Functional Annotation of Caenorhabditis elegans Genes by Analysis of Gene Co-Expression Networks
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Weighted Gene Co-Expression Network Analysis
2.3. Functional Annotation of the Modules
2.4. Comparison of Gene Prediction Using Published Data
3. Results and Discussion
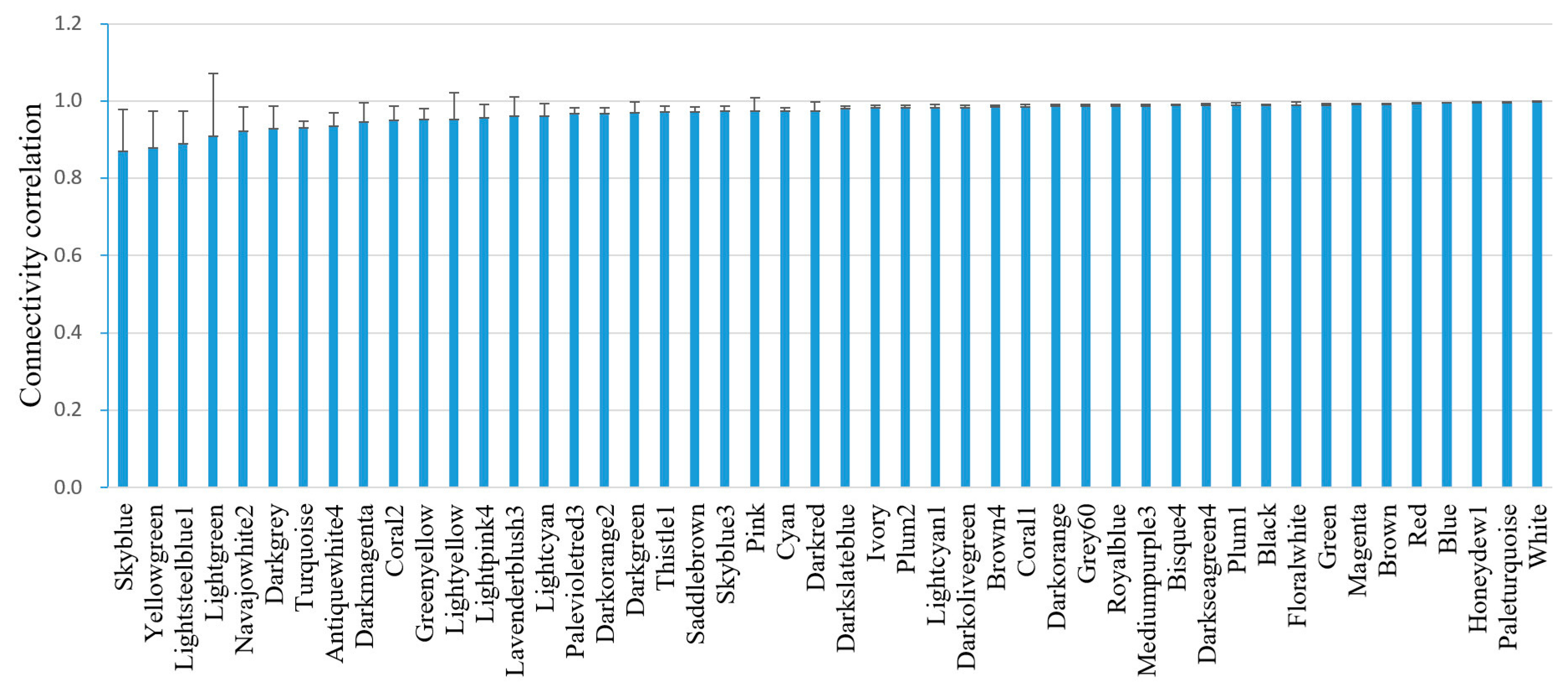
3.1. A Gene Coexpression Network of C. elegans Was Successfully Constructed
3.2. Gene Expression Variation in Modules
3.3. Modules That Correlate with Experimental Conditions
3.4. Modules That May Correlate with Molting
3.5. Genes Function Annotation
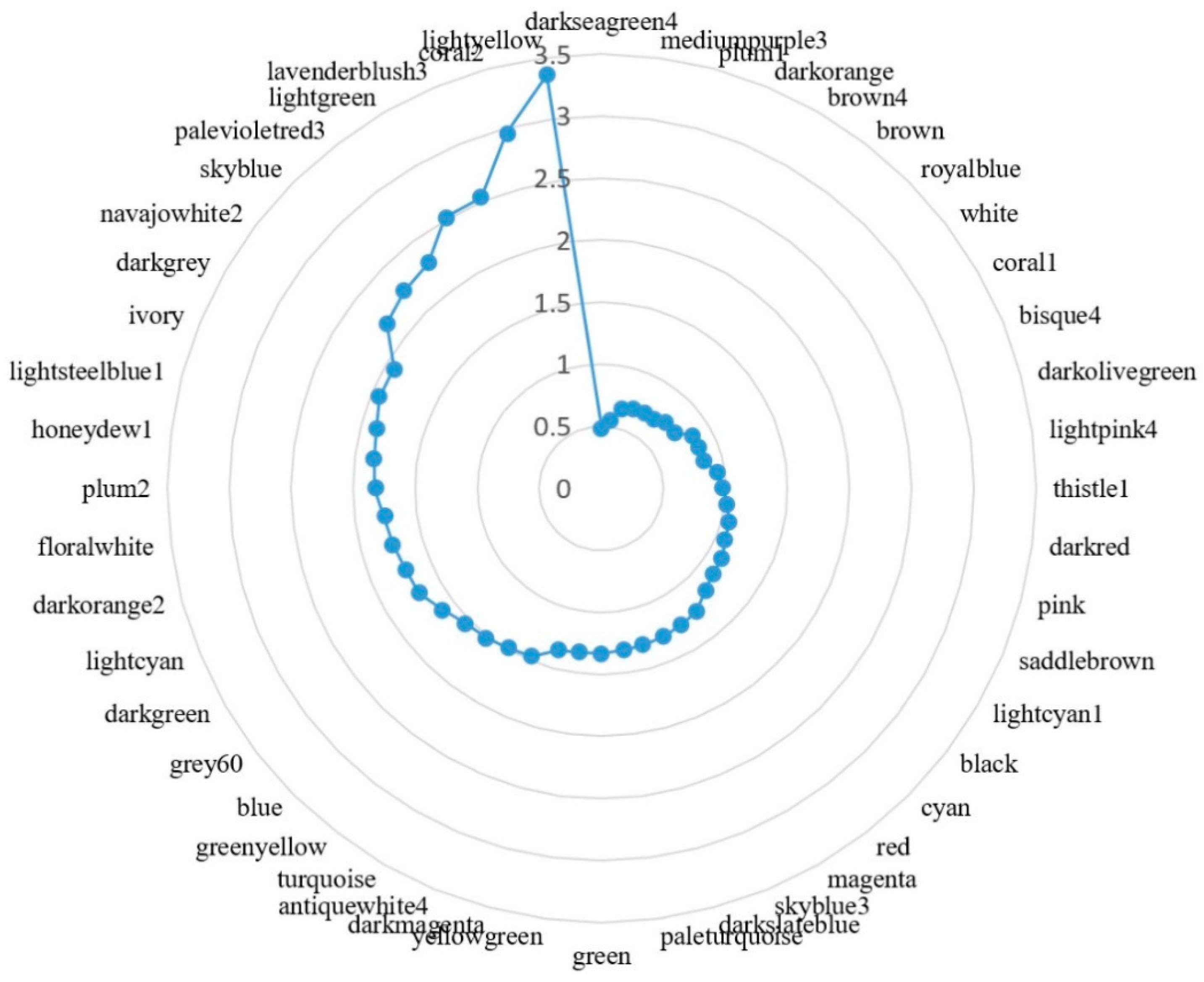
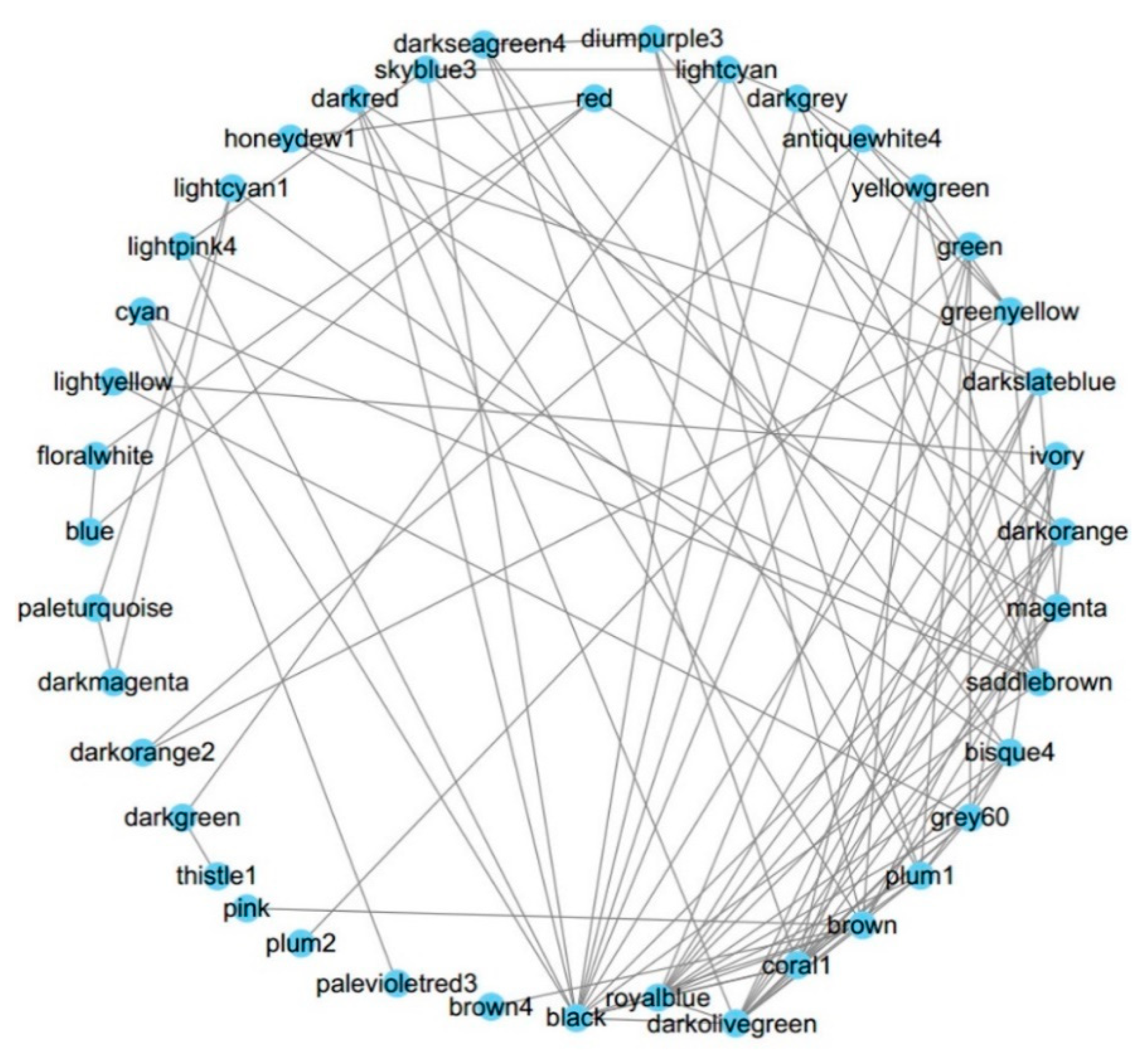
3.6. Higher Order Module Organization
3.7. Comparison with Previous C. elegans Networks
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, S.K.; Lund, J.; Kiraly, M.; Duke, K.; Jiang, M.; Stuart, J.M.; Eizinger, A.; Wylie, B.N.; Davidson, G.S. A gene expression map for Caenorhabditis elegans. Science 2001, 293, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Spencer, W.C.; Zeller, G.; Watson, J.D.; Henz, S.R.; Watkins, K.L.; McWhirter, R.D.; Petersen, S.; Sreedharan, V.T.; Widmer, C.; Jo, J.; et al. A spatial and temporal map of C. elegans gene expression. Genome Res. 2011, 21, 325–341. [Google Scholar] [CrossRef] [PubMed]
