The Architecture of the Rag GTPase Signaling Network
Abstract
:1. Introduction
2. The Rag GTPase Module
2.1. Structure and Function of the C-Terminal Domains of Gtr1 and Gtr2
2.2. Structure and Function of the Gtr1 G Domain
2.3. Structure and Function of the Gtr2 G Domain
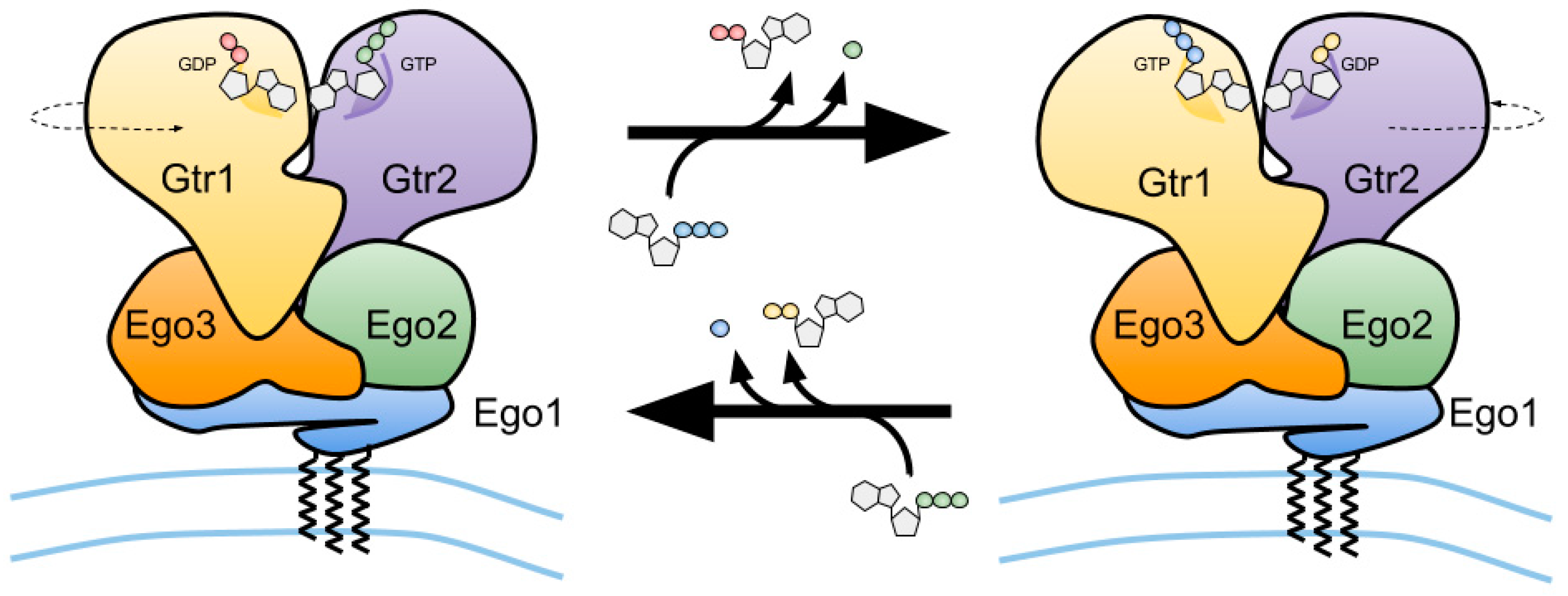
2.4. The Gtr1–Gtr2 Module and its Interactions with TORC1
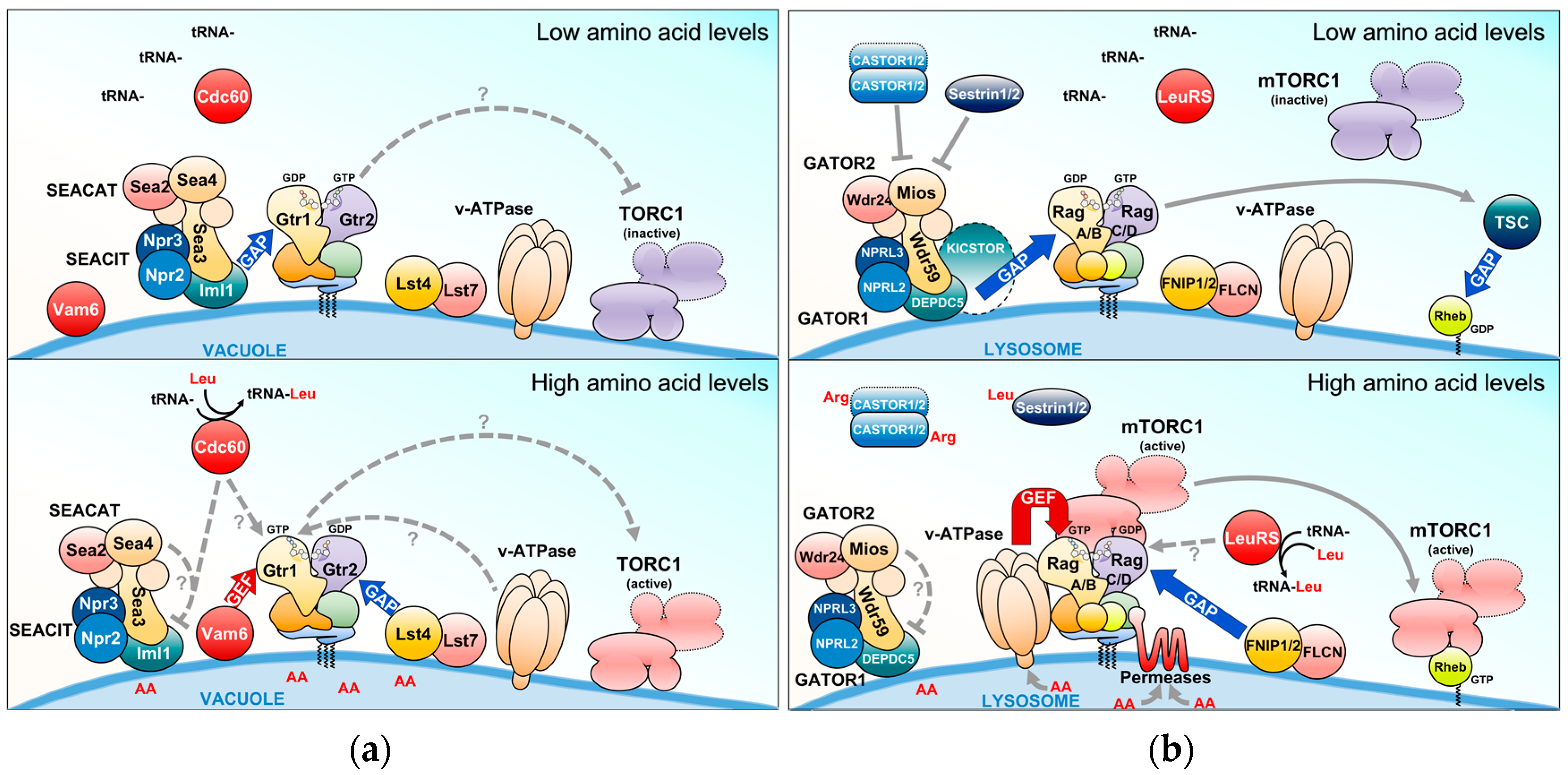
3. Regulators of Rag GTPases
3.1. The EGO-TC and Mammalian Ragulator
3.2. SEACIT and SEACAT and the Orthologous Mammalian GATOR Complexes
3.3. The Lst4–Lst7 and Orthologous FNIP1/2–FLCN Complexes
4. Amino Acids and Their Sensors Upstream of the Rag GTPases
4.1. Leucine
4.2. Glutamine
4.3. Arginine
4.4. The V-ATPase and Vacuolar/Lysosomal-Membrane Resident Amino Acid Permeases
5. Additional Signals and Modulators that Impinge on Rag GTPases
6. Final Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Kunz, J.; Henriquez, R.; Schneider, U.; Deuter-Reinhard, M.; Movva, N.R.; Hall, M.N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596. [Google Scholar] [CrossRef]
- Cafferkey, R.; Young, P.R.; McLaughlin, M.M.; Bergsma, D.J.; Koltin, Y.; Sathe, G.M.; Faucette, L.; Eng, W.K.; Johnson, R.K.; Livi, G.P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell. Biol. 1993, 13, 6012–6023. [Google Scholar] [CrossRef] [PubMed]
- Helliwell, S.B.; Wagner, P.; Kunz, J.; Deuter-Reinhard, M.; Henriquez, R.; Hall, M.N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 1994, 5, 105–118. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.J.; Albers, M.W.; Bum Shin, T.; Ichikawa, K.; Keith, C.T.; Lane, W.S.; Schreiber, S.L. A mammalian protein targeted by G1-arresting rapamycin–receptor complex. Nature 1994, 369, 756–758. [Google Scholar] [CrossRef] [PubMed]
- Chiu, M.I.; Katz, H.; Berlin, V. RAPT1, a mammalian homolog of yeast Tor, interacts with the FKBP12/rapamycin complex. Proc. Natl. Acad. Sci. USA 1994, 91, 12574–12578. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Sabers, C.J.; Martin, M.M.; Brunn, G.J.; Williams, J.M.; Dumont, F.J.; Wiederrecht, G.; Abraham, R.T. Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 1995, 270, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Jacinto, E.; Wullschleger, S.; Lorberg, A.; Crespo, J.L.; Bonenfant, D.; Oppliger, W.; Jenoe, P.; Hall, M.N. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol. Cell 2002, 10, 457–468. [Google Scholar] [CrossRef]
- Wullschleger, S.; Loewith, R.; Hall, M.N. TOR signaling in growth and metabolism. Cell 2006, 124, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Eltschinger, S.; Loewith, R. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016, 26, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR signaling in growth, metabolism, and disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Cornu, M.; Albert, V.; Hall, M.N. mTOR in aging, metabolism, and cancer. Curr. Opin. Genet. Dev. 2013, 23, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Sarbassov, D.D.; Ali, S.M.; Sabatini, D.M. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 2005, 17, 596–603. [Google Scholar] [CrossRef] [PubMed]
- Gaubitz, C.; Prouteau, M.; Kusmider, B.; Loewith, R. TORC2 structure and function. Trends Biochem. Sci. 2016, 41, 532–545. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Maruki, Y.; Long, X.; Yoshino, K.; Oshiro, N.; Hidayat, S.; Tokunaga, C.; Avruch, J.; Yonezawa, K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002, 110, 177–189. [Google Scholar] [CrossRef]
- Kim, D.-H.; Sarbassov, D.D.; Ali, S.M.; King, J.E.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002, 110, 163–175. [Google Scholar] [CrossRef]
- Wedaman, K.P.; Reinke, A.; Anderson, S.; Yates, J.; McCaffery, J.M.; Powers, T. Tor kinases are in distinct membrane-associated protein complexes in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 1204–1220. [Google Scholar] [CrossRef] [PubMed]
- Long, X.; Lin, Y.; Ortiz-Vega, S.; Yonezawa, K.; Avruch, J. Rheb Binds and Regulates the mTOR Kinase. Curr. Biol. 2005, 15, 702–713. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Thoreen, C.C.; Peterson, T.R.; Lindquist, R.A.; Kang, S.A.; Spooner, E.; Carr, S.A.; Sabatini, D.M. PRAS40 is an insulin-regulated inhibitor of the mTORC1 protein kinase. Mol. Cell 2007, 25, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Dibble, C.C.; Elis, W.; Menon, S.; Qin, W.; Klekota, J.; Asara, J.M.; Finan, P.M.; Kwiatkowski, D.J.; Murphy, L.O.; Manning, B.D. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol. Cell 2012, 47, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Inoki, K.; Zhu, T.; Guan, K.-L. TSC2 mediates cellular energy response to control cell growth and survival. Cell 2003, 115, 577–590. [Google Scholar] [CrossRef]
