Genetics of Lipodystrophy: Can It Help in Understanding the Pathophysiology of Metabolic Syndrome?
Abstract
1. Introduction
2. Discussions
3. Conclusions
Acknowledgments
Conflicts of Interest
References
- Patni, N.; Garg, A. Congenital generalized lipodystrophies-new insights into metabolic dysfunction. Nat. Rev. Endocrinol. 2015, 11, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Samuel, V.T.; Shulman, G.I. Mechanisms for insulin resistance: Common threads and missing links. Cell 2012, 148, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Shields, B.M.; Hicks, S.; Shepherd, M.H.; Colclough, K.; Hattersley, A.T.; Ellard, S. Maturity-onset diabetes of the young (MODY): How many cases are we missing? Diabetologia 2010, 53, 2504–2508. [Google Scholar] [CrossRef] [PubMed]
- Unnikrishnan, R.; Pradeepa, R.; Joshi, S.R.; Mohan, V. Type 2 Diabetes: Demystifying the Global Epidemic. Diabetes 2017, 66, 1432–1442. [Google Scholar] [CrossRef] [PubMed]
- Tallapragada, D.S.; Bhaskar, S.; Chandak, G.R. New insights from monogenic diabetes for “common” type 2 diabetes. Front. Genet. 2015, 6, 251. [Google Scholar] [CrossRef] [PubMed]
- Melvin, A.; O’Rahilly, S.; Savage, D.B. Genetic syndromes of severe insulin resistance. Curr. Opin. Genet. Dev. 2018, 50, 60–67. [Google Scholar] [CrossRef] [PubMed]
- Schleinitz, D. Genetic Determination of Serum Levels of Diabetes-Associated Adipokines. Rev. Diabet. Stud. 2015, 12, 277–298. [Google Scholar] [CrossRef] [PubMed]
- Hegele, R.A. Familial partial lipodystrophy: A monogenic form of the insulin resistance syndrome. Mol. Genet. Metab. 2000, 71, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Frucht, H. Activation of the PPAR pathway induces apoptosis and COX-2 inhibition in HT-29 human colon cancer cells. Carcinogenesis 2001, 22, 1379–1383. [Google Scholar] [CrossRef] [PubMed]
- Gomes, K.B.; Pardini, V.C.; Ferreira, A.C.; Fernandes, A.P. Phenotypic heterogeneity in biochemical parameters correlates with mutations in AGPAT2 or Seipin genes among Berardinelli-Seip congenital lipodystrophy patients. J. Inherit. Metab. Dis. 2005, 28, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Saxena, A.; Gupta, N.; Medicherla, K.M.; Suravajhala, P.; Mathur, S.K. Systems Genomics of Adipose Tissue among Type-2 Diabetics Revealed Distinct Protein Interaction Hubs. Front. Genet. 2018. (Under review). [Google Scholar]
- Nolis, T. Exploring the pathophysiology behind the more common genetic and acquired lipodystrophies. J. Hum. Genet. 2014, 59, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Putcharoen, O.; Wattanachanya, L.; Sophonphan, J.; Siwamogsatham, S.; Sapsirisavat, V.; Gatechompol, S.; Phonphithak, S.; Kerr, S.J.; Chattranukulchai, P.; Avihingsanon, Y.; et al. New-onset diabetes in HIV-treated adults: Predictors, long-term renal and cardiovascular outcomes. Aids 2017, 31, 1535–1543. [Google Scholar] [CrossRef] [PubMed]
- Lipodystrophy search in NCBI GenBank. Available online: https://www.ncbi.nlm.nih.gov/gene/?term=%22lipodystrophy%22++AND+Homo+sapiens+%5Borgn%5D (accessed on 19 June 2018).