- Reinke, V.; Smith, H.E.; Nance, J.; Wang, J.; Van Doren, C.; Begley, R.; Jones, S.J.; Davis, E.B.; Scherer, S.; Ward, S.; et al. A global profile of germline gene expression in C. elegans. Mol. Cell 2000, 6, 605–616. [Google Scholar] [CrossRef]
- McKay, S.J.; Johnsen, R.; Khattra, J.; Asano, J.; Baillie, D.L.; Chan, S.; Dube, N.; Fang, L.; Goszczynski, B.; Ha, E.; et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003, 68, 159–169. [Google Scholar] [CrossRef] [PubMed]
- Hill, A.A.; Hunter, C.P.; Tsung, B.T.; Tucker-Kellogg, G.; Brown, E.L. Genomic analysis of gene expression in C. elegans. Science 2000, 290, 809–812. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Suo, B.; Emmons, S.W. Gene function prediction based on developmental transcriptomes of the two sexes in C. elegans. Cell Rep. 2016, 17, 917–928. [Google Scholar] [CrossRef] [PubMed]
- Boeck, M.E.; Huynh, C.; Gevirtzman, L.; Thompson, O.A.; Wang, G.; Kasper, D.M.; Reinke, V.; Hillier, L.W.; Waterston, R.H. The time-resolved transcriptome of C. elegans. Genome Res. 2016, 26, 1441–1450. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.K.; Hsu, A.K.; Sajdak, J.; Qin, J.; Pavlidis, P. Coexpression analysis of human genes across many microarray data sets. Genome Res. 2004, 14, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Wang, A.; Huang, K.; Shen, Y.; Xue, Z.; Cai, C.; Horvath, S.; Fan, G. Functional modules distinguish human induced pluripotent stem cells from embryonic stem cells. Stem Cells Dev. 2011, 20, 1937–1950. [Google Scholar] [CrossRef] [PubMed]
- Langfelder, P.; Horvath, S. Wgcna: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of genechip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Oldham, M.C.; Konopka, G.; Iwamoto, K.; Langfelder, P.; Kato, T.; Horvath, S.; Geschwind, D.H. Functional organization of the transcriptome in human brain. Nat. Neurosci. 2008, 11, 1271–1282. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Angeles-Albores, D.; Lee, R.Y.N.; Chan, J.; Sternberg, P.W. Tissue enrichment analysis for C. elegans genomics. BMC Bioinform. 2016, 17, 366. [Google Scholar] [CrossRef] [PubMed]
- De Preter, K.; Barriot, R.; Speleman, F.; Vandesompele, J.; Moreau, Y. Positional gene enrichment analysis of gene sets for high-resolution identification of overrepresented chromosomal regions. Nucleic Acids Res. 2008, 36, e43. [Google Scholar] [CrossRef] [PubMed]
- NCBI PubMed. 2003. Available online: https://www.ncbi.nlm.nih.gov/pubmed (accessed on 27 July 2018).