- Tee, A.R.; Manning, B.D.; Roux, P.P.; Cantley, L.C.; Blenis, J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 2003, 13, 1259–1268. [Google Scholar] [CrossRef]
- Nakashima, N.; Noguchi, E.; Nishimoto, T. Saccharomyces cerevisiae putative G protein, Gtr1p, which forms complexes with itself and a novel protein designated as Gtr2p, negatively regulates the Ran/Gsp1p G protein cycle through Gtr2p. Genetics 1999, 152, 853–867. [Google Scholar] [PubMed]
- Sekiguchi, T.; Hirose, E.; Nakashima, N.; Ii, M.; Nishimoto, T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J. Biol. Chem. 2001, 276, 7246–7257. [Google Scholar] [CrossRef] [PubMed]
- Gong, R.; Li, L.; Liu, Y.; Wang, P.; Yang, H.; Wang, L.; Cheng, J.; Guan, K.-L.; Xu, Y. Crystal structure of the Gtr1p-Gtr2p complex reveals new insights into the amino acid-induced TORC1 activation. Genes Dev. 2011, 25, 1668–1673. [Google Scholar] [CrossRef] [PubMed]
- Schürmann, A.; Brauers, A.; Massmann, S.; Becker, W.; Joost, H.G. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J. Biol. Chem. 1995, 270, 28982–28988. [Google Scholar] [CrossRef] [PubMed]
- Wittinghofer, A.; Vetter, I.R. Structure-function relationships of the G domain, a canonical switch motif. Annu. Rev. Biochem. 2011, 80, 943–971. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, T.; Kamada, Y.; Furuno, N.; Funakoshi, M.; Kobayashi, H. Amino acid residues required for Gtr1p-Gtr2p complex formation and its interactions with the Ego1p-Ego3p complex and TORC1 components in yeast. Genes Cells 2014, 19, 449–463. [Google Scholar] [CrossRef] [PubMed]
- Bun-Ya, M.; Harashima, S.; Oshima, Y. Putative GTP-binding protein, Gtr1, associated with the function of the Pho84 inorganic phosphate transporter in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 2958–2966. [Google Scholar] [CrossRef] [PubMed]
- Binda, M.; Péli-Gulli, M.-P.; Bonfils, G.; Panchaud, N.; Urban, J.; Sturgill, T.W.; Loewith, R.; De Virgilio, C. The Vam6 GEF controls TORC1 by activating the EGO complex. Mol. Cell 2009, 35, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Demetriades, C.; Doumpas, N.; Teleman, A.A. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 2014, 156, 786–799. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Kaiser, C.A. A conserved GTPase-containing complex is required for intracellular sorting of the general amino-acid permease in yeast. Nat. Cell Biol. 2006, 8, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Goraksha-Hicks, P.; Li, L.; Neufeld, T.P.; Guan, K.-L. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 2008, 10, 935–945. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Kira, S.; Tabata, K.; Shirahama-Noda, K.; Nozoe, A.; Yoshimori, T.; Noda, T. Reciprocal conversion of Gtr1 and Gtr2 nucleotide-binding states by Npr2-Npr3 inactivates TORC1 and induces autophagy. Autophagy 2014, 10, 1565–1578. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Lambright, D.G. Invited review: Small GTPases and their GAPs. Biopolymers 2016, 105, 431–448. [Google Scholar] [CrossRef] [PubMed]
- Sancak, Y.; Bar-Peled, L.; Zoncu, R.; Markhard, A.L.; Nada, S.; Sabatini, D.M. Ragulator-Rag Complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 2010, 141, 290–303. [Google Scholar] [CrossRef] [PubMed]
- Powis, K.; Zhang, T.; Panchaud, N.; Wang, R.; De Virgilio, C.; Ding, J. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res. 2015, 25, 1043–1059. [Google Scholar] [CrossRef] [PubMed]
- Fawal, M.-A.; Brandt, M.; Djouder, N. MCRS1 binds and couples Rheb to amino acid-dependent mTORC1 activation. Dev. Cell 2015, 33, 67–81. [Google Scholar] [CrossRef] [PubMed]
- Averous, J.; Lambert-Langlais, S.; Carraro, V.; Gourbeyre, O.; Parry, L.; B’Chir, W.; Muranishi, Y.; Jousse, C.; Bruhat, A.; Maurin, A.-C.; et al. Requirement for lysosomal localization of mTOR for its activation differs between leucine and other amino acids. Cell. Signal. 2014, 26, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Sturgill, T.W.; Cohen, A.; Diefenbacher, M.; Trautwein, M.; Martin, D.E.; Hall, M.N. TOR1 and TOR2 have distinct locations in live cells. Eukaryot. Cell 2008, 7, 1819–1830. [Google Scholar] [CrossRef] [PubMed]
- Kira, S.; Kumano, Y.; Ukai, H.; Takeda, E.; Matsuura, A.; Noda, T. Dynamic relocation of the TORC1-Gtr1/2-Ego1/2/3 complex is regulated by Gtr1 and Gtr2. Mol. Biol. Cell 2016, 27, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-H.; Lee, K.-H.; Kim, Y.-M.; Kim, D.-H.; Oh, B.-H.; Kim, Y.-G. Crystal structure of the Gtr1pGTP-Gtr2pGDP protein complex reveals large structural rearrangements triggered by GTP-to-GDP conversion. J. Biol. Chem. 2012, 287, 29648–29653. [Google Scholar] [CrossRef] [PubMed]