- Heijmans, B.T.; Tobi, E.W.; Stein, A.D.; Putter, H.; Blauw, G.J.; Susser, E.S.; Slagboom, P.E.; Lumey, L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA 2008, 105, 17046–17049. [Google Scholar] [CrossRef] [PubMed]
- Vigouroux, C.; Guénantin, A.C.; Vatier, C.; Capel, E.; Le Dour, C.; Afonso, P.; Bidault, G.; Béréziat, V.; Lascols, O.; Capeau, J.; et al. Lipodystrophic syndromes due to LMNA mutations: Recent developments on biomolecular aspects, pathophysiological hypotheses and therapeutic perspectives. Nucleus 2018, 9, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010, 38 (Suppl. 2), W214–W220. [Google Scholar] [CrossRef] [PubMed]
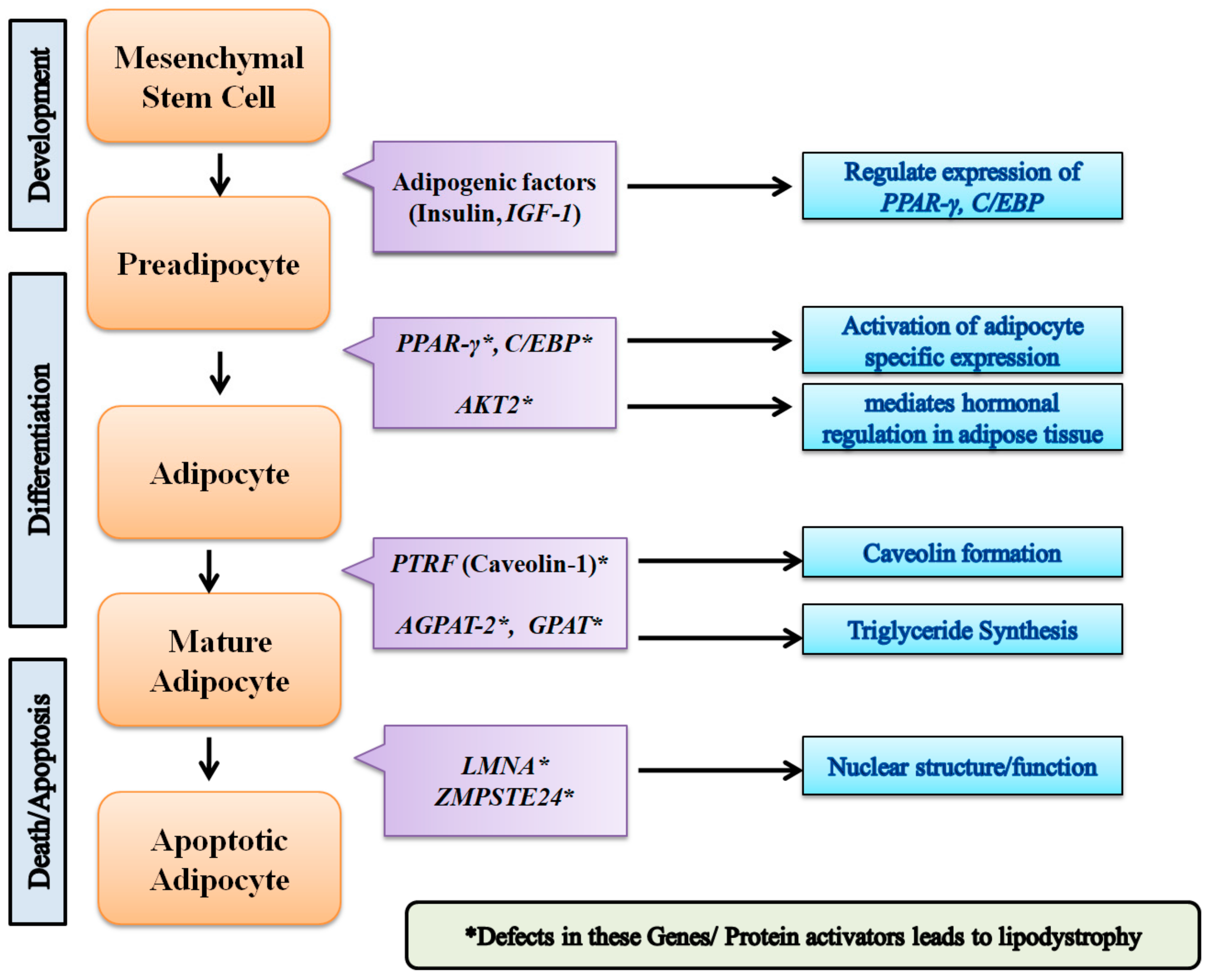
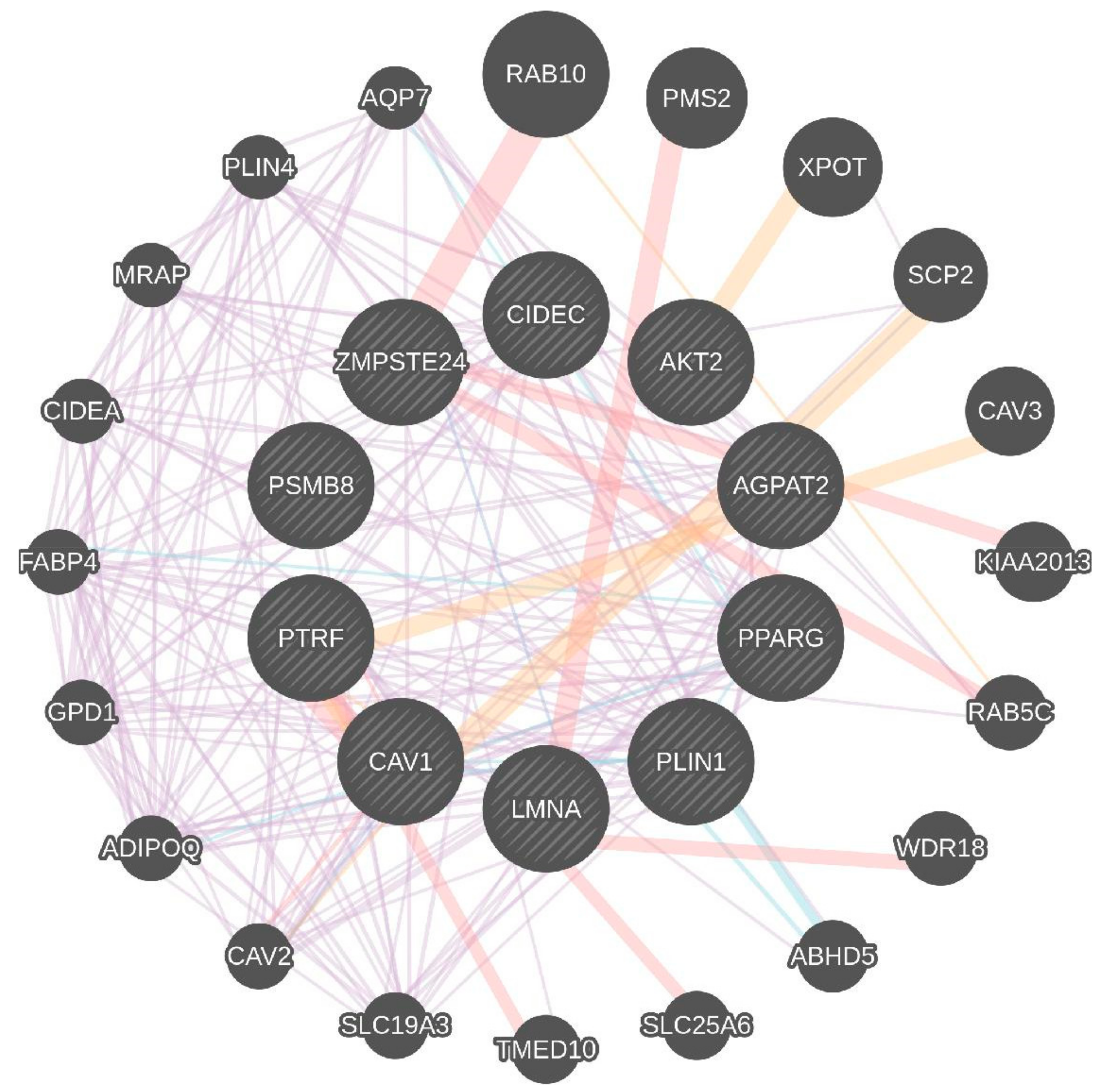
| Key Genes | Gene Function |
|---|---|
| AGPAT2 | AGPAT2 catalyzes formation of phosphatidic acid. |
| AKT2 | AKT2, also known as protein kinase B. Involved in adipocyte differentiation. Downstream insulin receptor signaling. |
| CAV1 | Caveolin 1 binds and transports fatty acids to lipid droplets. |
| LMNA | Lamins A and C are nuclear lamina proteins linked with the cytoskeleton. |
| PLIN1 | Perilipin 1 is an essential component for lipid storage. |
| PPAR-γ | PPARG is crucial for adipogenesis and for maintenance of the differentiation phase. |
| PTRF | Creates caveolae and regulates expression of caveolins 1 and 3. |
| ZMPSTE24 | A zinc metalloproteinase involved in the correct processing and maturation of lamin A (LMNA). |