- Cho, A.; Shin, J.; Hwang, S.; Kim, C.; Shim, H.; Kim, H.; Kim, H.; Lee, I. Wormnet v3: A network-assisted hypothesis-generating server for Caenorhabditis elegans. Nucleic Acids Res. 2014, 42, W76–W82. [Google Scholar] [CrossRef] [PubMed]
- Hibbs, M.A.; Hess, D.C.; Myers, C.L.; Huttenhower, C.; Li, K.; Troyanskaya, O.G. Exploring the functional landscape of gene expression: Directed search of large microarray compendia. Bioinformatics 2007, 23, 2692–2699. [Google Scholar] [CrossRef] [PubMed]
- Farber, C.R. Identification of a gene module associated with BMD through the integration of network analysis and genome-wide association data. J. Bone Miner. Res. 2010, 25, 2359–2367. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.H.; Hsiao, K.M.; Chou, C.C. Molecular characterization of toxicity mechanism of single-walled carbon nanotubes. Biomaterials 2013, 34, 5661–5669. [Google Scholar] [CrossRef] [PubMed]
- Kippenberger, S.; Bernd, A.; Loitsch, S.; Guschel, M.; Muller, J.; Bereiter-Hahn, J.; Kaufmann, R. Signaling of mechanical stretch in human keratinocytes via map kinases. J. Investig. Dermatol. 2000, 114, 408–412. [Google Scholar] [CrossRef] [PubMed]
- Page, A.P.; Johnstone, I.L. The Cuticle. 2007, pp. 1–15. Available online: http://www.wormbook.org/chapters/www_cuticle/cuticle.html WormBook (accessed on 27 July 2018).
