Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus
Abstract
:1. Introduction
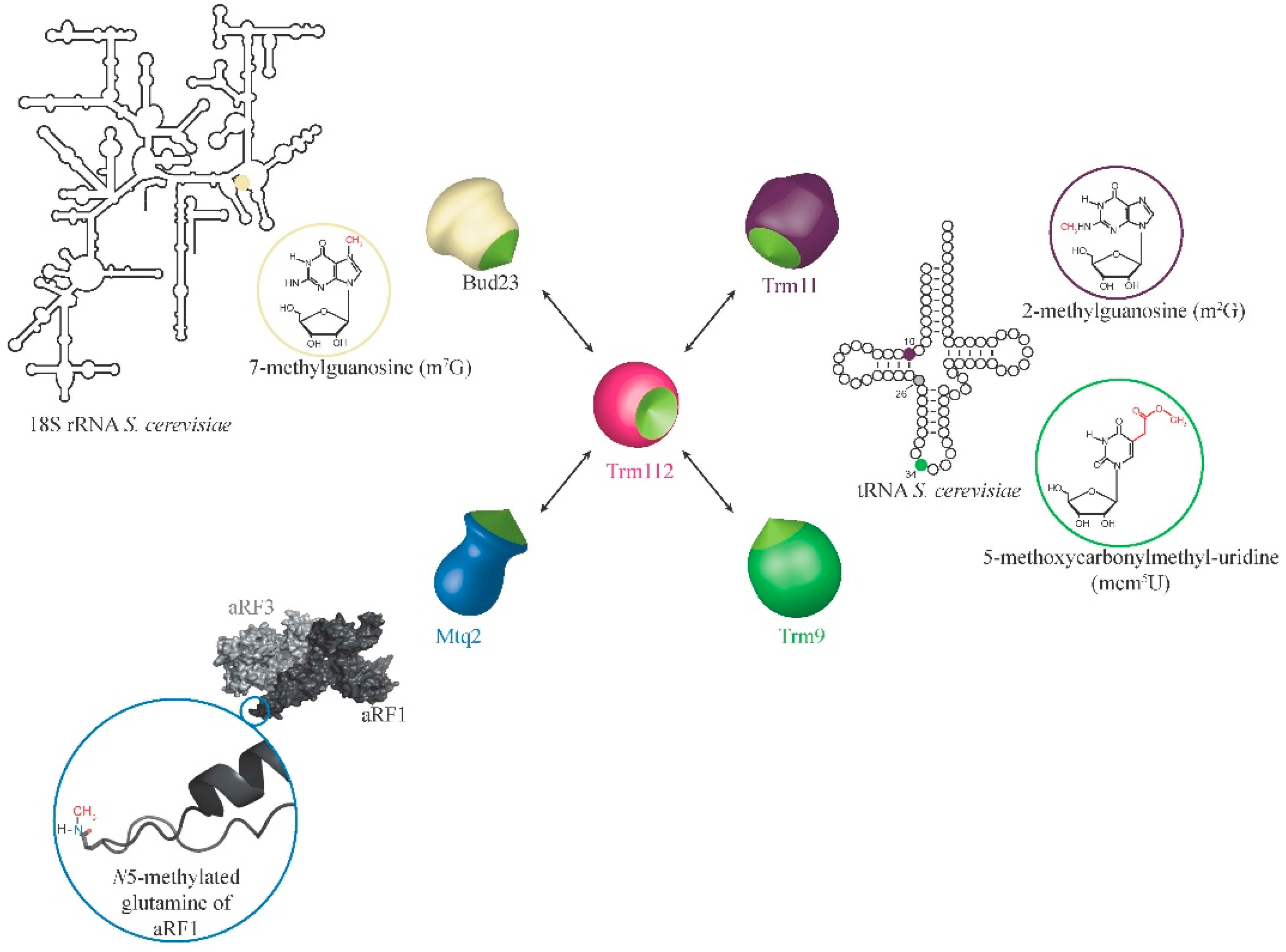
2. Eukaryotic Trm112 Network
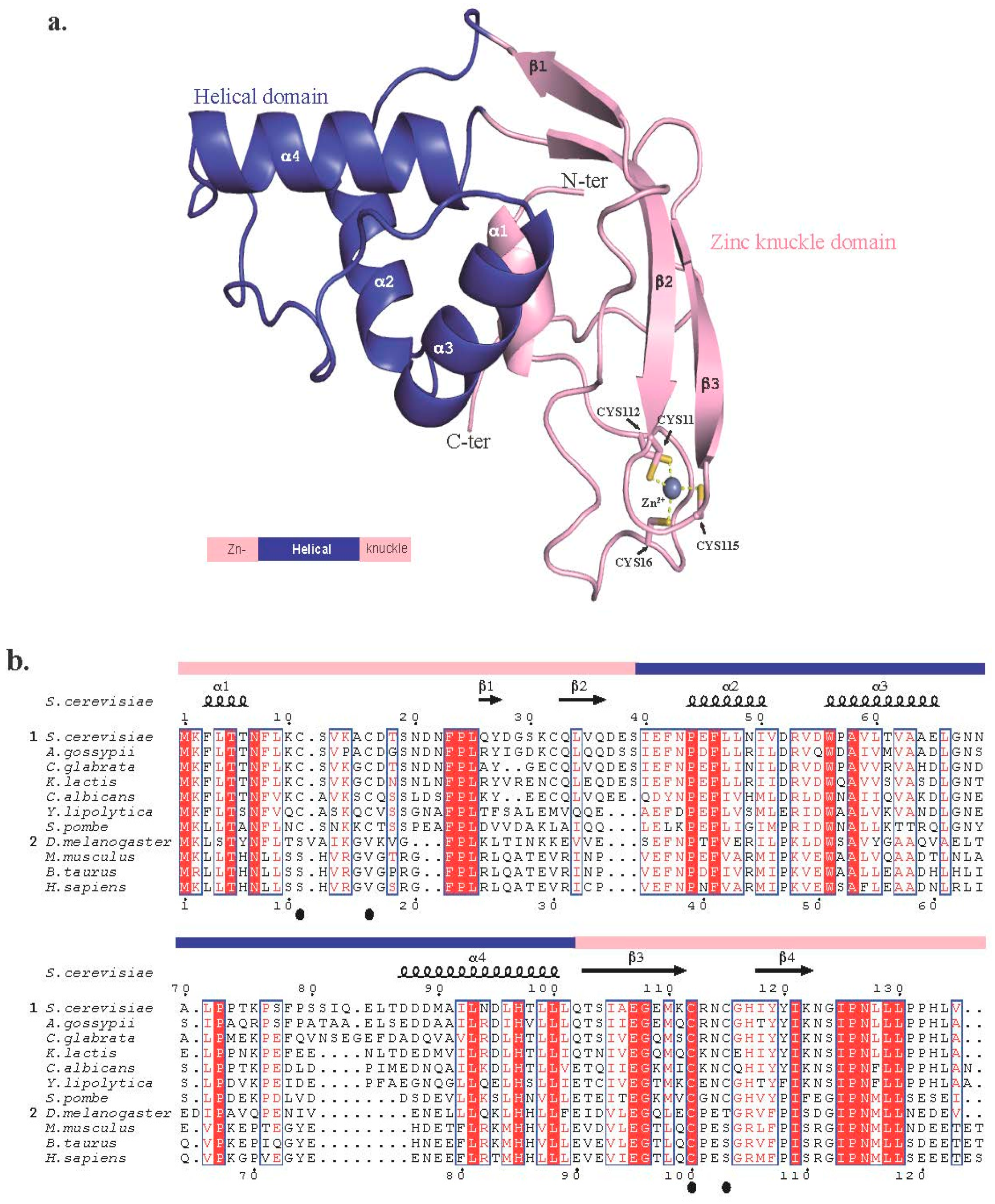
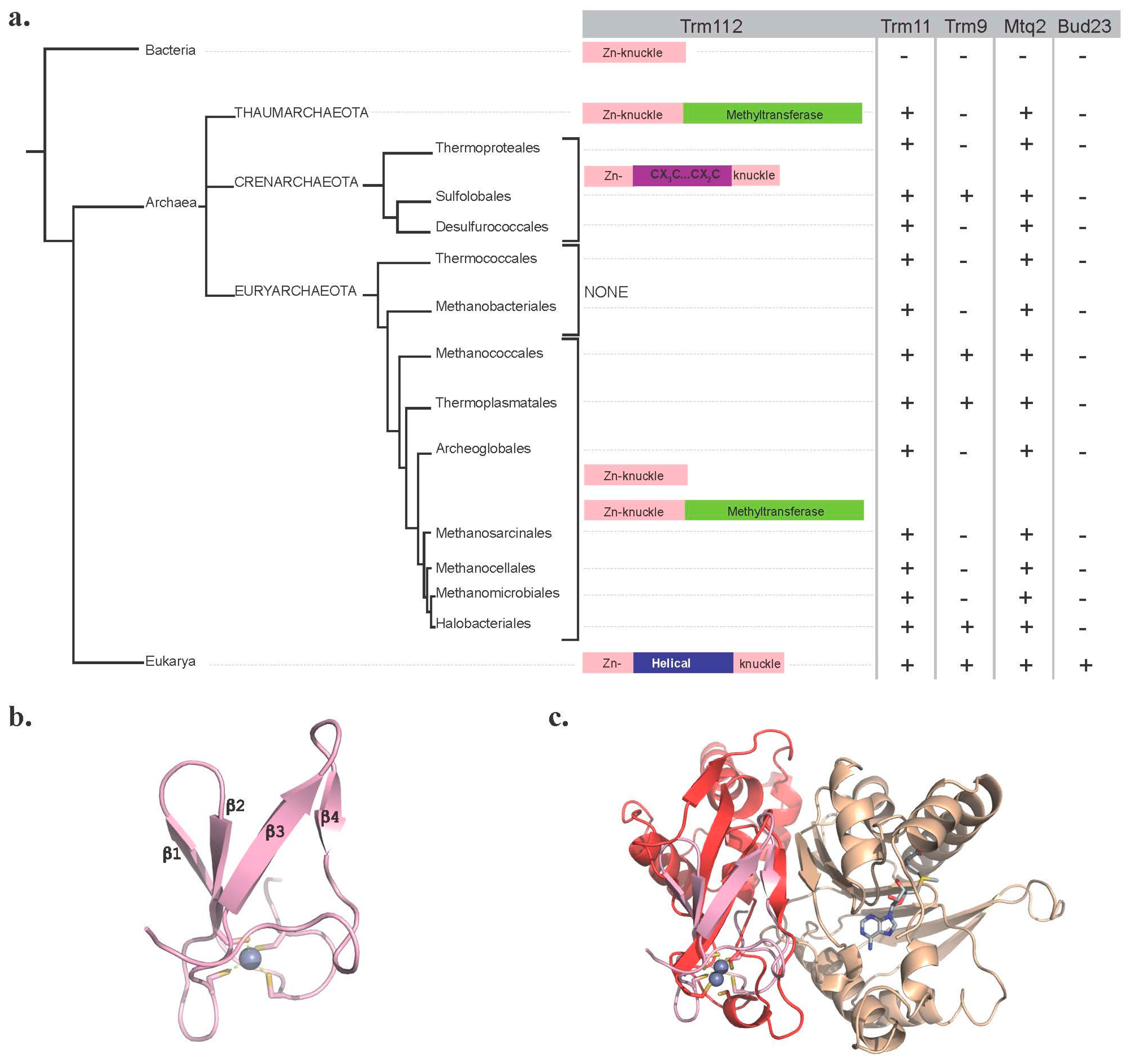
2.1. Trm112
2.2. Role of Trm112 in tRNA Modification
2.2.1. Trm11-Trm112
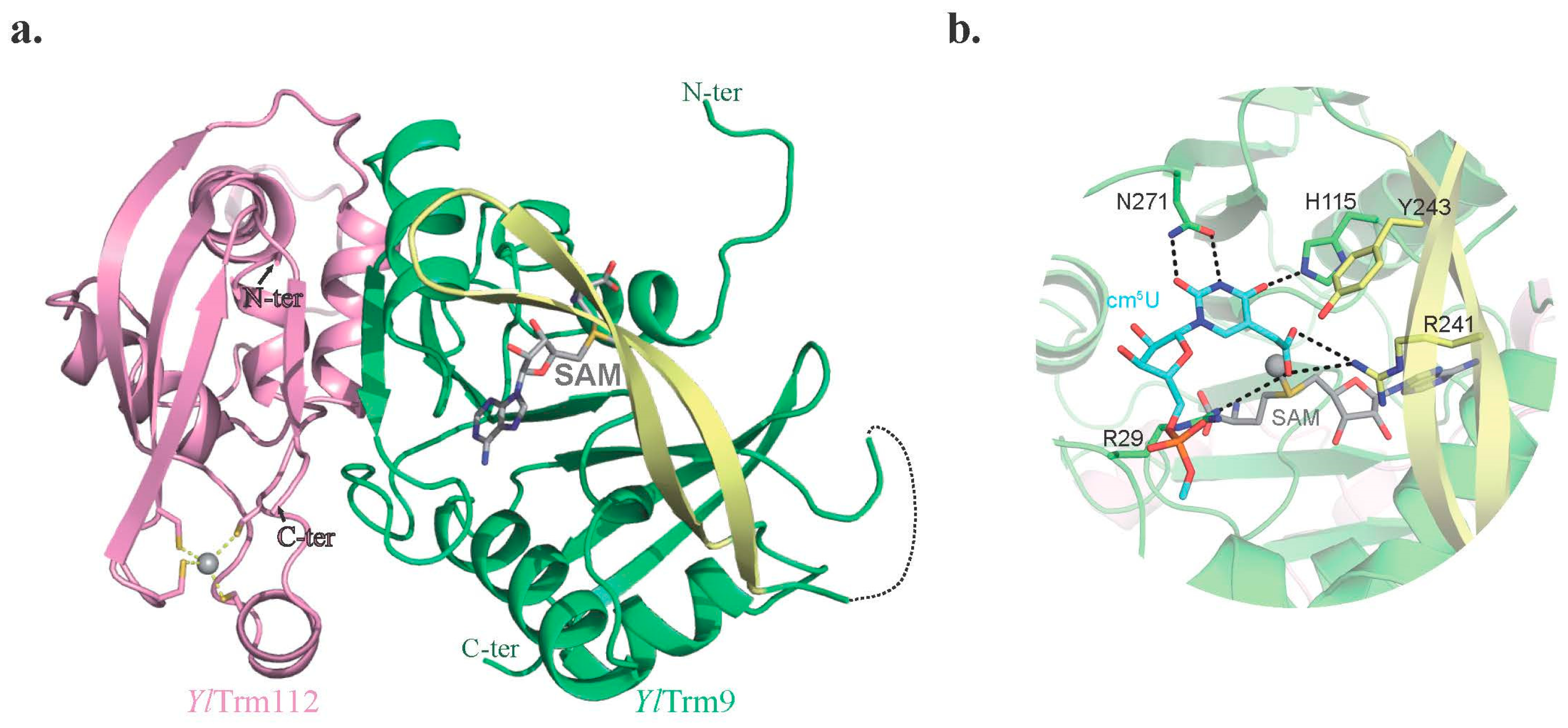
2.2.2. Trm9-Trm112
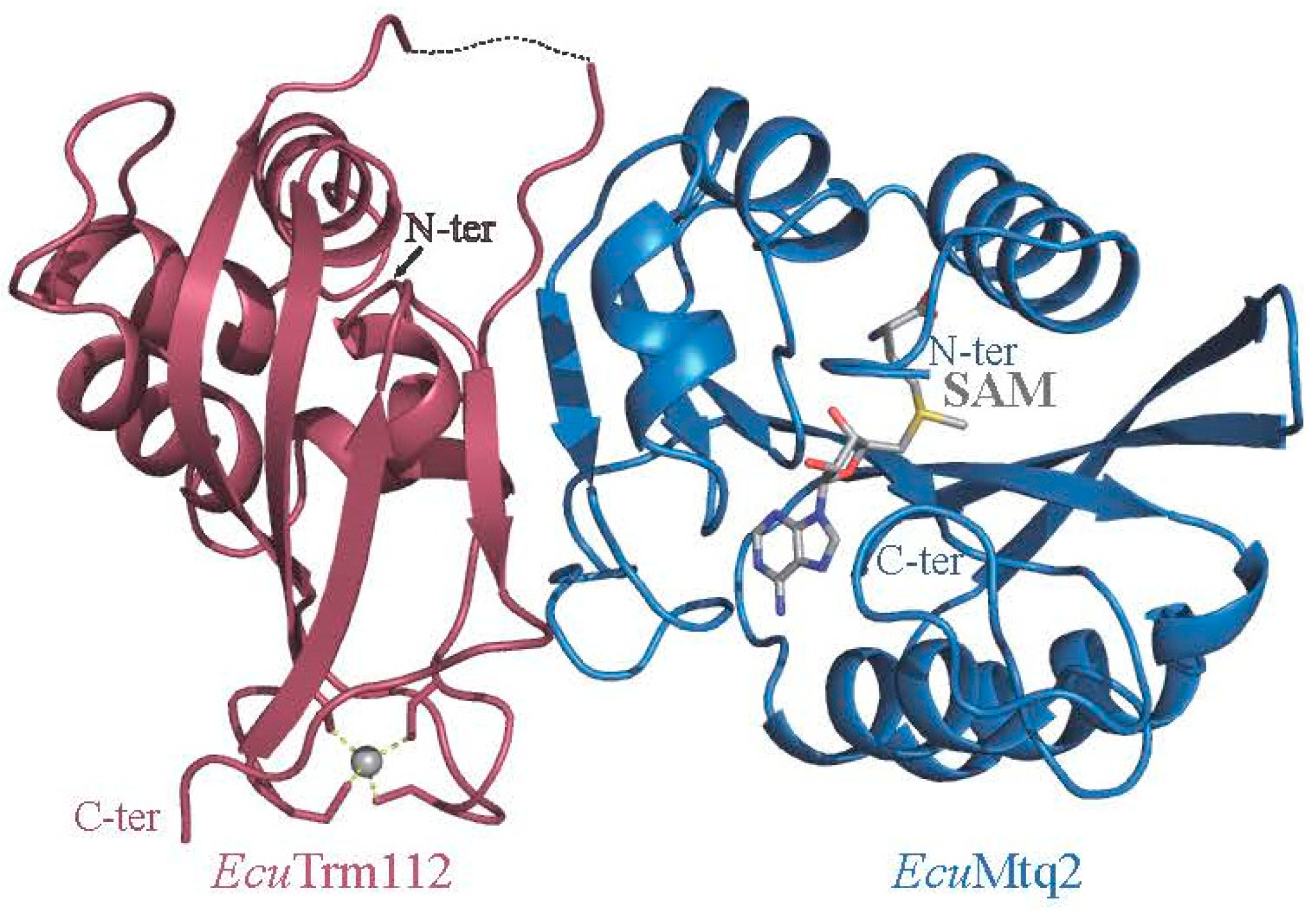
2.3. Role of Trm112 in Translation Termination: Mtq2-Trm112
2.4. Role of Trm112 in Ribosome Biogenesis
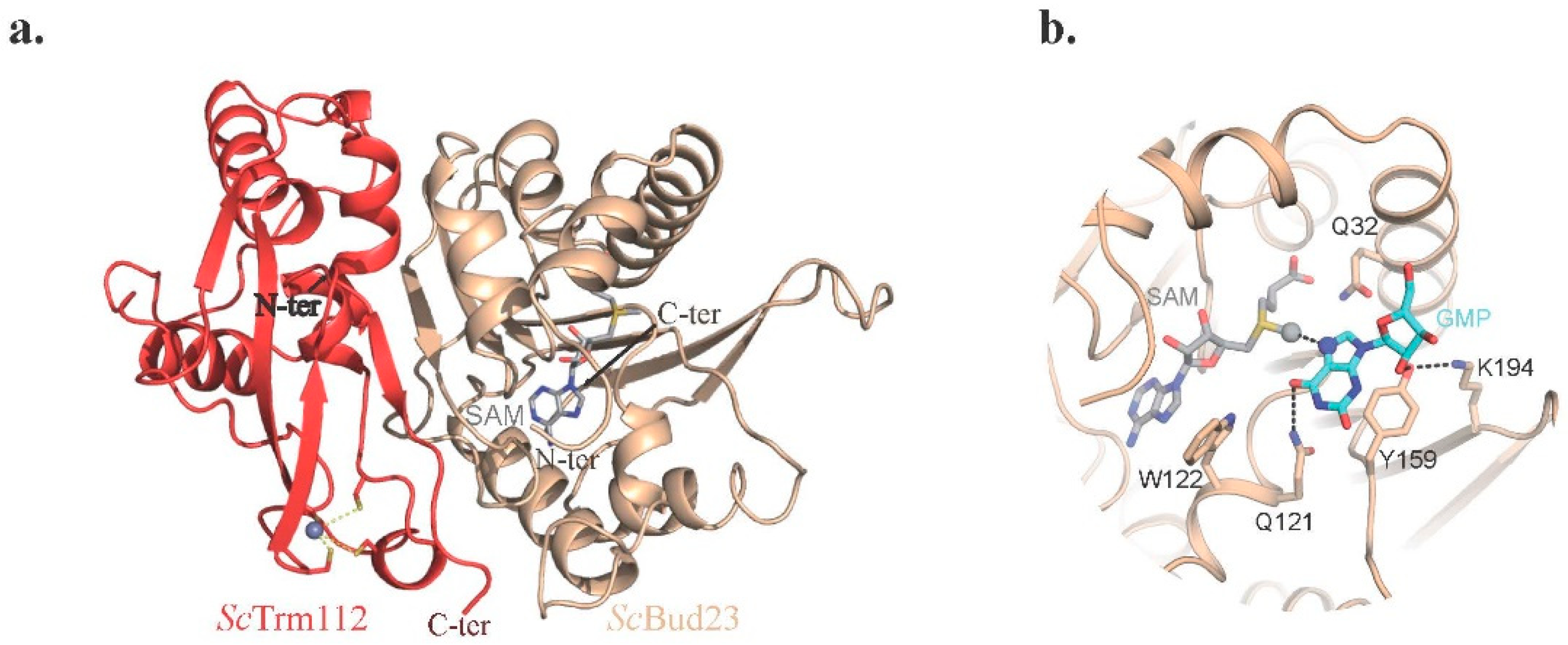
2.4.1. The Bud23-Trm112 Complex Is Involved in 40S Maturation
2.4.2. Trm112 also Influences 60S Formation
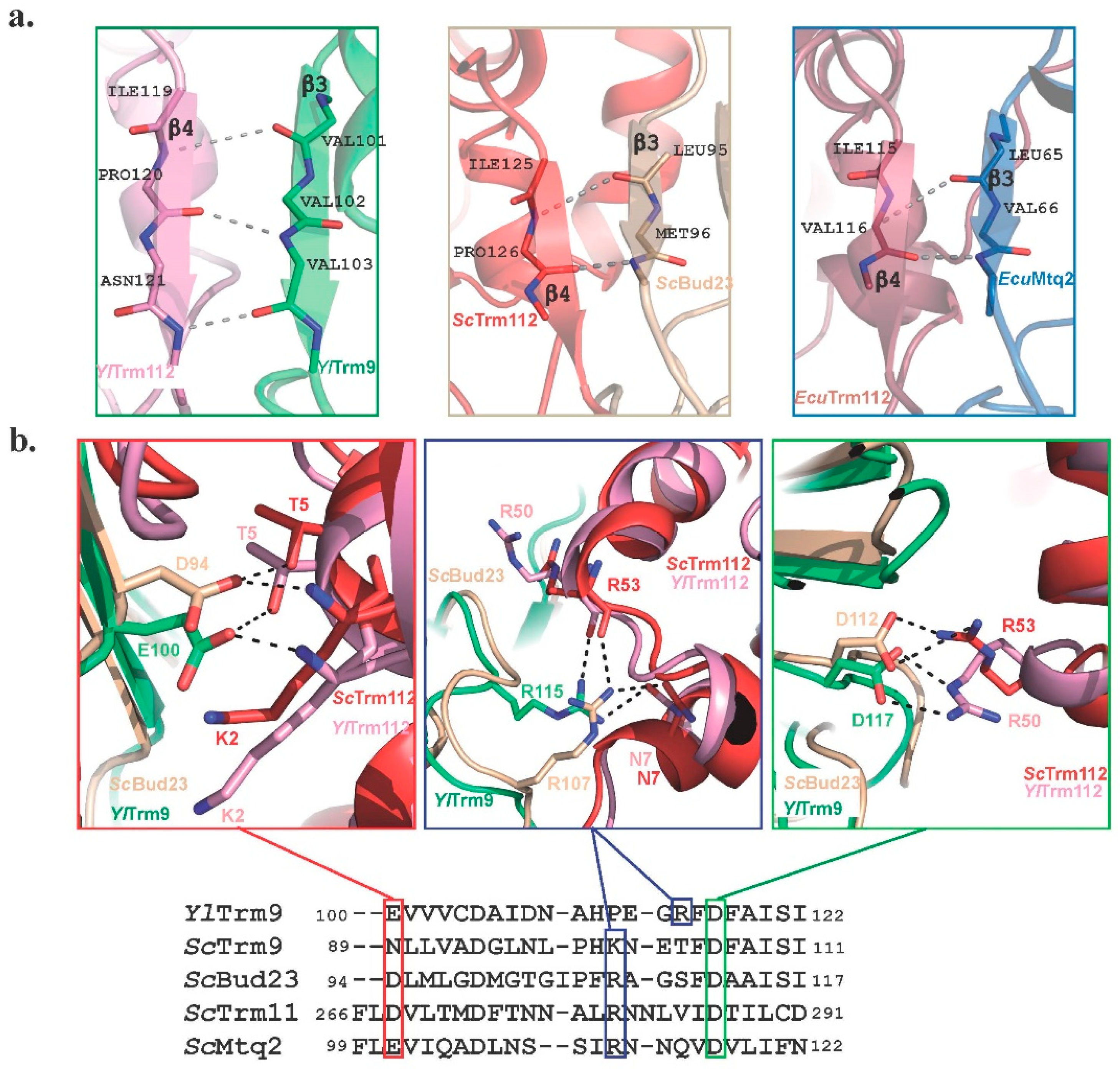
2.5. Common Themes in Recognition and Activation of These MTases by Trm112
3. Trm112 in Prokaryotes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef] [PubMed]
- McKenney, K.M.; Alfonzo, J.D. From Prebiotics to Probiotics: The Evolution and Functions of tRNA Modifications. Life 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Lafontaine, D.L. ‘View From A Bridge’: A New Perspective on Eukaryotic rRNA Base Modification. Trends Biochem. Sci. 2015, 40, 560–575. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m(6)A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Jaffrey, S.R. The dynamic epitranscriptome: N6-methyladenosine and gene expression control. Nat. Rev. Mol. Cell Biol. 2014, 15, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Nachtergaele, S.; Moshitch-Moshkovitz, S.; Peer, E.; Kol, N.; Ben-Haim, M.S.; Dai, Q.; Di Segni, A.; Salmon-Divon, M.; Clark, W.C.; et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016, 530, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, S.; Bernstein, D.A.; Mumbach, M.R.; Jovanovic, M.; Herbst, R.H.; Leon-Ricardo, B.X.; Engreitz, J.M.; Guttman, M.; Satija, R.; Lander, E.S.; et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014, 159, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Delatte, B.; Wang, F.; Ngoc, L.V.; Collignon, E.; Bonvin, E.; Deplus, R.; Calonne, E.; Hassabi, B.; Putmans, P.; Awe, S.; et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016, 351, 282–285. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Bell, T.A.; Schaening, C. Messenger RNA modifications: Form, distribution, and function. Science 2016, 352, 1408–1412. [Google Scholar] [CrossRef] [PubMed]
- Polevoda, B.; Sherman, F. Methylation of proteins involved in translation. Mol. Microbiol. 2007, 65, 590–606. [Google Scholar] [CrossRef] [PubMed]
- Giaever, G.; Chu, A.M.; Ni, L.; Connelly, C.; Riles, L.; Veronneau, S.; Dow, S.; Lucau-Danila, A.; Anderson, K.; Andre, B.; et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature 2002, 418, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Purushothaman, S.K.; Bujnicki, J.M.; Grosjean, H.; Lapeyre, B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell Biol. 2005, 25, 4359–4370. [Google Scholar] [CrossRef] [PubMed]
