facilitated rRNA Cleavage
4.3. The Relation of TtnuCD32 to snoRNA U14
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Henras, A.K.; Plisson-Chastang, C.; O’Donohue, M.F.; Chakraborty, A.; Gleizes, P.E. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 2015, 6, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Sloan, K.E.; Warda, A.S.; Sharma, S.; Entian, K.D.; Lafontaine, D.L.J.; Bohnsack, M.T. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017, 14, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Mullineux, S.T.; Lafontaine, D.L.J. Mapping the cleavage sites on mammalian pre-rRNAs: Where do we stand? Biochimie 2012, 94, 1521–1532. [Google Scholar] [CrossRef] [PubMed]
- Watkins, N.J.; Bohnsack, M.T. The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip. Rev. RNA 2012, 3, 397–414. [Google Scholar] [CrossRef] [PubMed]
- Ganot, P.; Caizergues-Ferrer, M.; Kiss, T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997, 11, 941–956. [Google Scholar] [CrossRef] [PubMed]
- Kiss-László, Z.; Henry, Y.; Bachellerie, J.P.; Caizergues-Ferrer, M.; Kiss, T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 1996, 85, 1077–1088. [Google Scholar] [CrossRef]
- Kiss, T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001, 20, 3617–3622. [Google Scholar] [CrossRef] [PubMed]
- Atzorn, V.; Fragapane, P.; Kiss, T. U17/snR30 Is a Ubiquitous snoRNA with Two Conserved Sequence Motifs Essential for 18S rRNA Production. Mol. Cell. Biol. 2004, 24, 1769–1778. [Google Scholar] [CrossRef] [PubMed]
- Hughes, J.M.; Ares, M. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991, 10, 4231–4239. [Google Scholar] [CrossRef] [PubMed]
- Li, H.D.; Zagorski, J.; Fournier, M.J. Depletion of U14 small nuclear RNA (snR128) disrupts production of 18S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1990, 10, 1145–1152. [Google Scholar] [CrossRef] [PubMed]
- Lygerou, Z.; Allmang, C.; Tollervey, D.; Séraphin, B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 1996, 272, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Peculis, B.A.; Steitz, J.A. Disruption of U8 nucleolar snRNA inhibits 5.8S and 28S rRNA processing in the Xenopus oocyte. Cell 1993, 73, 1233–1245. [Google Scholar] [CrossRef]
- Tollervey, D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J. 1987, 6, 4169–4175. [Google Scholar] [CrossRef] [PubMed]
- Tycowski, K.T.; Smith, C.M.; Shu, M.D.; Steitz, J.A. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. USA 1996, 93, 14480–14485. [Google Scholar] [CrossRef] [PubMed]
- Bally, M.; Hughes, J.; Cesareni, G. SnR30: A new, essential small nuclear RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1988, 16, 5291–5303. [Google Scholar] [CrossRef] [PubMed]
- Borovjagin, A.V.; Gerbi, S.A. U3 small nucleolar RNA is essential for cleavage at sites 1, 2 and 3 in pre-rRNA and determines which rRNA processing pathway is taken in xenopus oocytes. J. Mol. Biol. 1999, 286, 1347–1363. [Google Scholar] [CrossRef] [PubMed]
- Decatur, W.A.; Fournier, M.J. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002, 27, 344–351. [Google Scholar] [CrossRef]
- Esguerra, J.; Warringer, J.; Blomberg, A. Functional importance of individual rRNA 2′-O-ribose methylation revealed by high-resolution phenotyping. RNA 2008, 14, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Erales, J.; Marchand, V.; Panthu, B.; Gillot, S.; Belin, S.; Ghayad, S.E.; Garcia, M.; Laforêts, F.; Marcel, V.; Baudin-Baillieu, A.; et al. Evidence for rRNA 2′-O-methylation plasticity: Control of intrinsic translational capabilities of human ribosomes. Proc. Natl. Acad. Sci. USA 2017. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Liu, Q.; Fournier, M.J. rRNA Modifications in an Intersubunit Bridge of the Ribosome Strongly Affect Both Ribosome Biogenesis and Activity. Mol. Cell 2007, 28, 965–977. [Google Scholar] [CrossRef] [PubMed]
- Borovjagin, A.V.; Gerbi, S.A. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005, 33, 4995–5005. [Google Scholar] [CrossRef] [PubMed]
- Dunbar, D.A.; Baserga, S.J. The U14 snoRNA is required for 2′-O-methylation of the pre-18S rRNA in Xenopus oocytes. RNA 1998, 4, 195–204. [Google Scholar] [PubMed]