- Chappie, J.S.; Acharya, S.; Leonard, M.; Schmid, S.L.; Dyda, F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature 2010, 465, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Focia, P.J.; Shepotinovskaya, I.V.; Seidler, J.A.; Freymann, D.M. Heterodimeric GTPase core of the SRP targeting complex. Science 2004, 303, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; von der Malsburg, A.; Paeschke, S.; Behlke, J.; Haller, O.; Kochs, G.; Daumke, O. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 2010, 465, 502–506. [Google Scholar] [CrossRef] [PubMed]
- Low, H.H.; Sachse, C.; Amos, L.A.; Löwe, J. Structure of a Bacterial Dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 2009, 139, 1342–1352. [Google Scholar] [CrossRef] [PubMed]
- Sengottaiyan, P.; Spetea, C.; Lagerstedt, J.O.; Samyn, D.; Andersson, M.; Ruiz-Pavón, L.; Persson, B.L. The intrinsic GTPase activity of the Gtr1 protein from Saccharomyces cerevisiae. BMC Biochem. 2012, 13, 11. [Google Scholar] [CrossRef] [PubMed]
- Kleuss, C.; Raw, A.S.; Lee, E.; Sprang, S.R.; Gilman, A.G. Mechanism of GTP hydrolysis by G-protein alpha subunits. Proc. Natl. Acad. Sci. USA 1994, 91, 9828–9831. [Google Scholar] [CrossRef] [PubMed]
- Buhrman, G.; Holzapfel, G.; Fetics, S.; Mattos, C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc. Natl. Acad. Sci. USA 2010, 107, 4931–4936. [Google Scholar] [CrossRef] [PubMed]
- Brondyk, W.H.; McKiernan, C.J.; Burstein, E.S.; Macara, I.G. Mutants of Rab3A analogous to oncogenic Ras mutants. Sensitivity to Rab3A-GTPase activating protein and Rab3A-guanine nucleotide releasing factor. J. Biol. Chem. 1993, 268, 9410–9415. [Google Scholar] [PubMed]
- Barbacid, M. ras GENES. Annu. Rev. Biochem. 1987, 56, 779–827. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, N.; Péli-Gulli, M.-P.; De Virgilio, C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 2013, 6, ra42. [Google Scholar] [CrossRef] [PubMed]
- Péli-Gulli, M.-P.; Sardu, A.; Panchaud, N.; Raucci, S.; De Virgilio, C. Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-Activating protein complex for the Rag family GTPase Gtr2. Cell Rep. 2015, 13, 1–7. [Google Scholar] [CrossRef] [PubMed]
- John, J.; Rensland, H.; Schlichting, I.; Vetter, I.; Borasio, G.D.; Goody, R.S.; Wittinghofer, A. Kinetic and structural analysis of the Mg2+-binding site of the guanine nucleotide-binding protein p21H−ras. J. Biol. Chem. 1993, 268, 923–929. [Google Scholar] [PubMed]
- Nassar, N.; Singh, K.; Garcia-Diaz, M. Structure of the dominant negative S17N mutant of Ras. Biochemistry 2010, 49, 1970–1974. [Google Scholar] [CrossRef] [PubMed]
- Farnsworth, C.L.; Feig, L.A. Dominant inhibitory mutations in the Mg2+-binding site of RasH prevent its activation by GTP. Mol. Cell. Biol. 1991, 11, 4822–4829. [Google Scholar] [CrossRef] [PubMed]
- Rensland, H.; Lautwein, A.; Wittinghofer, A.; Goody, R.S. Is there a rate-limiting step before GTP cleavage by H-ras p21? Biochemistry 1991, 30, 11181–11185. [Google Scholar] [CrossRef] [PubMed]
- Simon, I.; Zerial, M.; Goody, R.S. Kinetics of interaction of Rab5 and Rab7 with nucleotides and magnesium ions. J. Biol. Chem. 1996, 271, 20470–20478. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Kurihara, Y.; Sato, T.; Toh, H.; Kobayashi, H.; Sekiguchi, T. Gtr1p differentially associates with Gtr2p and Ego1p. Gene 2009, 437, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Trahey, M.; McCormick, F. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 1987, 238, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Rudoni, S.; Colombo, S.; Coccetti, P.; Martegani, E. Role of guanine nucleotides in the regulation of the Ras/cAMP pathway in Saccharomyces cerevisiae. Biochim. Biophys. Acta 2001, 1538, 181–189. [Google Scholar] [CrossRef]
- Coleman, D.E.; Berghuis, A.M.; Lee, E.; Linder, M.E.; Gilman, A.G.; Sprang, S.R. Structures of active conformations of Gi alpha 1 and the mechanism of GTP hydrolysis. Science 1994, 265, 1405–1412. [Google Scholar] [CrossRef] [PubMed]
- Tsun, Z.-Y.; Bar-Peled, L.; Chantranupong, L.; Zoncu, R.; Wang, T.; Kim, C.; Spooner, E.; Sabatini, D.M. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol. Cell 2013, 52, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Dubouloz, F.; Deloche, O.; Wanke, V.; Cameroni, E.; De Virgilio, C. The TOR and EGO protein complexes orchestrate microautophagy in yeast. Mol. Cell 2005, 19, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Schweitzer, L.D.; Zoncu, R.; Sabatini, D.M. Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 2012, 150, 1196–1208. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Péli-Gulli, M.-P.; Yang, H.; De Virgilio, C.; Ding, J. Ego3 Functions as a homodimer to mediate the interaction between Gtr1-Gtr2 and Ego1 in the EGO complex to activate TORC1. Structure 2012, 20, 2151–2160. [Google Scholar] [CrossRef] [PubMed]