- Mazauric, M.H.; Dirick, L.; Purushothaman, S.K.; Bjork, G.R.; Lapeyre, B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J. Biol. Chem. 2010, 285, 18505–18515. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Huang, B.; Anderson, J.T.; Bystrom, A.S. Unexpected accumulation of ncm5U and ncm5S2 (U) in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5S2U. PLoS ONE 2011, 6, e20783. [Google Scholar]
- Figaro, S.; Wacheul, L.; Schillewaert, S.; Graille, M.; Huvelle, E.; Mongeard, R.; Zorbas, C.; Lafontaine, D.L.; Heurgue-Hamard, V. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol. Cell Biol. 2012, 32, 2254–2267. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Qin, Z.; Wang, M.; Xu, C.; Feng, G.; Liu, J.; Meng, Z.; Hu, Y. The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J. 2010, 61, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Gu, T.; He, H.; Zhang, Y.; Han, Z.; Hou, G.; Zeng, T.; Liu, Q.; Wu, Q. Trmt112 gene expression in mouse embryonic development. Acta Histochem. Cytochem. 2012, 45, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Heurgue-Hamard, V.; Graille, M.; Scrima, N.; Ulryck, N.; Champ, S.; Van Tilbeurgh, H.; Buckingham, R.H. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 2006, 281, 36140–36148. [Google Scholar] [CrossRef] [PubMed]
- Liger, D.; Mora, L.; Lazar, N.; Figaro, S.; Henri, J.; Scrima, N.; Buckingham, R.H.; Van Tilbeurgh, H.; Heurgue-Hamard, V.; Graille, M. Mechanism of activation of methyltransferases involved in translation by the Trm112 ‘hub’ protein. Nucleic Acids Res. 2011, 39, 6249–6259. [Google Scholar] [CrossRef] [PubMed]
- Letoquart, J.; Huvelle, E.; Wacheul, L.; Bourgeois, G.; Zorbas, C.; Graille, M.; Heurgue-Hamard, V.; Lafontaine, D.L. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc. Natl. Acad. Sci. USA 2014, 111, E5518–E5526. [Google Scholar] [CrossRef] [PubMed]
- Letoquart, J.; Tran, N.V.; Caroline, V.; Aleksandrov, A.; Lazar, N.; van Tilbeurgh, H.; Liger, D.; Graille, M. Insights into molecular plasticity in protein complexes from Trm9-Trm112 tRNA modifying enzyme crystal structure. Nucleic Acids Res. 2015, 43, 10989–11002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Halbach, F.; Rode, M.; Conti, E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA 2012, 18, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Cantara, W.A.; Crain, P.F.; Rozenski, J.; McCloskey, J.A.; Harris, K.A.; Zhang, X.; Vendeix, F.A.; Fabris, D.; Agris, P.F. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Res. 2011, 39, D195–D201. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Saito, K.; Ishitani, R.; Ito, K.; Nureki, O. Structural basis for translation termination by archaeal RF1 and GTP-bound EF1alpha complex. Nucleic Acids Res. 2012, 40, 9319–9328. [Google Scholar] [CrossRef] [PubMed]