- Thierry-Mieg, D.; Thierry-Mieg, J. Aceview: A comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006, 7, S12. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Dierking, K.; Schulenburg, H. Wormexp: A web-based application for a Caenorhabditis elegans-specific gene expression enrichment analysis. Bioinformatics 2016, 32, 943–945. [Google Scholar] [CrossRef] [PubMed]
- Youngman, M.J.; Rogers, Z.N.; Kim, D.H. A decline in p38 mapk signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet. 2011, 7, e1002082. [Google Scholar] [CrossRef] [PubMed]
- Taffoni, C.; Pujol, N. Mechanisms of innate immunity in C. elegans epidermis. Tissue Barriers 2015, 3, e1078432. [Google Scholar] [CrossRef] [PubMed]
- Tong, A.; Lynn, G.; Ngo, V.; Wong, D.; Moseley, S.L.; Ewbank, J.J.; Goncharov, A.; Wu, Y.-C.; Pujol, N.; Chisholm, A.D. Negative regulation of Caenorhabditis elegans epidermal damage responses by death-associated protein kinase. Proc. Natl. Acad. Sci. USA 2009, 106, 1457–1461. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; Lu, Z.J.; Zhong, M.; Sarov, M.; Murray, J.I.; Brdlik, C.M.; Janette, J.; Chen, C.; Alves, P.; Preston, E.; et al. Diverse transcription factor binding features revealed by genome-wide chip-seq in C. elegans. Genome Res. 2011, 21, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.-F.; Cho, C.-Y.; Cheng, Y.-T.; Huang, J.-W.; Wu, Y.-Z.; Yeh, A.Y.-C.; Nishiwaki, K.; Chang, S.-C.; Wu, Y.-C. Blmp-1/blimp-1 regulates the spatiotemporal cell migration pattern in C. elegans. PLoS Genet. 2014, 10, e1004428. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.N.; Fields, C.; Kramer, J.M.; Rosenzweig, B.; Hirsh, D. Sequence comparisons of developmentally regulated collagen genes of Caenorhabditis elegans. Gene 1989, 76, 331–344. [Google Scholar] [CrossRef]
- Sebastiano, M.; Lassandro, F.; Bazzicalupo, P. Cut-1 a Caenorhabditis elegans gene coding for a dauer-specific noncollagenous component of the cuticle. Dev. Biol. 1991, 146, 519–530. [Google Scholar] [CrossRef]
- Frand, A.R.; Russel, S.; Ruvkun, G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005, 3, e312. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Cheng, J.; Allaire, J.J.; Xie, Y.; McPherson, J. Shiny: Web Application Framework for R. R Foundation for Statistical Computing, Vienna, Austria 2015. R Package Version 0.11. Available online: http://CRAN.R-project.org/package=shiny (accessed on 27 July 2018).