- Doerder, F.P.; Deak, J.C.; Lief, J.H. Rate of phenotypic assortment in Tetrahymena thermophila. Dev. Genet. 1992, 13, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Kapler, G.M. Developmentally regulated processing and replication of the Tetrahymena rDNA minichromosome. Curr. Opin. Genet. Dev. 1993, 3, 730–735. [Google Scholar] [CrossRef]
- Kister, K.P.; Muller, B.; Eckert, W.A. Complex endonudeolytic cleavage pattern during early events in the processing of pre-rRNA in the lower eukaryote, Tetrahymena thermophila. Nucleic Acids Res. 1983, 11, 3487–3502. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Shalem, B.; Hury, A.; Tkacz, I.D.; Liang, X.H.; Uliel, S.; Myslyuk, I.; Doniger, T.; Salmon-Divon, M.; Unger, R.; et al. Elucidating the role of C/D snoRNA in rRNA processing and modification in Trypanosoma brucei. Eukaryot. Cell 2008, 7, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Ørum, H.; Nielsen, H.; Engberg, J. Structural organization of the genes encoding the small nuclear RNAs U1 to U6 of Tetrahymena thermophila is very similar to that of plant small nuclear RNA genes. J. Mol. Biol. 1992, 227, 114–121. [Google Scholar] [CrossRef]
- Andersen, K.L.; Nielsen, H. Experimental identification and analysis of macronuclear non-coding RNAs from the ciliate Tetrahymena thermophila. Nucleic Acids Res. 2012, 40. [Google Scholar] [CrossRef] [PubMed]
- Ørum, H.; Nielsen, H.; Engberg, J. Sequence of a new snRNA from the ciliate Tetrahymena thermophila. Nucleic Acids Res. 1992, 20, 3518. [Google Scholar] [CrossRef] [PubMed]
- Cassidy-Hanley, D.; Bowen, J.; Lee, J.H.; Cole, E.; VerPlank, L.A.; Gaertig, J.; Gorovsky, M.A.; Bruns, P.J. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics 1997, 146, 135–147. [Google Scholar] [PubMed]
- Lowe, T.M.; Eddy, S.R. A computational screen for methylation guide snoRNAs in yeast. Science 1999, 283, 1168–1171. [Google Scholar] [CrossRef] [PubMed]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, 1620–1642. [Google Scholar] [CrossRef] [PubMed]
- Maden, B.E.H. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 2001, 25, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36. [Google Scholar] [CrossRef] [PubMed]
- Notredame, C.; Higgins, D.G.; Heringa, J.; Notredame, C.; Higgins, D.G.; Heringa, J. T-coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef] [PubMed]
- Chevenet, F.; Brun, C.; Bañuls, A.L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Kiss-László, Z.; Henry, Y.; Kiss, T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998, 17, 797–807. [Google Scholar] [CrossRef] [PubMed]
- Birkedal, U.; Christensen-Dalsgaard, M.; Krogh, N.; Sabarinathan, R.; Gorodkin, J.; Nielsen, H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew. Chem. Int. Ed. 2015, 54, 451–455. [Google Scholar] [CrossRef]
- Andersen, K.L.; Birkedal, U.; Nielsen, J.M.; Kirpekar, F.; Nielsen, H. High-throughput RiboMeth-seq profiling of ribose 2′-O-methylations in rRNA and snRNAs reveal conditional alteration of rRNA modification patters during cell stress. unpublished. 2018; manuscript in preparation. [Google Scholar]
- Engberg, J.; Nielsen, H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990, 18, 6915–6919. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, M.M.; Donti, T.R.; Sebastian Yakisich, J.; Smith, A.G.; Kapler, G.M. Tetrahymena ORC contains a ribosomal RNA fragment that participates in rDNA origin recognition. EMBO J. 2007, 26, 5048–5060. [Google Scholar] [CrossRef] [PubMed]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3. [Google Scholar] [CrossRef]
- Liang, W.Q.; Fournier, M.J. U14 base-pairs with 18S rRNA: A novel snoRNA interaction required for rRNA processing. Genes Dev. 1995, 9, 2433–2443. [Google Scholar] [CrossRef] [PubMed]
- Yuan, G.; Klämbt, C.; Bachellerie, J.P.; Brosius, J.; Hüttenhofer, A. RNomics in Drosophila melanogaster: Identification of 66 candidates for novel non-messenger RNAs. Nucleic Acids Res. 2003, 31, 2495–2507. [Google Scholar] [CrossRef] [PubMed]
- Narla, A.; Ebert, B.L. Ribosomopathies: Human disorders of ribosome dysfunction. Blood 2010, 115, 3196–3205. [Google Scholar] [CrossRef] [PubMed]