- Aravind, L.; Koonin, E.V. THUMP—A predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001, 26, 215–217. [Google Scholar] [CrossRef]
- Bujnicki, J.M.; Droogmans, L.; Grosjean, H.; Purushothaman, S.K.; Lapeyre, B. Bioinformatics-guided identification and experimental characterization of novel RNA methyltransferases. In Practical Bioinformatics; Bujnicki, J.M., Ed.; Springer: Heidelberg, Germany, 2004; Volume 15, pp. 139–168. [Google Scholar]
- Hirata, A.; Nishiyama, S.; Tamura, T.; Yamauchi, A.; Hori, H. Structural and functional analyses of the archaeal tRNA m2G/m22G10 methyltransferase aTrm11 provide mechanistic insights into site specificity of a tRNA methyltransferase that contains common RNA-binding modules. Nucleic Acids Res. 2016, 44, 6377–6390. [Google Scholar] [CrossRef] [PubMed]
- Menezes, S.; Gaston, K.W.; Krivos, K.L.; Apolinario, E.E.; Reich, N.O.; Sowers, K.R.; Limbach, P.A.; Perona, J.J. Formation of m2G6 in Methanocaldococcus jannaschii tRNA catalyzed by the novel methyltransferase Trm14. Nucleic Acids Res. 2011, 39, 7641–7655. [Google Scholar] [CrossRef] [PubMed]
- Fislage, M.; Roovers, M.; Tuszynska, I.; Bujnicki, J.M.; Droogmans, L.; Versees, W. Crystal structures of the tRNA: m2G6 methyltransferase Trm14/TrmN from two domains of life. Nucleic Acids Res. 2012, 40, 5149–5161. [Google Scholar] [CrossRef] [PubMed]
- Neumann, P.; Lakomek, K.; Naumann, P.T.; Erwin, W.M.; Lauhon, C.T.; Ficner, R. Crystal structure of a 4-thiouridine synthetase-RNA complex reveals specificity of tRNA U8 modification. Nucleic Acids Res. 2014, 42, 6673–6685. [Google Scholar] [CrossRef] [PubMed]
- Armengaud, J.; Urbonavicius, J.; Fernandez, B.; Chaussinand, G.; Bujnicki, J.M.; Grosjean, H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J. Biol. Chem. 2004, 279, 37142–37152. [Google Scholar] [CrossRef] [PubMed]
- Okada, K.; Muneyoshi, Y.; Endo, Y.; Hori, H. Production of yeast (m2G10) methyltransferase (Trm11 and Trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp. Ser. (Oxf.) 2009, 53, 303–304. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, G.; Marcoux, J.; Saliou, J.M.; Cianférani, S.; Graille, M. Activation mode of the eukaryotic m2G10 tRNA methyltransferase Trm11 by its partner protein Trm112. Nucleic Acids Res. 2017, in press. [Google Scholar]
- Kohli, M.; Riska, S.M.; Mahoney, D.W.; Chai, H.S.; Hillman, D.W.; Rider, D.N.; Costello, B.A.; Qin, R.; Lamba, J.; Sahasrabudhe, D.M.; et al. Germline predictors of androgen deprivation therapy response in advanced prostate cancer. Mayo Clin. Proc. 2012, 87, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.P.; Ding, Y.; Chen, Z.; Liu, S.; Michalopoulos, A.; Chen, R.; Gulzar, Z.G.; Yang, B.; Cieply, K.M.; Luvison, A.; et al. Novel fusion transcripts associate with progressive prostate cancer. Am. J. Pathol. 2014, 184, 2840–2849. [Google Scholar] [CrossRef] [PubMed]