- Uygun, S.; Peng, C.; Lehti-Shiu, M.D.; Last, R.L.; Shiu, S.-H. Utility and limitations of using gene expression data to identify functional associations. PLoS Comput. Biol. 2016, 12, e1005244. [Google Scholar] [CrossRef] [PubMed]
| Module (No. of Genes) | Biological Process | Cellular Component | Molecule Function | Chromosome |
|---|---|---|---|---|
| Antiquewhite4 (35) | Ion transport (6 × 10−7) | Acetylcholine-gated channel complex (9 × 10−5) | Ion channel activity (2 × 10−4) | |
| Bisque4 (212) | Proteolysis (2 × 10−9) | Membrane raft (8 × 10−10) | Serine-type carboxypeptidase activity (3 × 10−12) | X (2 × 10−5) |
| Black (1173) | Ion transport (2 × 10−16) | Plasma membrane (5 × 10−14) | Signal transducer activity (1 × 10−12) | X (2 × 10−13) |
| Blue (930) | Regulation of cell shape (3 × 10−25) | Extrinsic component of cytoplasmic side of plasma membrane (8 × 10−14) | Protein kinase activity (1 × 10−22) | IV (1 × 10−9) |
| Brown (933) | Embryo development ending in birth or egg hatching (8 × 10−41) | Nucleus (3 × 10−11) | Protein binding (3 × 10−8) | I (1 × 10−15) |
| Brown4 (137) | Embryo development ending in birth or egg hatching (8 × 10−14) | Cytoplasm (2 × 10−15) | Protein binding (7 × 10−5) | III (4 × 10−3) |
| Coral1 (153) | Embryo development ending in birth or egg hatching (5 × 10−11) | Nucleus (1 × 10−10) | Protein binding (5 × 10−2) | I (3 × 10−2) |
| Coral2 (34) | Body morphogenesis (2 × 10−2) | Collagen trimer (2 × 10−25) | Structural constituent of cuticle (8 × 10−26) | |
| Cyan (219) | Axon guidance (2 × 10−7) | Axon (7 × 10−3) | Protein binding (3 × 10−6) | X (2 × 10−20) |
| Darkgreen (160) | Reproduction (8 × 10−5) | Cytoplasm (2 × 10−2) | Nucleotide binding (2 × 10−4) | III (1 × 10−6) |
| Darkgrey (136) | Neuropeptide signaling pathway (1 × 10−9) | Heterotrimeric G-protein complex (5 × 10−5) | Calcium ion binding (4 × 10−4) | X (2 × 10−2) |
| Darkmagenta (109) | Extracellular space (2 × 10−4) | Iron ion binding (3 × 10−2) | ||
| Darkolivegreen (163) | Embryo development ending in birth or egg hatching (8 × 10−3) | Nucleus (1 × 10−2) | Protein binding (6 × 10−4) | III (8 × 10−3) |
| Darkorange (367) | Embryo development ending in birth or egg hatching (2 × 10−9) | Mitochondrion (6 × 10−4) | III (2 × 10−9) | |
| Darkorange2 (221) | Innate immune response (4 × 10−4) | V (3 × 10−4) | ||
| Darkred (167) | Embryo development ending in birth or egg hatching (8 × 10−11) | P granule (8 × 10−4) | I (2 × 10−3) | |
| Darkseagreen4 (41) | Nematode larval development (1 × 10−7) | Mitochondrion (6 × 10−16) | NADH dehydrogenase (ubiquinone) activity (4 × 10−7) | |
| Darkslateblue (127) | Endoplasmic reticulum unfolded protein response (1 × 10−4) | Collagen trimer (8 × 10−34) | Structural constituent of cuticle (9 × 10−34) | |
| Floralwhite (86) | Pseudopodium (3 × 10−3) | IV (1 × 10−4) | ||
| Green (602) | Embryo development ending in birth or egg hatching (3 × 10−5) | Nucleolus (6 × 10−8) | RNA binding (2 × 10−6) | I (4 × 10−2) |
| Greenyellow (389) | Nonmotile primary cilium assembly (1 × 10−29) | Ciliary basal body (2 × 10−19) | G-protein coupled receptor activity (5 × 10−11) | X (7 × 10−4) |
| Grey60 (522) | Embryo development ending in birth or egg hatching (8 × 10−6) | Nucleus (4 × 10−14) | Protein binding (9 × 10−3) | X (2 × 10−18) |