- Nadolski, M.J.; Linder, M.E. Molecular recognition of the palmitoylation substrate Vac8 by its palmitoyltransferase Pfa3. J. Biol. Chem. 2009, 284, 17720–17730. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.F.; Wan, J.; Bailey, A.O.; Sun, B.; Kuchar, J.A.; Green, W.N.; Phinney, B.S.; Yates, J.R.; Davis, N.G. Global analysis of protein palmitoylation in yeast. Cell 2006, 125, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Nada, S.; Hondo, A.; Kasai, A.; Koike, M.; Saito, K.; Uchiyama, Y.; Okada, M. The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK–ERK pathway to late endosomes. EMBO J. 2009, 28, 477–489. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, N.; Guan, K.-L.; Moreno, S. The Vam6 and Gtr1-Gtr2 pathway activates TORC1 in response to amino acids in fission yeast. J. Cell Sci. 2012, 125, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirata, A.; Ohsumi, Y.; Wada, Y. Vam2/Vps41p and Vam6/Vps39p are components of a protein complex on the vacuolar membranes and involved in the vacuolar assembly in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 1997, 272, 11344–11349. [Google Scholar] [CrossRef] [PubMed]
- Ostrowicz, C.W.; Meiringer, C.T.A.; Ungermann, C. Yeast vacuole fusion: A model system for eukaryotic endomembrane dynamics. Autophagy 2008, 4, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Kümmel, D.; Ungermann, C. Principles of membrane tethering and fusion in endosome and lysosome biogenesis. Curr. Opin. Cell Biol. 2014, 29, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, J.M.; Sen, N.D.; Maeda, T.; Heitman, J.; Cardenas, M.E. Endolysosomal membrane trafficking complexes drive nutrient-dependent TORC1 signaling to control cell growth in Saccharomyces cerevisiae. Genetics 2014, 196, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Zurita-Martinez, S.A.; Puria, R.; Pan, X.; Boeke, J.D.; Cardenas, M.E. Efficient Tor signaling requires a functional class C Vps protein complex in Saccharomyces cerevisiae. Genetics 2007, 176, 2139–2150. [Google Scholar] [CrossRef] [PubMed]
- Elbaz-Alon, Y.; Rosenfeld-Gur, E.; Shinder, V.; Futerman, A.H.; Geiger, T.; Schuldiner, M. A Dynamic interface between vacuoles and mitochondria in yeast. Dev. Cell 2014, 30, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Hönscher, C.; Mari, M.; Auffarth, K.; Bohnert, M.; Griffith, J.; Geerts, W.; van der Laan, M.; Cabrera, M.; Reggiori, F.; Ungermann, C. Cellular metabolism regulates contact sites between vacuoles and mitochondria. Dev. Cell 2014, 30, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Caplan, S.; Hartnell, L.M.; Aguilar, R.C.; Naslavsky, N.; Bonifacino, J.S. Human Vam6p promotes lysosome clustering and fusion in vivo. J. Cell Biol. 2001, 154, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Felici, A.; Wurthner, J.U.; Parks, W.T.; Giam, L.R.; Reiss, M.; Karpova, T.S.; McNally, J.G.; Roberts, A.B. TLP, a novel modulator of TGF-signaling, has opposite effects on Smad2- and Smad3-dependent signaling. EMBO J. 2003, 22, 4465–4477. [Google Scholar] [CrossRef] [PubMed]
- Messler, S.; Kropp, S.; Episkopou, V.; Felici, A.; Würthner, J.; Lemke, R.; Jerabek-Willemsen, M.; Willecke, R.; Scheu, S.; Pfeffer, K.; et al. The TGF-β signaling modulators TRAP1/TGFBRAP1 and VPS39/Vam6/TLP are essential for early embryonic development. Immunobiology 2011, 216, 343–350. [Google Scholar] [CrossRef] [PubMed]
- Lachmann, J.; Glaubke, E.; Moore, P.S.; Ungermann, C. The Vps39-like TRAP1 is an effector of Rab5 and likely the missing Vps3 subunit of human CORVET. Cell. Logist. 2014, 4, e970840. [Google Scholar] [CrossRef] [PubMed]
- Flinn, R.J.; Yan, Y.; Goswami, S.; Parker, P.J.; Backer, J.M. The late endosome is essential for mTORC1 signaling. Mol. Biol. Cell 2010, 21, 833–841. [Google Scholar] [CrossRef] [PubMed]
- Neklesa, T.K.; Davis, R.W. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 Complex. PLoS Genet. 2009, 5, e1000515. [Google Scholar] [CrossRef] [PubMed]
- Dokudovskaya, S.; Rout, M.P. A novel coatomer-related SEA complex dynamically associates with the vacuole in yeast and is implicated in the response to nitrogen starvation. Autophagy 2011, 7, 1392–1393. [Google Scholar] [CrossRef] [PubMed]
- Panchaud, N.; Péli-Gulli, M.-P.; De Virgilio, C. SEACing the GAP that nEGOCiates TORC1 activation: Evolutionary conservation of Rag GTPase regulation. Cell Cycle 2013, 12, 2948–2952. [Google Scholar] [CrossRef] [PubMed]
- Sutter, B.M.; Wu, X.; Laxman, S.; Tu, B.P. Methionine inhibits autophagy and promotes growth by inducing the SAM-responsive methylation of PP2A. Cell 2013, 154, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Chantranupong, L.; Cherniack, A.D.; Chen, W.W.; Ottina, K.A.; Grabiner, B.C.; Spear, E.D.; Carter, S.L.; Meyerson, M.; Sabatini, D.M. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 2013, 340, 1100–1106. [Google Scholar] [CrossRef] [PubMed]