- Lunardi, A.; Di Minin, G.; Provero, P.; Dal Ferro, M.; Carotti, M.; Del Sal, G.; Collavin, L. A genome-scale protein interaction profile of Drosophila p53 uncovers additional nodes of the human p53 network. Proc. Natl. Acad. Sci. USA 2010, 107, 6322–6327. [Google Scholar] [CrossRef] [PubMed]
- Gavin, A.C.; Bosche, M.; Krause, R.; Grandi, P.; Marzioch, M.; Bauer, A.; Schultz, J.; Rick, J.M.; Michon, A.M.; Cruciat, C.M.; et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 2002, 415, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Kalhor, H.R.; Clarke, S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell Biol. 2003, 23, 9283–9292. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Lu, J.; Bystrom, A.S. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008, 14, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Letoquart, J.; Faux, C.; Taylor, N.M.; Seraphin, B.; Muller, C.W. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat. Struct. Mol. Biol. 2012, 19, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Glatt, S.; Zabel, R.; Kolaj-Robin, O.; Onuma, O.F.; Baudin, F.; Graziadei, A.; Taverniti, V.; Lin, T.-Y.; Baymann, F.; Seraphin, B.; et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat. Struct. Mol. Biol. 2016, 23, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Noma, A.; Sakaguchi, Y.; Suzuki, T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009, 37, 1335–1352. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Huang, B.; Esberg, A.; Johansson, M.J.; Bystrom, A.S. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 2005, 11, 1648–1654. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Dyavaiah, M.; Patil, A.; Rooney, J.P.; DiRenzo, D.; Young, C.M.; Conklin, D.S.; Zitomer, R.S.; Begley, T.J. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 2007, 28, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Babu, I.R.; Su, D.; Yin, S.; Begley, T.J.; Dedon, P.C. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet. 2015, 11, e1005706. [Google Scholar] [CrossRef] [PubMed]
- Patil, A.; Chan, C.T.; Dyavaiah, M.; Rooney, J.P.; Dedon, P.C.; Begley, T.J. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol. 2012, 9, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Brophy, J.A.; Chan, C.T.; Atmore, K.A.; Begley, U.; Paules, R.S.; Dedon, P.C.; Begley, T.J.; Samson, L.D. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol. Cell Biol. 2010, 30, 2449–2459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, Y.; Dai, Q.; Zhang, W.; Ren, J.; Pan, T.; He, C. The AlkB domain of mammalian ABH8 catalyzes hydroxylation of 5-methoxycarbonylmethyluridine at the wobble position of tRNA. Angew. Chem. Int. Ed. Engl. 2010, 49, 8885–8888. [Google Scholar] [CrossRef] [PubMed]
- Pastore, C.; Topalidou, I.; Forouhar, F.; Yan, A.C.; Levy, M.; Hunt, J.F. Crystal structure and RNA binding properties of the RNA recognition motif (RRM) and AlkB domains in human AlkB homolog 8 (ABH8), an enzyme catalyzing tRNA hypermodification. J. Biol. Chem. 2012, 287, 2130–2143. [Google Scholar] [CrossRef] [PubMed]
