Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide
Abstract
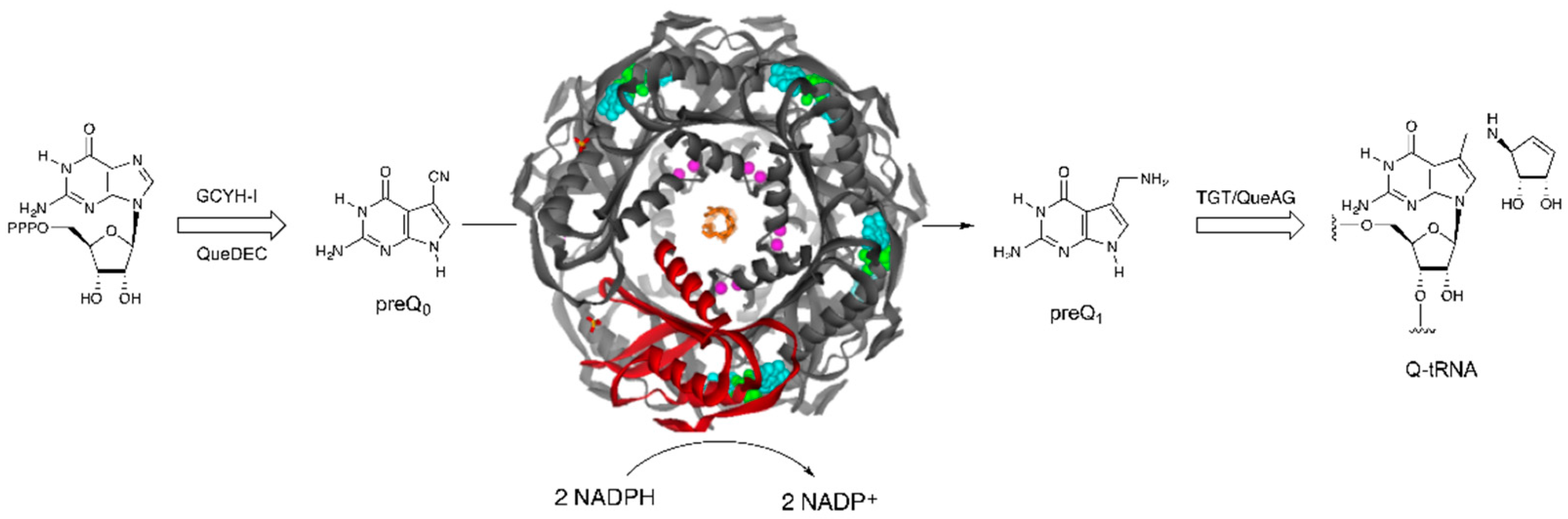
:1. Introduction
2. Results and Discussion
2.1. Activity of B. subtilis QueF Mutants
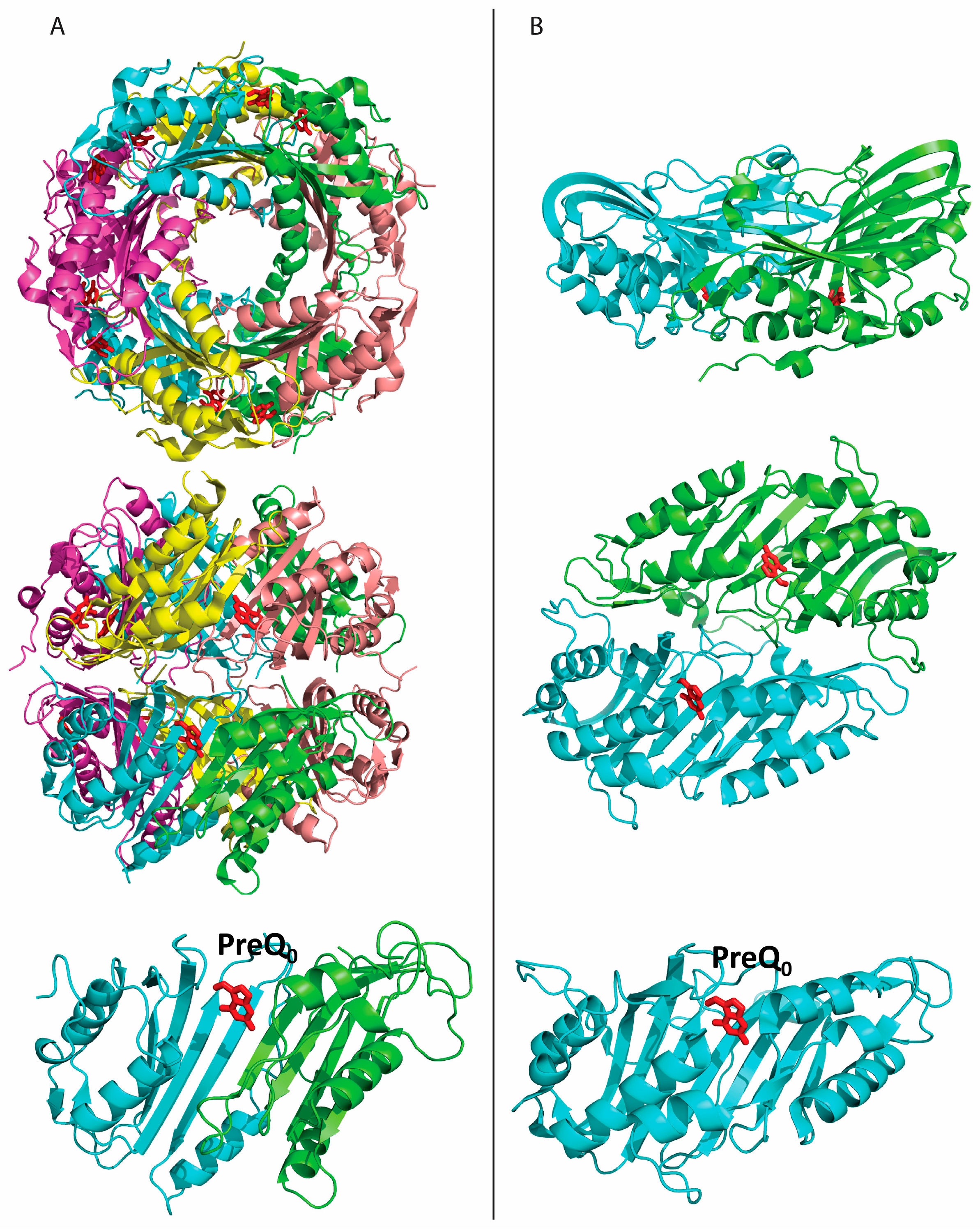