| CIDEC | A cell death-inducing DNA fragmentation factor-like effector family with important roles in apoptosis, regulated by insulin and its expression is positively correlated with insulin sensitivity. |
| PSMB8 | A proteasome subunit beta type-8 as known as 20S proteasome shapes the antigenic repertoire presented on major histocompatibility complex (MHC) class I molecules. |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mathur, S.K.; Tiwari, P.; Gupta, S.; Gupta, N.; Nimesh, S.; Medicherla, K.M.; Suravajhala, P. Genetics of Lipodystrophy: Can It Help in Understanding the Pathophysiology of Metabolic Syndrome? Biomolecules 2018, 8, 47. https://doi.org/10.3390/biom8030047
Mathur SK, Tiwari P, Gupta S, Gupta N, Nimesh S, Medicherla KM, Suravajhala P. Genetics of Lipodystrophy: Can It Help in Understanding the Pathophysiology of Metabolic Syndrome? Biomolecules. 2018; 8(3):47. https://doi.org/10.3390/biom8030047
Chicago/Turabian StyleMathur, Sandeep Kumar, Pradeep Tiwari, Sonal Gupta, Nidhi Gupta, Surendra Nimesh, Krishna Mohan Medicherla, and Prashanth Suravajhala. 2018. "Genetics of Lipodystrophy: Can It Help in Understanding the Pathophysiology of Metabolic Syndrome?" Biomolecules 8, no. 3: 47. https://doi.org/10.3390/biom8030047
APA StyleMathur, S. K., Tiwari, P., Gupta, S., Gupta, N., Nimesh, S., Medicherla, K. M., & Suravajhala, P. (2018). Genetics of Lipodystrophy: Can It Help in Understanding the Pathophysiology of Metabolic Syndrome? Biomolecules, 8(3), 47. https://doi.org/10.3390/biom8030047