- Shimada, K.; Nakamura, M.; Anai, S.; De Velasco, M.; Tanaka, M.; Tsujikawa, K.; Ouji, Y.; Konishi, N. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 2009, 69, 3157–3164. [Google Scholar] [CrossRef] [PubMed]
- Begley, U.; Sosa, M.S.; Avivar-Valderas, A.; Patil, A.; Endres, L.; Estrada, Y.; Chan, C.T.; Su, D.; Dedon, P.C.; Aguirre-Ghiso, J.A.; et al. A human tRNA methyltransferase 9-like protein prevents tumour growth by regulating LIN9 and HIF1-alpha. EMBO Mol. Med. 2013, 5, 366–383. [Google Scholar] [CrossRef] [PubMed]
- Leihne, V.; Kirpekar, F.; Vagbo, C.B.; Van den Born, E.; Krokan, H.E.; Grini, P.E.; Meza, T.J.; Falnes, P.O. Roles of Trm9- and ALKBH8-like proteins in the formation of modified wobble uridines in Arabidopsis tRNA. Nucleic Acids Res. 2011, 39, 7688–7701. [Google Scholar] [CrossRef] [PubMed]
- Klaholz, B.P. Molecular recognition and catalysis in translation termination complexes. Trends Biochem. Sci. 2011, 36, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Shao, S.; Murray, J.; Hegde, R.S.; Ramakrishnan, V. Structural basis for stop codon recognition in eukaryotes. Nature 2015, 524, 493–496. [Google Scholar] [CrossRef] [PubMed]
- Pavlov, M.Y.; Freistroffer, D.V.; Dincbas, V.; MacDougall, J.; Buckingham, R.H.; Ehrenberg, M. A direct estimation of the context effect on the efficiency of termination. J. Mol. Biol. 1998, 284, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Dincbas-Renqvist, V.; Engstrom, A.; Mora, L.; Heurgue-Hamard, V.; Buckingham, R.; Ehrenberg, M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000, 19, 6900–6907. [Google Scholar] [CrossRef] [PubMed]
- Heurgué-Hamard, V.; Champ, S.; Engstöm, Å.; Ehrenberg, M.; Buckingham, R.H. The hemK gene in Escherichia coli encodes the N5-glutamine methyltransferase that modifies peptide release factors. EMBO J. 2002, 21, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Nakahigashi, K.; Kubo, N.; Narita, S.; Shimaoka, T.; Goto, S.; Oshima, T.; Mori, H.; Maeda, M.; Wada, C.; Inokuchi, H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc. Natl. Acad. Sci. USA 2002, 99, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Graille, M.; Heurgue-Hamard, V.; Champ, S.; Mora, L.; Scrima, N.; Ulryck, N.; van Tilbeurgh, H.; Buckingham, R.H. Molecular basis for bacterial class I release factor methylation by PrmC. Mol. Cell 2005, 20, 917–927. [Google Scholar] [CrossRef] [PubMed]
- Mora, L.; Heurgue-Hamard, V.; de Zamaroczy, M.; Kervestin, S.; Buckingham, R.H. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 2007, 282, 35638–35645. [Google Scholar] [CrossRef] [PubMed]
- Heurgue-Hamard, V.; Champ, S.; Mora, L.; Merkulova-Rainon, T.; Kisselev, L.L.; Buckingham, R.H. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J. Biol. Chem. 2005, 280, 2439–2445. [Google Scholar] [CrossRef] [PubMed]