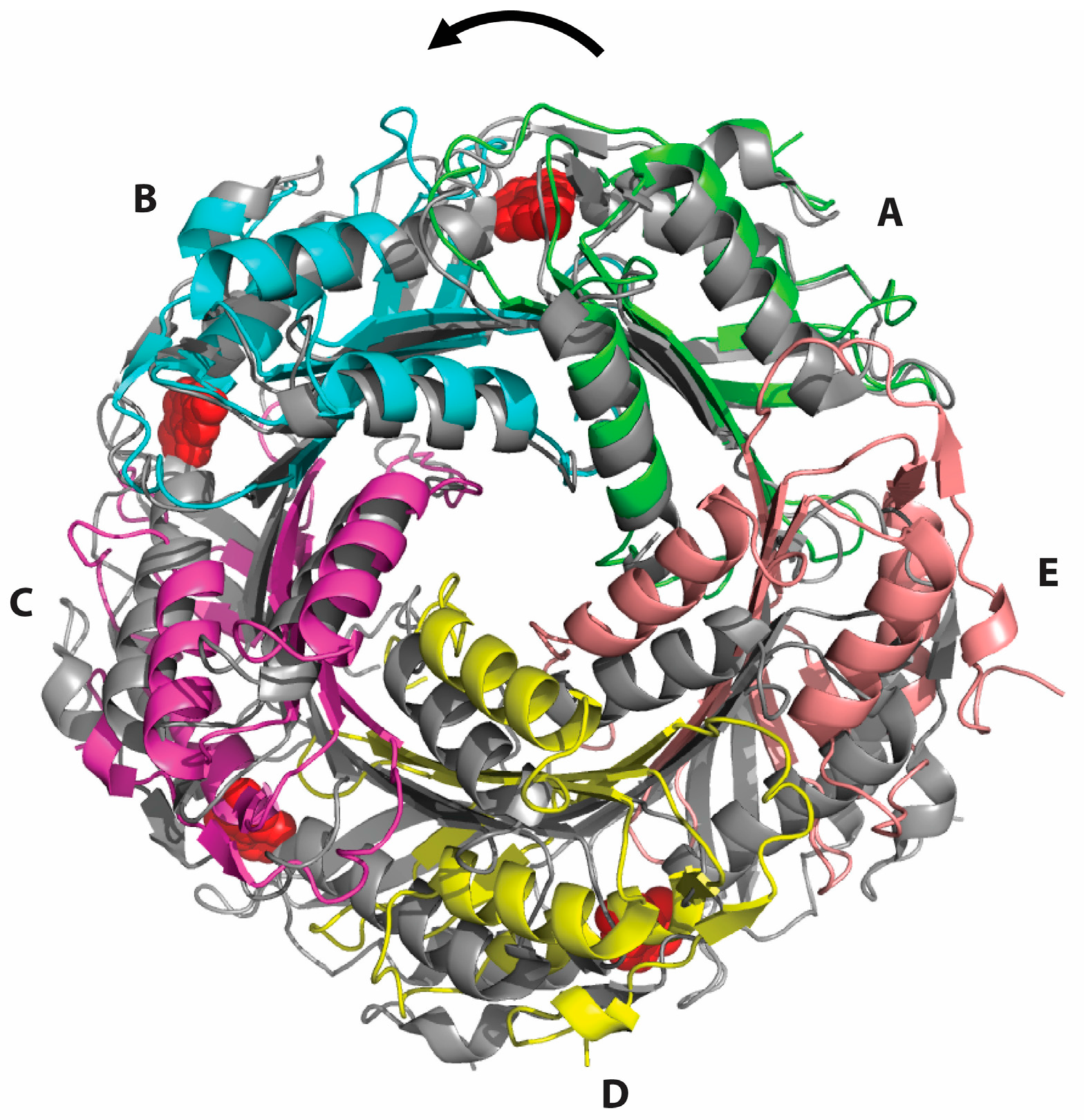
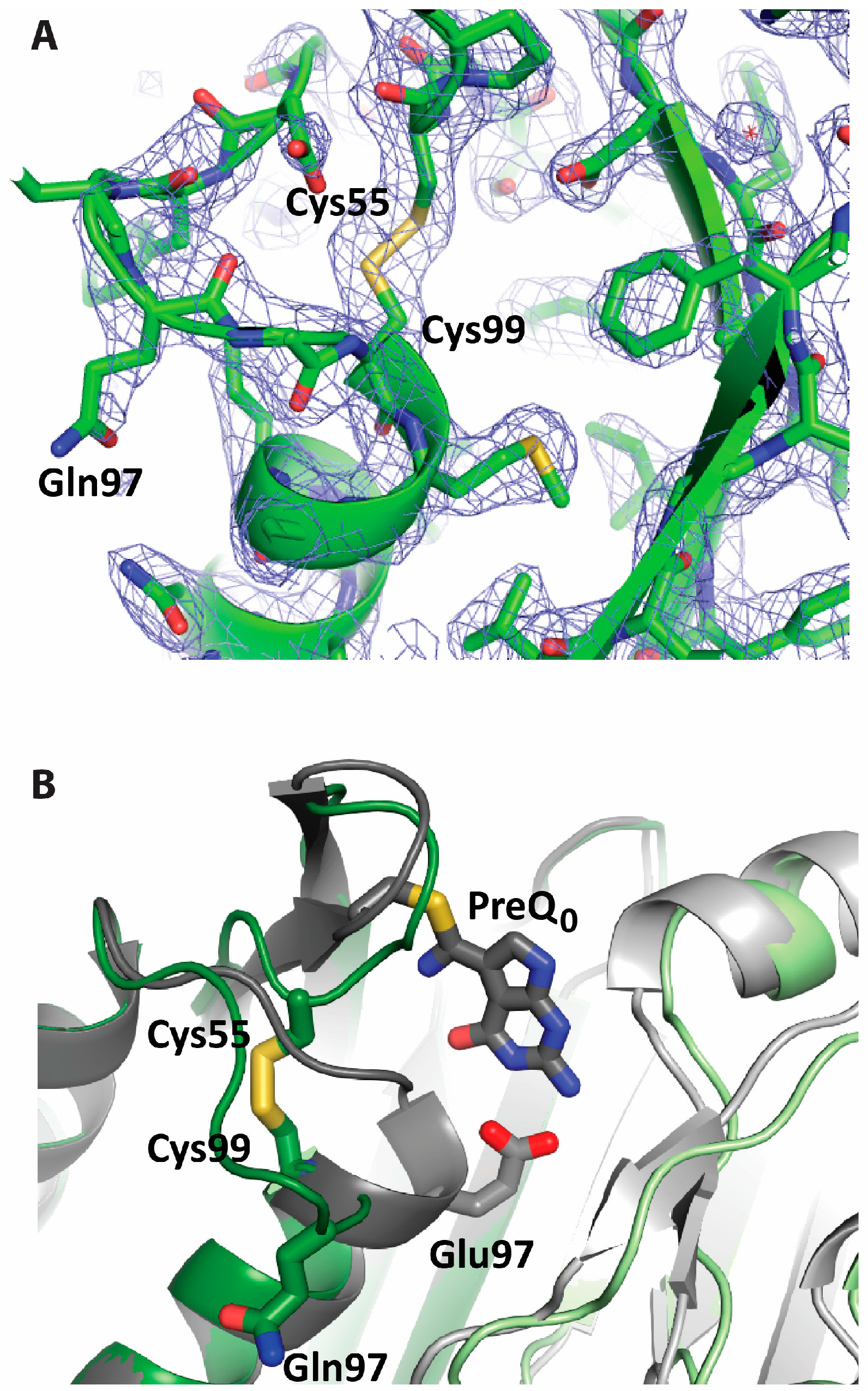
2.2. Formation of an Active-Site Disulfide in a Substrate-Free Mutant of B. subtilis QueF
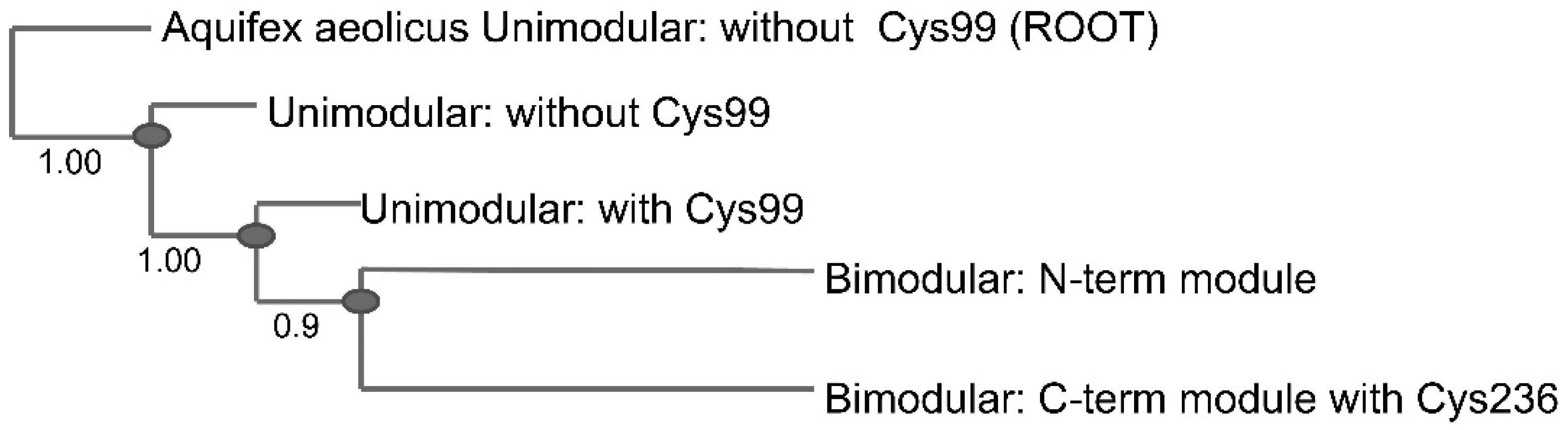