| Honeydew1 (44) | Collagen and cuticulin-based cuticle development (6 × 10−4) | Collagen trimer (5 × 10−24) | Structural constituent of cuticle (4 × 10−27) | |
| Ivory (95) | 3′-UTR-mediated mRNA destabilization (2 × 10−6) | Nucleus (2 × 10−6) | mRNA 3′-UTR binding (1 × 10−4) | |
| Lavenderblush3 (46) | Cul3-RING ubiquitin ligase complex (4 × 10−2) | V (4 × 10−2) | ||
| Lightcyan (213) | Locomotion (6 × 10−5) | Axon (1 × 10−4) | Kinase activity (6 × 10−5) | X (8 × 10−6) |
| Lightcyan1 (98) | Innate immune response (2 × 10-38) | Membrane raft (4 × 10−17) | Carbohydrate binding (1 × 10−5) | IV (8 × 10−3) |
| Lightgreen (189) | Striated muscle dense body (8 × 10−5) | I (3 × 10−3) | ||
| Lightpink4 (49) | Maturation of LSU-rRNA (1 × 10−3) | Nucleolus (6 × 10−10) | ||
| Lightsteelblue1 (98) | Zinc ion binding (3 × 10−2) | |||
| Lightyellow (184) | II (4 × 10−7) | |||
| Magenta (592) | Molting cycle, collagen and cuticulin-based cuticle (2 × 10−17) | Extracellular region (1 × 10−9) | Structural constituent of cuticle (5 × 10−7) | X (4 × 10−12) |
| Mediumpurple3 (100) | Embryo development ending in birth or egg hatching (2 × 10−15) | Mitochondrion (7 × 10−18) | Threonine-type endopeptidase activity (6 × 10−12) | III (1 × 10−2) |
| Navajowhite2 (51) | Proteolysis (7 × 10−3) | Carbohydrate binding (1 × 10−10) | ||
| Paleturquoise (114) | Lipid transport (2 × 10−2) | Extracellular region (1 × 10−14) | Structural constituent of cuticle (5 × 10−11) | X (3 × 10−3) |
| Palevioletred3 (55) | Membrane (4 × 10−2) | |||
| Pink (306) | Endoplasmic reticulum (2 × 10−2) | III (9 × 10−4) | ||
| Plum1 (102) | ATP hydrolysis coupled proton transport (2 × 10−11) | Mitochondrion (8 × 10−8) | Proton-transporting atpase activity, rotational mechanism (4 × 10−10) | X (1 × 10−2) |
| Plum2 (65) | ||||
| Red (591) | Regulation of cell shape (2 × 10−11) | Integral component of membrane (5 × 10−4) | Phosphoprotein phosphatase activity (2 × 10−8) | IV (8 × 10−4) |
| Royalblue (292) | Embryo development ending in birth or egg hatching (4 × 10−39) | Nucleus (2 × 10−31) | Helicase activity (1 × 10−7) | III (3 × 10−8) |
| Saddlebrown (123) | Transforming growth factor beta receptor signaling pathway (2 × 10−3) | Axon (2 × 10−6) | Protein binding (3 × 10−5) | X (1 × 10−5) |
| Skyblue (128) | Nucleosome assembly (5 × 10−6) | Nucleosome (2 × 10−6) | Protein heterodimerization activity (5 × 10−4) | |
| Skyblue3 (104) | ||||
| Thistle1 (58) | Reproduction (1 × 10−4) | Nucleolus (4 × 10−2) | I (1 × 10−2) | |
| Turquoise (3211) | G-protein coupled receptor signaling pathway (4 × 10−205) | Integral component of membrane (6 × 10−296) | G-protein coupled olfactory receptor activity (6 × 10−139) | V (4 × 10−99) |
| White (131) | Translation (3 × 10−91) | Ribosome (3 × 10−122) | Structural constituent of ribosome (4 × 10−109) | I (3 × 10−3) |
| Yellowgreen (104) | Ion transport (4 × 10−5) | Striated muscle dense body (1 × 10−6) | Actin binding (4 × 10−4) |
| Module (No. Genes) | Biological Process | Tissue |
|---|---|---|
| Grey60 (522) | Embryo development ending in birth or egg hatching (8 × 10−6) | Caap |
| Darkred (167) | Embryo development ending in birth or egg hatching (8 × 10−11) | Psub1 |
| Brown4 (137) | Embryo development ending in birth or egg hatching (8 × 10−14) | AWB |