- Polevoda, B.; Span, L.; Sherman, F. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J. Biol. Chem. 2006, 281, 2562–2571. [Google Scholar] [CrossRef] [PubMed]
- Figaro, S.; Scrima, N.; Buckingham, R.H.; Heurgue-Hamard, V. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 2008, 582, 2352–2356. [Google Scholar] [CrossRef] [PubMed]
- Nie, D.S.; Liu, Y.B.; Lu, G.X. Cloning and primarily function study of two novel putative N5-glutamine methyltransferase (Hemk) splice variants from mouse stem cells. Mol. Biol. Rep. 2009, 36, 2221–2228. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Nie, S.; Li, B.; Yang, Z.Q.; Xu, Z.M.; Fei, J.; Lin, C.; Zeng, R.; Xu, G.L. Deficiency in a glutamine-specific methyltransferase for the release factor causes mouse embryonic lethality. Mol. Cell Biol. 2010, 30, 4245–4253. [Google Scholar] [CrossRef] [PubMed]
- Kusevic, D.; Kudithipudi, S.; Jeltsch, A. Substrate Specificity of the HEMK2 Protein Glutamine Methyltransferase and Identification of Novel Substrates. J. Biol. Chem. 2016, 291, 6124–6133. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Johnson, A.W. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol. Biol. Cell 2012, 23, 4313–4322. [Google Scholar] [CrossRef] [PubMed]
- White, J.; Li, Z.; Sardana, R.; Bujnicki, J.M.; Marcotte, E.M.; Johnson, A.W. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell Biol. 2008, 28, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; White, J.P.; Johnson, A.W. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA 2013, 19, 828–840. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, A.A.; McCarthy, J.G. The structure of two N-methyltransferases from the caffeine biosynthetic pathway. Plant Physiol. 2007, 144, 879–889. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Zhu, J.; Gill, M.; Johnson, A.W. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol. Cell Biol. 2014, 34, 2208–2220. [Google Scholar] [CrossRef] [PubMed]
- Sardana, R.; Liu, X.; Granneman, S.; Zhu, J.; Gill, M.; Papoulas, O.; Marcotte, E.M.; Tollervey, D.; Correll, C.C.; Johnson, A.W. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol. 2015, 13, e1002083. [Google Scholar] [CrossRef] [PubMed]
- Ounap, K.; Leetsi, L.; Matsoo, M.; Kurg, R. The Stability of Ribosome Biogenesis Factor WBSCR22 Is Regulated by Interaction with TRMT112 via Ubiquitin-Proteasome Pathway. PLoS ONE 2015, 10, e0133841. [Google Scholar] [CrossRef] [PubMed]
- Zorbas, C.; Nicolas, E.; Wacheul, L.; Huvelle, E.; Heurgue-Hamard, V.; Lafontaine, D.L. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol. Biol. Cell 2015, 26, 2080–2095. [Google Scholar] [CrossRef] [PubMed]
- Ounap, K.; Kasper, L.; Kurg, A.; Kurg, R. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS ONE 2013, 8, e75686. [Google Scholar] [CrossRef] [PubMed]