- Bar-Peled, L.; Sabatini, D.M. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014, 24, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Wei, Y.; Jarnik, M.; Reich, J.; Lilly, M.A. The GATOR2 Component Wdr24 Regulates TORC1 Activity and Lysosome Function. PLoS Genet. 2016, 12, e1006036. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Wyant, G.A.; Gu, X.; Orozco, J.M.; Shen, K.; Condon, K.J.; Petri, S.; Kedir, J.; Scaria, S.M.; et al. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature 2017, 543, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Li, M.O. SZT2 dictates GATOR control of mTORC1 signalling. Nature 2017, 543, 433–437. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Jiang, C.; Chen, L.; Jin, J.; Wei, J.; Zhao, L.; Chen, M.; Pan, W.; Xu, Y.; Chu, H.; et al. The ubiquitination of rag A GTPase by RNF152 negatively regulates mTORC1 activation. Mol. Cell 2015, 58, 804–818. [Google Scholar] [CrossRef] [PubMed]
- Jin, G.; Lee, S.-W.; Zhang, X.; Cai, Z.; Gao, Y.; Chou, P.-C.; Rezaeian, A.H.; Han, F.; Wang, C.-Y.; Yao, J.-C.; et al. Skp2-mediated RagA ubiquitination elicits a negative feedback to prevent amino-acid-dependent mTORC1 hyperactivation by recruiting GATOR1. Mol. Cell 2015, 58, 989–1000. [Google Scholar] [CrossRef] [PubMed]
- Petit, C.S.; Roczniak-Ferguson, A.; Ferguson, S.M. Recruitment of folliculin to lysosomes supports the amino acid–dependent activation of Rag GTPases. J. Cell Biol. 2013, 202, 1107–1122. [Google Scholar] [CrossRef] [PubMed]
- Martina, J.A.; Diab, H.I.; Lishu, L.; Jeong-A, L.; Patange, S.; Raben, N.; Puertollano, R. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 2014, 7, ra9. [Google Scholar] [CrossRef] [PubMed]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [PubMed]
- Stracka, D.; Jozefczuk, S.; Rudroff, F.; Sauer, U.; Hall, M.N. Nitrogen source activates TOR (target of rapamycin) complex 1 via glutamine and independently of Gtr/Rag proteins. J. Biol. Chem. 2014, 289, 25010–25020. [Google Scholar] [CrossRef] [PubMed]
- Kingsbury, J.M.; Sen, N.D.; Cardenas, M.E. Branched-chain aminotransferases control TORC1 signaling in Saccharomyces cerevisiae. PLoS Genet. 2015, 11, e1005714. [Google Scholar] [CrossRef] [PubMed]
- Echols, N.; Harrison, P.; Balasubramanian, S.; Luscombe, N.M.; Bertone, P.; Zhang, Z.; Gerstein, M. Comprehensive analysis of amino acid and nucleotide composition in eukaryotic genomes, comparing genes and pseudogenes. Nucleic Acids Res. 2002, 30, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Avruch, J.; Long, X.; Ortiz-Vega, S.; Rapley, J.; Papageorgiou, A.; Dai, N. Amino acid regulation of TOR complex 1. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E592–E602. [Google Scholar] [CrossRef] [PubMed]
- Bonfils, G.; Jaquenoud, M.; Bontron, S.; Ostrowicz, C.; Ungermann, C.; De Virgilio, C. Leucyl-tRNA synthetase controls TORC1 via the EGO complex. Mol. Cell 2012, 46, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Han, J.M.; Jeong, S.J.; Park, M.C.; Kim, G.; Kwon, N.H.; Kim, H.K.; Ha, S.H.; Ryu, S.H.; Kim, S. Leucyl-tRNA synthetase is an intracellular leucine sensor for the mTORC1-signaling pathway. Cell 2012, 149, 410–424. [Google Scholar] [CrossRef] [PubMed]
- Yoon, M.-S.; Son, K.; Arauz, E.; Han, J.M.; Kim, S.; Chen, J. Leucyl-tRNA synthetase activates Vps34 in amino acid-sensing mTORC1 signaling. Cell Rep. 2016, 16, 1510–1517. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Wolfson, R.L.; Orozco, J.M.; Saxton, R.A.; Scaria, S.M.; Bar-Peled, L.; Spooner, E.; Isasa, M.; Gygi, S.P.; Sabatini, D.M. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep. 2014, 9, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Ro, S.-H.; Kim, M.; Park, H.-W.; Semple, I.A.; Park, H.; Cho, U.-S.; Wang, W.; Guan, K.-L.; Karin, M.; et al. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci. Rep. 2015, 5, 9502. [Google Scholar] [CrossRef] [PubMed]
- Parmigiani, A.; Nourbakhsh, A.; Ding, B.; Wang, W.; Kim, Y.C.; Akopiants, K.; Guan, K.-L.; Karin, M.; Budanov, A.V. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014, 9, 1281–1291. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Yin, N.; Li, M.O. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell 2014, 159, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Wolfson, R.L.; Chantranupong, L.; Saxton, R.A.; Shen, K.; Scaria, S.M.; Cantor, J.R.; Sabatini, D.M. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science 2015, 351, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Knockenhauer, K.E.; Wolfson, R.L.; Chantranupong, L.; Pacold, M.E.; Wang, T.; Schwartz, T.U.; Sabatini, D.M. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science 2015, 351, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Knockenhauer, K.E.; Schwartz, T.U.; Sabatini, D.M. The apo-structure of the leucine sensor Sestrin2 is still elusive. Sci. Signal. 2016, 9, ra92. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; An, S.; Ro, S.-H.; Teixeira, F.; Park, G.J.; Kim, C.; Cho, C.-S.; Kim, J.-S.; Jakob, U.; Lee, J.H.; et al. Janus-faced Sestrin2 controls ROS and mTOR signalling through two separate functional domains. Nat. Commun. 2015, 6, 10025. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Cho, U.-S.; Karin, M. Sestrin regulation of TORC1: Is Sestrin a leucine sensor? Sci. Signal. 2016, 9, re5. [Google Scholar] [CrossRef] [PubMed]