- Langhendries, J.-L.; Nicolas, E.; Doumont, G.; Goldman, S.; Lafontaine, D.L.J.; Langhendries, J.-L.; Nicolas, E.; Doumont, G.; Goldman, S.; Lafontaine, D.L.J. The human box C/D snoRNAs U3 and U8 are required for pre-rRNA processing and tumorigenesis. Oncotarget 2016, 5, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Pelletier, J.; Thomas, G.; Volarevi, S. Ribosome biogenesis in cancer: New players and therapeutic avenues. Nat. Rev. Cancer 2017, 18, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Dieci, G.; Preti, M.; Montanini, B. Eukaryotic snoRNAs: A paradigm for gene expression flexibility. Genomics 2009, 94, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Bounova, G.; Purdom, E.; Speed, T.P.; Collins, K. A Tetrahymena Piwi Bound to Mature tRNA 3′ Fragments Activates the Exonuclease Xrn2 for RNA Processing in the Nucleus. Mol. Cell 2012, 48, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Petfalski, E.; Tollervey, D. The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 1996, 10, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Khade, P.K.; Sanbonmatsu, K.Y.; Joseph, S. Functional role of the sarcin-ricin loop of the 23s rRNA in the elongation cycle of protein synthesis. J. Mol. Biol. 2012, 419, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, L.; Lambert, N.J.; Maklan, E.J.; Horan, L.H.; Noller, H.F. The sarcin-ricin loop of 23S rRNA is essential for assembly of the functional core of the 50S ribosomal subunit. RNA 2008, 14, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.A.; Bruzik, J.P.; Steitz, J.A. An in vitro interaction between the human U3 snRNP and 28S rRNA sequences near the α-sarcin site. Nucleic Acids Res. 1988, 16, 10493–10509. [Google Scholar] [CrossRef] [PubMed]
- Röther, S.; Meister, G. Small RNAs derived from longer non-coding RNAs. Biochimie 2011, 93, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Collins, K. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J. Biol. Chem. 2005, 280, 42744–42749. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.L.; Collins, K. Several RNase T2 enzymes function in induced tRNA and rRNA turnover in the ciliate Tetrahymena. Mol. Biol. Cell 2012, 23. [Google Scholar] [CrossRef] [PubMed]
- Couvillion, M.T.; Sachidanandam, R.; Collins, K. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev. 2010, 24, 2742–2747. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Sun, Y.; Yang, X.; Wu, Z.; Guo, K.; Niu, X.; Wang, Q.; Ruan, J.; Bu, W.; Gao, S. Two featured series of rRNA-derived RNA fragments (rRFs) constitute a novel class of small RNAs. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, H.; Sun, Q.; Yao, Y. Characterization of small RNAs derived from tRNAs, rRNAs and snoRNAs and their response to heat stress in wheat seedlings. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Samarsky, D.A.; Schneider, G.S.; Fournier, M.J. An essential domain in Saccharomyces cerevisiae U14 snoRNA is absent in vertebrates, but conserved in other yeasts. Nucleic Acids Res. 1996, 24, 2059–2066. [Google Scholar] [CrossRef] [PubMed]
- Enright, C.A.; Maxwell, E.S.; Eliceiri, G.L.; Sollner-Webb, B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA 1996, 2, 1094–1099. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andersen, K.L.; Nielsen, H. Knock-Down of a Novel snoRNA in Tetrahymena Reveals a Dual Role in 5.8S rRNA Processing and Generation of a 26S rRNA Fragment. Biomolecules 2018, 8, 128. https://doi.org/10.3390/biom8040128
Andersen KL, Nielsen H. Knock-Down of a Novel snoRNA in Tetrahymena Reveals a Dual Role in 5.8S rRNA Processing and Generation of a 26S rRNA Fragment. Biomolecules. 2018; 8(4):128. https://doi.org/10.3390/biom8040128
Chicago/Turabian StyleAndersen, Kasper L., and Henrik Nielsen. 2018. "Knock-Down of a Novel snoRNA in Tetrahymena Reveals a Dual Role in 5.8S rRNA Processing and Generation of a 26S rRNA Fragment" Biomolecules 8, no. 4: 128. https://doi.org/10.3390/biom8040128
APA StyleAndersen, K. L., & Nielsen, H. (2018). Knock-Down of a Novel snoRNA in Tetrahymena Reveals a Dual Role in 5.8S rRNA Processing and Generation of a 26S rRNA Fragment. Biomolecules, 8(4), 128. https://doi.org/10.3390/biom8040128