- Doll, A.; Grzeschik, K.H. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet. Cell Genet. 2001, 95, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Merla, G.; Ucla, C.; Guipponi, M.; Reymond, A. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum. Genet. 2002, 110, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, Y.; Arai, H.; Fujita, N. The novel metastasis promoter Merm1/Wbscr22 enhances tumor cell survival in the vasculature by suppressing Zac1/p53-dependent apoptosis. Cancer Res. 2011, 71, 1146–1155. [Google Scholar] [CrossRef] [PubMed]
- Tiedemann, R.E.; Zhu, Y.X.; Schmidt, J.; Shi, C.X.; Sereduk, C.; Yin, H.; Mousses, S.; Stewart, A.K. Identification of molecular vulnerabilities in human multiple myeloma cells by RNA interference lethality screening of the druggable genome. Cancer Res. 2012, 72, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Stefanska, B.; Cheishvili, D.; Suderman, M.; Arakelian, A.; Huang, J.; Hallett, M.; Han, Z.G.; Al-Mahtab, M.; Akbar, S.M.; Khan, W.A.; et al. Genome-wide study of hypomethylated and induced genes in patients with liver cancer unravels novel anticancer targets. Clin. Cancer Res. 2014, 20, 3118–3132. [Google Scholar] [CrossRef] [PubMed]
- Jangani, M.; Poolman, T.M.; Matthews, L.; Yang, N.; Farrow, S.N.; Berry, A.; Hanley, N.; Williamson, A.J.; Whetton, A.D.; Donn, R.; et al. The Methyltransferase WBSCR22/Merm1 Enhances Glucocorticoid Receptor Function and is Regulated in Lung Inflammation and Cancer. J. Biol. Chem. 2014, 289, 8931–8946. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Yang, J.; Watzinger, P.; Kotter, P.; Entian, K.D. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013, 41, 9062–9076. [Google Scholar] [CrossRef] [PubMed]
- Bourgeois, G.; Ney, M.; Gaspar, I.; Aigueperse, C.; Schaefer, M.; Kellner, S.; Helm, M.; Motorin, Y. Eukaryotic rRNA Modification by Yeast 5-Methylcytosine-Methyltransferases and Human Proliferation-Associated Antigen p120. PLoS ONE 2015, 10, e0133321. [Google Scholar] [CrossRef] [PubMed]
- Studte, P.; Zink, S.; Jablonowski, D.; Bar, C.; Von der Haar, T.; Tuite, M.F.; Schaffrath, R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol. 2008, 69, 1266–1277. [Google Scholar] [CrossRef] [PubMed]
- Armengod, M.E.; Moukadiri, I.; Prado, S.; Ruiz-Partida, R.; Benitez-Paez, A.; Villarroya, M.; Lomas, R.; Garzon, M.J.; Martinez-Zamora, A.; Meseguer, S.; et al. Enzymology of tRNA modification in the bacterial MnmEG pathway. Biochimie 2012, 94, 1510–1520. [Google Scholar] [CrossRef] [PubMed]
- Isaksson, L.A.; Phillips, J.H. Studies on microbial RNA. V. A comparison of the in vivo methylated components of ribosomal RNA from Escherichia coli and Saccharomyces cerevisiae. Biochim. Biophys. Acta 1968, 155, 63–71. [Google Scholar] [CrossRef]