- Budanov, A.V.; Karin, M. p53 Target Genes Sestrin1 and Sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Pavlova, N.N.; Thompson, C.B. Cancer cell metabolism: The essential role of the nonessential amino acid, glutamine. EMBO J. 2017, e201696151. [Google Scholar] [CrossRef] [PubMed]
- Durán, R.V.; Oppliger, W.; Robitaille, A.M.; Heiserich, L.; Skendaj, R.; Gottlieb, E.; Hall, M.N. Glutaminolysis activates Rag-mTORC1 signaling. Mol. Cell 2012, 47, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Sener, A.; Malaisse, W.J. L-leucine and a nonmetabolized analogue activate pancreatic islet glutamate dehydrogenase. Nature 1980, 288, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, C.; Allen, A.; Stanley, C.A.; Smith, T.J. The structure and allosteric regulation of glutamate dehydrogenase. Neurochem. Int. 2011, 59, 445–455. [Google Scholar] [CrossRef] [PubMed]
- Nicklin, P.; Bergman, P.; Zhang, B.; Triantafellow, E.; Wang, H.; Nyfeler, B.; Yang, H.; Hild, M.; Kung, C.; Wilson, C.; et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell 2009, 136, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Milkereit, R.; Persaud, A.; Vanoaica, L.; Guetg, A.; Verrey, F.; Rotin, D. LAPTM4b recruits the LAT1–4F2hc Leu transporter to lysosomes and promotes mTORC1 activation. Nat. Commun. 2015, 6, 7250. [Google Scholar] [CrossRef] [PubMed]
- Jewell, J.L.; Kim, Y.C.; Russell, R.C.; Yu, F.-X.; Park, H.W.; Plouffe, S.W.; Tagliabracci, V.S.; Guan, K.-L. Differential regulation of mTORC1 by leucine and glutamine. Science 2015, 347, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.; Cunningham, K.W. A LAPF/phafin1-like protein regulates TORC1 and lysosomal membrane permeabilization in response to endoplasmic reticulum membrane stress. Mol. Biol. Cell 2015, 26, 4631–4645. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.H.; Hatakeyama, R.; Kimmig, P.; Arter, M.; Peter, M.; Matos, J.; De Virgilio, C.; Kornmann, B. Functional mapping of yeast genomes by saturated transposition. eLife 2017, 6, e23570. [Google Scholar] [CrossRef] [PubMed]
- Tanigawa, M.; Maeda, T. An in vitro TORC1 kinase assay that recapitulates the Gtr-independent glutamine-responsive TORC1 activation mechanism on yeast vacuoles. Mol. Cell. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tsun, Z.-Y.; Wolfson, R.L.; Shen, K.; Wyant, G.A.; Plovanich, M.E.; Yuan, E.D.; Jones, T.D.; Chantranupong, L.; Comb, W.; et al. Metabolism. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015, 347, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Rebsamen, M.; Pochini, L.; Stasyk, T.; de Araújo, M.E.G.; Galluccio, M.; Kandasamy, R.K.; Snijder, B.; Fauster, A.; Rudashevskaya, E.L.; Bruckner, M.; et al. SLC38A9 is a component of the lysosomal amino acid sensing machinery that controls mTORC1. Nature 2015, 519, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.; Genau, H.M.; Behrends, C. Amino acid-dependent mTORC1 regulation by the lysosomal membrane protein SLC38A9. Mol. Cell. Biol. 2015, 35, 2479–2494. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Scaria, S.M.; Saxton, R.A.; Gygi, M.P.; Shen, K.; Wyant, G.A.; Wang, T.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 2016, 165, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Chantranupong, L.; Knockenhauer, K.E.; Schwartz, T.U.; Sabatini, D.M. Mechanism of arginine sensing by CASTOR1 upstream of mTORC1. Nature 2016, 536, 229–233. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Wang, R.; Zhang, T.; Ding, J. Structural insight into the arginine-binding specificity of CASTOR1 in amino acid-dependent mTORC1 signaling. Cell Discov. 2016, 2, 16035. [Google Scholar] [CrossRef] [PubMed]
- Gai, Z.; Wang, Q.; Yang, C.; Wang, L.; Deng, W.; Wu, G. Structural mechanism for the arginine sensing and regulation of CASTOR1 in the mTORC1 signaling pathway. Cell Discov. 2016, 2, 16051. [Google Scholar] [CrossRef] [PubMed]