2.3. Conservation of Disulfide-Forming Cysteines in QueF Proteins
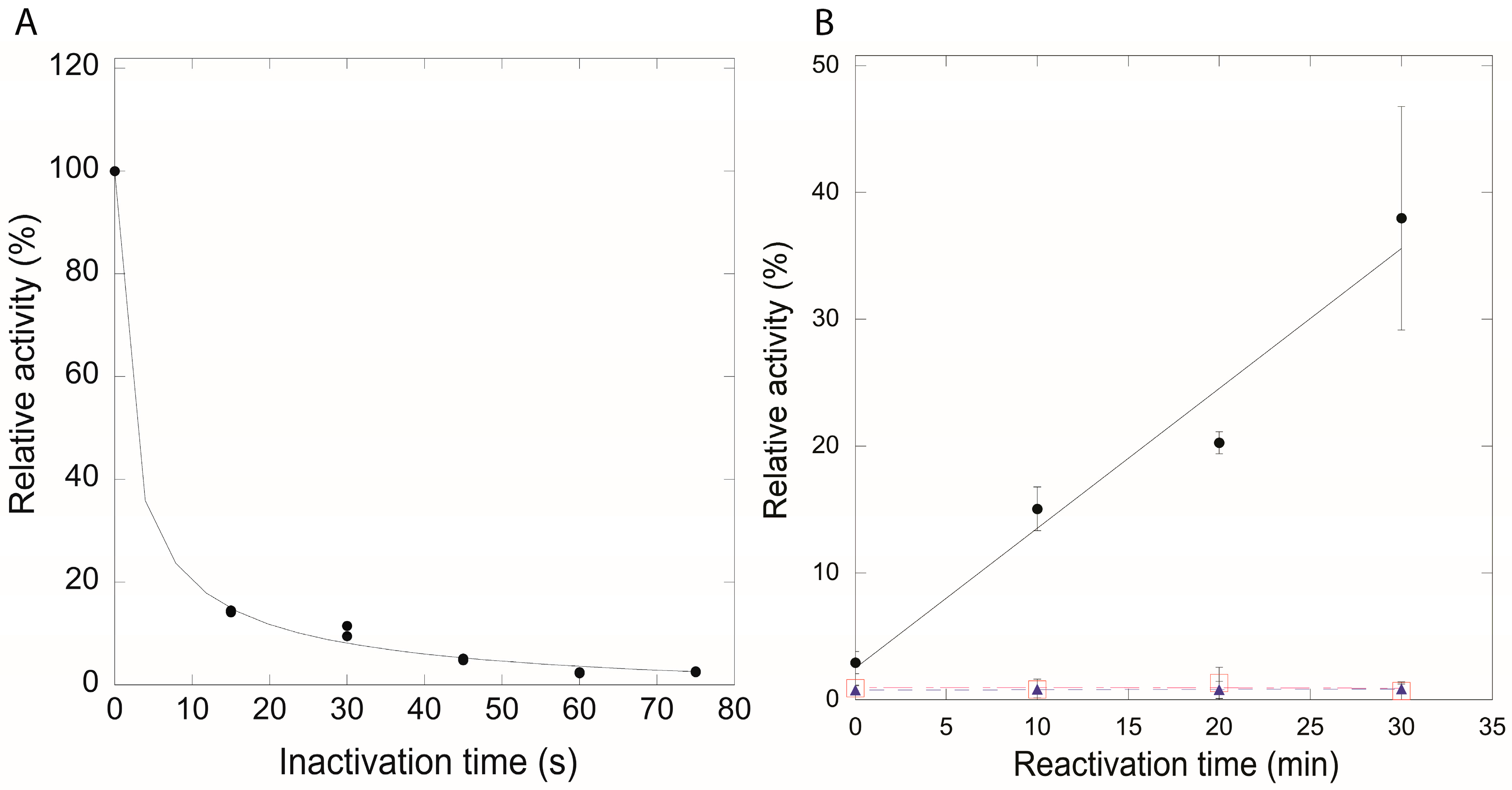
2.4. Disulfide-Mediated Protection of QueF from Irreversible Oxidation In Vitro
3. Materials and Methods
3.1. Mutagenesis of QueF
- E97Q(sense)
- 5’-GTTCATGATGATATTCATGCAGTCCTTGTGGAAGTCAC-3’
- E97Q(antisense)
- 5’-GGTGACTTCCACCAGGACTGCATGAATATCATCATGAACG-3’
- C99S(sense)
- 5’-GGTGACTTCCACGAGGACAGCATGAATATCATCATGAACG-3’
- C99S(antisense)
- 5’- CGTTCATGATGATATTCATGCTGTCCTCGTGGAAGTCACC-3’
- C99A(sense)
- 5’-GGTGACTTCCACGAGGACGCCATGAATATCATCATGAACG-3’
- C99A(antisense)
- 5’- CGTTCATGATGATATTCATGGCGTCCTCGTGGAAGTCACC-3’.