- Nishimura, K.; Hosaka, T.; Tokuyama, S.; Okamoto, S.; Ochi, K. Mutations in rsmG, encoding a 16S rRNA methyltransferase, result in low-level streptomycin resistance and antibiotic overproduction in Streptomyces coelicolor A3(2). J. Bacteriol. 2007, 189, 3876–3883. [Google Scholar] [CrossRef] [PubMed]
- Brochier-Armanet, C.; Forterre, P.; Gribaldo, S. Phylogeny and evolution of the Archaea: One hundred genomes later. Curr. Opin. Microbiol. 2011, 14, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Cox, C.J.; Foster, P.G.; Hirt, R.P.; Harris, S.R.; Embley, T.M. The archaebacterial origin of eukaryotes. Proc. Natl. Acad. Sci. USA 2008, 105, 20356–20361. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Mugnier, P.; Das, A.K.; Webb, H.M.; Evans, D.R.; Tuite, M.F.; Hemmings, B.A.; Barford, D. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 2000, 100, 311–321. [Google Scholar] [CrossRef]
- Saito, K.; Kobayashi, K.; Wada, M.; Kikuno, I.; Takusagawa, A.; Mochizuki, M.; Uchiumi, T.; Ishitani, R.; Nureki, O.; Ito, K. Omnipotent role of archaeal elongation factor 1 alpha (EF1alpha in translational elongation and termination, and quality control of protein synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 19242–19247. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984, 259, 9461–9471. [Google Scholar] [PubMed]
- Tomikawa, C.; Ohira, T.; Inoue, Y.; Kawamura, T.; Yamagishi, A.; Suzuki, T.; Hori, H. Distinct tRNA modifications in the thermo-acidophilic archaeon, Thermoplasma acidophilum. FEBS Lett. 2013, 587, 3575–3580. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Gaspin, C.; Marck, C.; Decatur, W.A.; De Crecy-Lagard, V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: Identification of RNA modification genes. BMC Genom. 2008, 9, 470. [Google Scholar] [CrossRef] [PubMed]
- Phillips, G.; de Crécy-Lagard, V. Biosynthesis and function of tRNA modifications in Archaea. Curr. Opin. Microbiol. 2011, 14, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Selvadurai, K.; Wang, P.; Seimetz, J.; Huang, R.H. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat. Chem. Biol. 2014, 10, 810–812. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bourgeois, G.; Létoquart, J.; Van Tran, N.; Graille, M. Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus. Biomolecules 2017, 7, 7. https://doi.org/10.3390/biom7010007
Bourgeois G, Létoquart J, Van Tran N, Graille M. Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus. Biomolecules. 2017; 7(1):7. https://doi.org/10.3390/biom7010007
Chicago/Turabian StyleBourgeois, Gabrielle, Juliette Létoquart, Nhan Van Tran, and Marc Graille. 2017. "Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus" Biomolecules 7, no. 1: 7. https://doi.org/10.3390/biom7010007
APA StyleBourgeois, G., Létoquart, J., Van Tran, N., & Graille, M. (2017). Trm112, a Protein Activator of Methyltransferases Modifying Actors of the Eukaryotic Translational Apparatus. Biomolecules, 7(1), 7. https://doi.org/10.3390/biom7010007