- Parsons, A.B.; Brost, R.L.; Ding, H.; Li, Z.; Zhang, C.; Sheikh, B.; Brown, G.W.; Kane, P.M.; Hughes, T.R.; Boone, C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004, 22, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Sekito, T.; Fujiki, Y.; Ohsumi, Y.; Kakinuma, Y. Novel families of vacuolar amino acid transporters. IUBMB Life 2008, 60, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I.; Fuchs, R.; Helenius, A. Acidification of the endocytic and exocytic pathways. Annu. Rev. Biochem. 1986, 55, 663–700. [Google Scholar] [CrossRef] [PubMed]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar] [CrossRef] [PubMed]
- Balgi, A.D.; Diering, G.H.; Donohue, E.; Lam, K.K.Y.; Fonseca, B.D.; Zimmerman, C.; Numata, M.; Roberge, M. Regulation of mTORC1 signaling by pH. PLoS ONE 2011, 6, e21549. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, B.D.; Diering, G.H.; Bidinosti, M.A.; Dalal, K.; Alain, T.; Balgi, A.D.; Forestieri, R.; Nodwell, M.; Rajadurai, C.V.; Gunaratnam, C.; et al. Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 2012, 287, 17530–17545. [Google Scholar] [CrossRef] [PubMed]
- Zoncu, R.; Bar-Peled, L.; Efeyan, A.; Wang, S.; Sancak, Y.; Sabatini, D.M. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011, 334, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Dechant, R.; Saad, S.; Ibáñez, A.J.; Peter, M. Cytosolic pH regulates cell growth through distinct GTPases, Arf1 and Gtr1, to promote Ras/PKA and TORC1 activity. Mol. Cell 2014, 55, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Heublein, S.; Kazi, S.; Ögmundsdóttir, M.H.; Attwood, E.V.; Kala, S.; Boyd, C.A.R.; Wilson, C.; Goberdhan, D.C.I. Proton-assisted amino-acid transporters are conserved regulators of proliferation and amino-acid-dependent mTORC1 activation. Oncogene 2010, 29, 4068–4079. [Google Scholar] [CrossRef] [PubMed]
- Ögmundsdóttir, M.H.; Heublein, S.; Kazi, S.; Reynolds, B.; Visvalingam, S.M.; Shaw, M.K.; Goberdhan, D.C.I. Proton-assisted amino acid transporter PAT1 complexes with Rag GTPases and activates TORC1 on late endosomal and lysosomal membranes. PLoS ONE 2012, 7, e36616. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Shimabukuro-Demoto, S.; Yoshida-Sugitani, R.; Furuyama-Tanaka, K.; Karyu, H.; Sugiura, Y.; Shimizu, Y.; Hosaka, T.; Goto, M.; Kato, N.; et al. The histidine transporter SLC15A4 coordinates mTOR-dependent inflammatory responses and pathogenic antibody production. Immunity 2014, 41, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.-S.; Jiang, B.; Li, M.; Zhu, M.; Peng, Y.; Zhang, Y.-L.; Wu, Y.-Q.; Li, T.Y.; Liang, Y.; Lu, Z.; et al. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014, 20, 526–540. [Google Scholar] [CrossRef] [PubMed]
- Efeyan, A.; Zoncu, R.; Chang, S.; Gumper, I.; Snitkin, H.; Wolfson, R.L.; Kirak, O.; Sabatini, D.D.; Sabatini, D.M. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 2013, 493, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. State transitions in the TORC1 signaling pathway and information processing in Saccharomyces cerevisiae. Genetics 2014, 198, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Hughes Hallett, J.E.; Luo, X.; Capaldi, A.P. Snf1/AMPK promotes the formation of Kog1/raptor-bodies to increase the activation threshold of TORC1 in budding yeast. Elife 2015, 4, e09181. [Google Scholar] [CrossRef] [PubMed]
- Castellano, B.M.; Thelen, A.M.; Moldavski, O.; Feltes, M.; van der Welle, R.E.N.; Mydock-McGrane, L.; Jiang, X.; van Eijkeren, R.J.; Davis, O.B.; Louie, S.M.; et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 2017, 355, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Bryk, B.; Hahn, K.; Cohen, S.M.; Teleman, A.A. MAP4K3 regulates body size and metabolism in Drosophila. Dev. Biol. 2010, 344, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Findlay, G.M.; Yan, L.; Procter, J.; Mieulet, V.; Lamb, R.F. A MAP4 kinase related to Ste20 is a nutrient-sensitive regulator of mTOR signalling. Biochem. J. 2007, 403, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Mieulet, V.; Burgess, D.; Findlay, G.M.; Sully, K.; Procter, J.; Goris, J.; Janssens, V.; Morrice, N.A.; Lamb, R.F. PP2AT61ε is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol. Cell 2010, 37, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Duran, A.; Amanchy, R.; Linares, J.F.; Joshi, J.; Abu-Baker, S.; Porollo, A.; Hansen, M.; Moscat, J.; Diaz-Meco, M.T. p62 is a key regulator of nutrient sensing in the mTORC1 pathway. Mol. Cell 2011, 44, 134–146. [Google Scholar] [CrossRef] [PubMed]