| Darkorange (367) | Embryo development ending in birth or egg hatching (2 × 10−9) | Reproductive system, thermosensory neuron |
| Green (602) | Embryo development ending in birth or egg hatching (3 × 10−5) | Reproductive system, hermaphrodite distal tip cell |
| Darkolivegreen (163) | Embryo development ending in birth or egg hatching (8 × 10−3) | Reproductive system, pharyngeal interneuron |
| Royalblue (292) | Embryo development ending in birth or egg hatching (4 × 10−39) | Reproductive system, Capp |
| Mediumpurple3 (100) | Embryo development ending in birth or egg hatching (2 × 10−15) | Reproductive system, anal depressor muscle |
| Brown (933) | Embryo development ending in birth or egg hatching (8 × 10−41) | Reproductive system, Psub1 |
| Coral1 (153) | Embryo development ending in birth or egg hatching (5 × 10−11) | Reproductive system, AVA |
| Darkorange2 (221) | Innate immune response (4 × 10−4) | AVA, pharyngeal interneuron |
| Lightcyan1 (98) | Innate immune response (2 × 10−38) | Intestine, outer labial sensillum, PVD |
| Black (1173) | Ion transport (2 × 10−16) | Ventral nerve cord, FLP, tail |
| Yellowgreen (104) | Ion transport (4 × 10−5) | Striated muscle |
| Antiquewhite4 (35) | Ion transport (6 × 10−7) | Pharyngeal interneuron, retrovesicular ganglion |
| Paleturquoise (114) | Lipid transport (5 × 10−4) | Cephalic sheath cell, hermaphrodite |
| Bisque4 (212) | Proteolysis (2 × 10−10) | Intestine, PVD |
| Navajowhite2 (51) | Proteolysis (6 × 10−3) | Male, reproductive system |
| Thistle1 (58) | Reproduction (3 × 10−5) | AVA |
| Darkgreen (160) | Reproduction (3 × 10−5) | Reproductive system |
| Module | Tissue Enrichment (p Value) | Phenotype Enrichment (p Value) | Stage | Hub Gene |
|---|---|---|---|---|
| Coral2 | Amphid socket cell (2 × 10−4) | Dumpy (3 × 10−5) | Dauer | col-2 |
| Darkslateblue | Gonadal primordium (4 × 10−7) | Dumpy (7 × 10−7) | Embryo | ZK662.2 |
| Honeydew1 | Hyp7 syncytium (4 × 10−10) | Blistered (3 × 10−6) | L4 | col-138 |
| Magenta | Outer labial sensillum (2 × 10−13) | Molt variant (3 × 10−20), paralyzed (1 × 10−11) | L1 | abu-13 |
| Paleturquoise | Cephalic sheath cell (8 × 10−27) | Pathogen susceptibility increased (2 × 10−6) | Adult | C10G8.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Li, L.; He, Y.; Cai, S.; Zhao, W.; Zheng, H.; Zhong, Y.; Wang, S.; Zou, Y.; Xu, Z.; et al. Functional Annotation of Caenorhabditis elegans Genes by Analysis of Gene Co-Expression Networks. Biomolecules 2018, 8, 70. https://doi.org/10.3390/biom8030070
Liu W, Li L, He Y, Cai S, Zhao W, Zheng H, Zhong Y, Wang S, Zou Y, Xu Z, et al. Functional Annotation of Caenorhabditis elegans Genes by Analysis of Gene Co-Expression Networks. Biomolecules. 2018; 8(3):70. https://doi.org/10.3390/biom8030070
Chicago/Turabian StyleLiu, Wei, Ling Li, Yiruo He, Sen Cai, Wenjie Zhao, Hao Zheng, Yuexian Zhong, Shaobo Wang, Yang Zou, Zhenhua Xu, and et al. 2018. "Functional Annotation of Caenorhabditis elegans Genes by Analysis of Gene Co-Expression Networks" Biomolecules 8, no. 3: 70. https://doi.org/10.3390/biom8030070
APA StyleLiu, W., Li, L., He, Y., Cai, S., Zhao, W., Zheng, H., Zhong, Y., Wang, S., Zou, Y., Xu, Z., Zhang, Y., & Tu, W. (2018). Functional Annotation of Caenorhabditis elegans Genes by Analysis of Gene Co-Expression Networks. Biomolecules, 8(3), 70. https://doi.org/10.3390/biom8030070