- Kwon, O.; Kwak, D.; Ha, S.H.; Jeon, H.; Park, M.; Chang, Y.; Suh, P.-G.; Ryu, S.H. Nudix-type motif 2 contributes to cancer proliferation through the regulation of Rag GTPase-mediated mammalian target of rapamycin complex 1 localization. Cell. Signal. 2017, 32, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-M.; Stone, M.; Hwang, T.H.; Kim, Y.-G.; Dunlevy, J.R.; Griffin, T.J.; Kim, D.-H. SH3BP4 is a negative regulator of amino acid-Rag GTPase-mTORC1 signaling. Mol. Cell 2012, 46, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, L.D.; Comb, W.C.; Bar-Peled, L.; Sabatini, D.M. Disruption of the Rag-Ragulator complex by c17orf59 inhibits mTORC1. Cell Rep. 2015, 12, 1445–1455. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Wang, J.; Yoshida, S.; Nada, S.; Okada, M.; Inoki, K. Role of Ragulator in the regulation of mechanistic target of rapamycin signaling in podocytes and glomerular function. J. Am. Soc. Nephrol. 2016, 27, 3653–3665. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Guo, S.; Gao, J.; Zhong, M.; Yan, G.; Wu, W.; Chao, Y.; Jiang, Y. General control nonderepressible 2 (GCN2) kinase inhibits target of rapamycin complex 1 in response to amino acid starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2017, 292, 2660–2669. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.D.; Zhang, Y.-J.; Wei, Y.-H.; Cho, J.-H.; Morris, L.E.; Wang, H.-Y.; Zheng, X.F.S. Rab1A is an mTORC1 activator and a colorectal oncogene. Cancer Cell 2014, 26, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Oshiro, N.; Rapley, J.; Avruch, J. Amino acids activate mammalian target of rapamycin (mTOR) complex 1 without changing Rag GTPase guanyl nucleotide charging. J. Biol. Chem. 2014, 289, 2658–2674. [Google Scholar] [CrossRef] [PubMed]
- Chantranupong, L.; Wolfson, R.L.; Sabatini, D.M. Nutrient-sensing mechanisms across evolution. Cell 2015, 161, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Hatakeyama, R.; De Virgilio, C. Unsolved mysteries of Rag GTPase signaling in yeast. Small GTPases 2016, 7, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Powis, K.; De Virgilio, C. Conserved regulators of Rag GTPases orchestrate amino acid-dependent TORC1 signaling. Cell Discov. 2016, 2, 15049. [Google Scholar] [CrossRef] [PubMed]
- Liko, D.; Hall, M.N. mTOR in health and in sickness. J. Mol. Med. 2015, 93, 1061–1073. [Google Scholar] [CrossRef] [PubMed]
- Aylett, C.H.S.; Sauer, E.; Imseng, S.; Boehringer, D.; Hall, M.N.; Ban, N.; Maier, T. Architecture of human mTOR complex 1. Science 2016, 351, 48–52. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Rudge, D.G.; Koos, J.D.; Vaidialingam, B.; Yang, H.J.; Pavletich, N.P. mTOR kinase structure, mechanism and regulation. Nature 2013, 497, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Yip, C.K.; Murata, K.; Walz, T.; Sabatini, D.M.; Kang, S.A. Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 2010, 38, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Baretić, D.; Berndt, A.; Ohashi, Y.; Johnson, C.M.; Williams, R.L. Tor forms a dimer through an N-terminal helical solenoid with a complex topology. Nat. Commun. 2016, 7, 11016. [Google Scholar] [CrossRef] [PubMed]
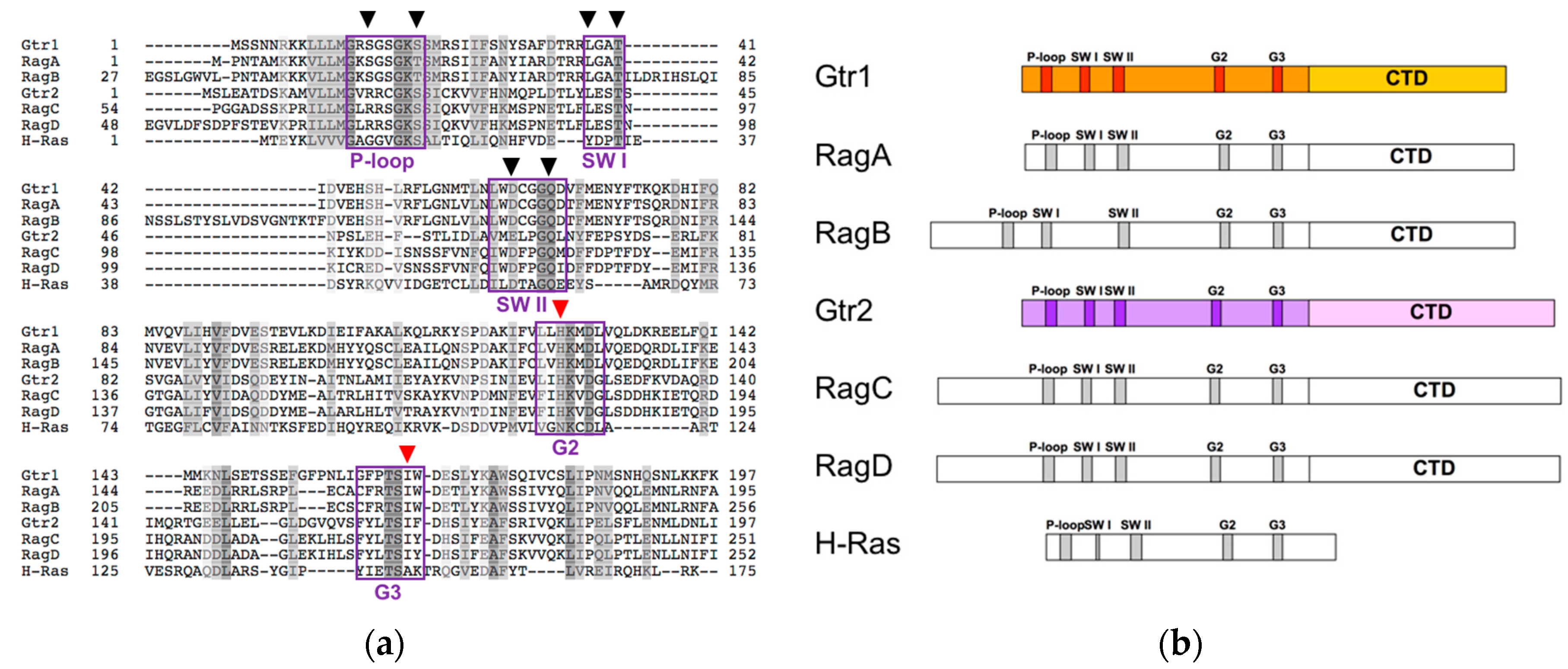
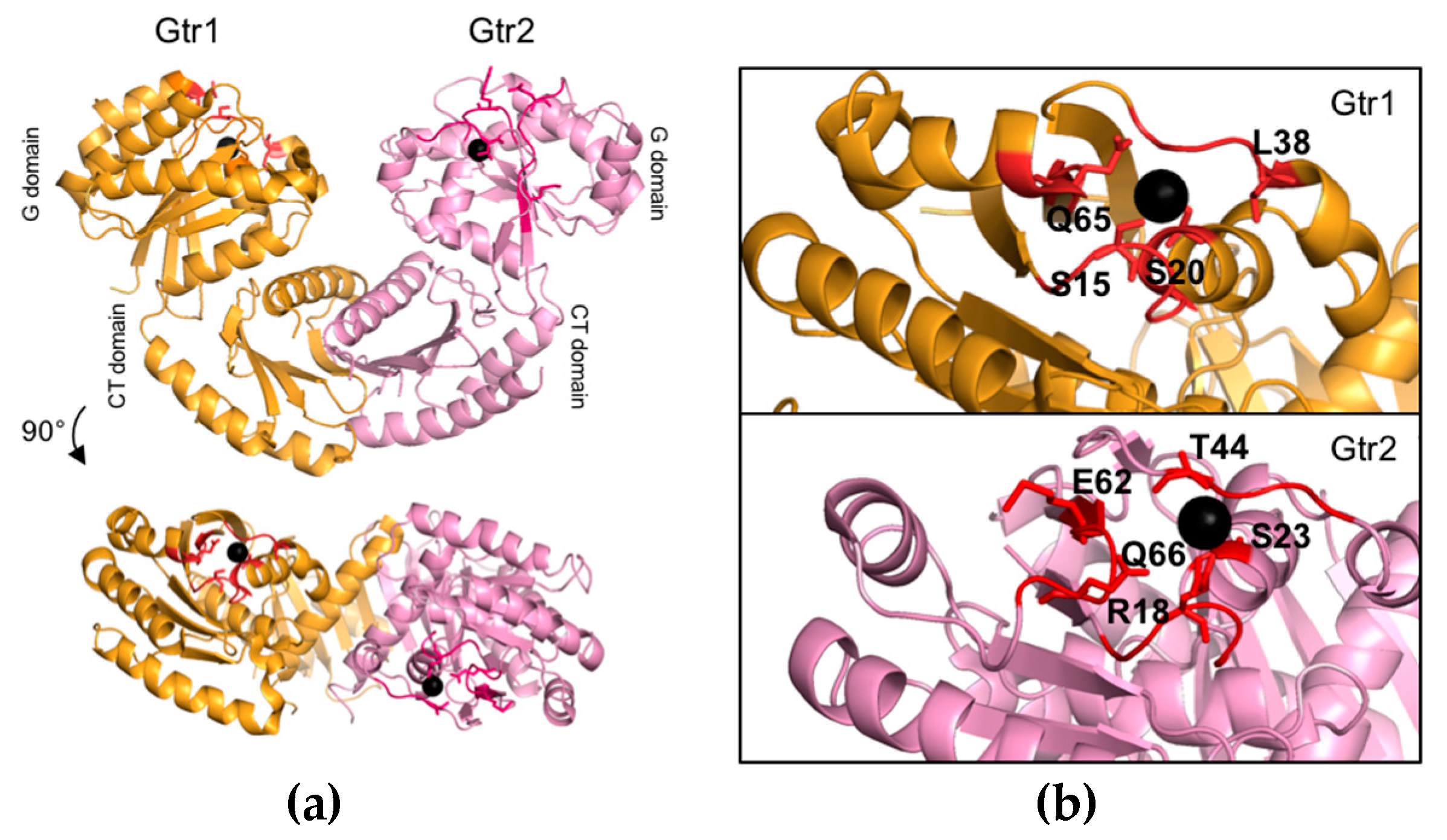
| Protein | Residue | Function | Orthologous Site in RagA/RagB |
| Gtr1 | S15 | Possibly impairs GTP hydrolysis | S16/S49 |
| Gtr1 | S20 | Mg2+ coordination | T21/T54 |
| Gtr1 | L38 | Does not favor GTP hydrolysis | L39/L72 |
| Gtr1 | Q65 | Stabilizes the transition state | Q66/Q127 |
| Gtr1 | L207 | Dimerization | L205/L266 |
| Protein | Residue | Function | Orthologous Site in RagC/RagD |
| Gtr2 | R18 | Stabilizes the transition state | R70/R71 |
| Gtr2 | S23 | Mg2+ coordination | S75/S76 |
| Gtr2 | T44 | Mg2+ coordination | T96/T97 |
| Gtr2 | E62 | Mg2+ coordination | D116/D117 |
| Gtr2 | Q66 | Stabilizes the transition state | Q120/Q121 |
| Gtr2 | L207 | Dimerization | L261/L262 |
| Gtr2 | I214 | G domain stabilization | L268/L269 |
| Protein | Residue(s) | Function | Orthologous Site(s) in Gtr1 |
| RagA | R24/S25/N30/Y31 | Interaction with Raptor | R23/S24/N29/Y30 |
| RagA | R34/D35/R37/R38 | Interaction with Raptor | -/D34/R36/R37 |
| RagA | H47/H49/R51 | Interaction with Raptor | H46/H48/R50 |
| RagA | N55/V57/N59/W61 | Interaction with Raptor | N54/-/N58/W60 |
| RagA | K230 | Ubiquitinated, regulates interaction with GATOR1 | K245 |
| RagA | K244 | Ubiquitinated, regulates interaction with GATOR1 | K259 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nicastro, R.; Sardu, A.; Panchaud, N.; De Virgilio, C. The Architecture of the Rag GTPase Signaling Network. Biomolecules 2017, 7, 48. https://doi.org/10.3390/biom7030048
Nicastro R, Sardu A, Panchaud N, De Virgilio C. The Architecture of the Rag GTPase Signaling Network. Biomolecules. 2017; 7(3):48. https://doi.org/10.3390/biom7030048
Chicago/Turabian StyleNicastro, Raffaele, Alessandro Sardu, Nicolas Panchaud, and Claudio De Virgilio. 2017. "The Architecture of the Rag GTPase Signaling Network" Biomolecules 7, no. 3: 48. https://doi.org/10.3390/biom7030048
APA StyleNicastro, R., Sardu, A., Panchaud, N., & De Virgilio, C. (2017). The Architecture of the Rag GTPase Signaling Network. Biomolecules, 7(3), 48. https://doi.org/10.3390/biom7030048