3.2. Activity Assays of Glu97Gln and Cys99Ala/Ser Mutants
3.3. Crystallization, X-ray Data Collection and Crystal Structure Determination
3.4. Sequence Analysis
3.5. H2O2 Oxidation of Wild-Type QueF and Cys99Ala/Ser Mutants
3.6. Activity Recovery of Oxidized QueF Enzymes
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Van Lanen, S.G.; Reader, J.S.; Swairjo, M.A.; de Crecy-Lagard, V.; Lee, B.; Iwata-Reuyl, D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. USA 2005, 102, 4264–4269. [Google Scholar] [CrossRef] [PubMed]
- Harada, F.; Nishimura, S. Possible anticodon sequences of tRNA His, tRNA Asm, and tRNA Asp from Escherichia coli B. Universal presence of nucleoside Q in the first position of the anticondons of these transfer ribonucleic acids. Biochemistry 1972, 11, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Noguchi, S.; Nishimura, Y.; Hirota, Y.; Nishimura, S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine-transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 1982, 257, 6544–6550. [Google Scholar] [PubMed]
- Iwata-Reuyl, D.; de Crécy-Lagard, V. Enzymatic formation of the 7-deazaguanosine hypermodified nucleosides of tRNA. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution; Grosjean, H., Ed.; Landes Bioscience: New York, NY, USA, 2009; pp. 379–394. [Google Scholar]
- Iwata-Reuyl, D. Biosynthesis of the 7-deazaguanosine hypermodified nucleosides of transfer RNA. Bioorg. Chem. 2003, 31, 24–43. [Google Scholar] [CrossRef]
- Vinayak, M.; Pathak, C. Queuosine modification of tRNA: Its divergent role in cellular machinery. Biosci. Rep. 2010, 30, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Marks, T.; Farkas, W.R. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem. Biophys. Res. Commun. 1997, 230, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Meier, F.; Suter, B.; Grosjean, H.; Keith, G.; Kubli, E. Queuosine modification of the wobble base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J. 1985, 4, 823–827. [Google Scholar] [PubMed]
- Rakovich, T.; Boland, C.; Bernstein, I.; Chikwana, V.M.; Iwata-Reuyl, D.; Kelly, V.P. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J. Biol. Chem. 2011, 286, 19354–19363. [Google Scholar] [CrossRef] [PubMed]
- Reader, J.; Metzgar, D.; Schimmel, P.; de Crécy-lagard, V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004, 279, 6280–6285. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.W.; Lanen, S.G.; Iwata-Reuyl, D. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in queuosine biosynthesis. Biochemistry 2007, 46, 12844–12854. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Zhou, M.; Moy, S.; Morales, J.; Cunningham, M.A.; Joachimiak, A. High-resolution structure of the nitrile reductase quef combined with molecular simulations provide insight into enzyme mechanism. J. Mol. Biol. 2010, 404, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Wilding, B.; Winkler, M.; Petschacher, B.; Kratzer, R.; Glieder, A.; Klempier, N. Nitrile reductase from Geobacillus kaustophilus: A potential catalyst for a new nitrile biotransformation reaction. Adv. Synth. Catal. 2012, 354, 2191–2198. [Google Scholar] [CrossRef]
- Wilding, B.; Winkler, M.; Petschacher, B.; Kratzer, R.; Egger, S.; Steinkellner, G.; Lyskowski, A.; Nidetzky, B.; Gruber, K.; Klempier, N. Targeting the substrate binding site of E. Coli nitrile reductase QueF by modeling, substrate and enzyme engineering. Chemistry 2013, 19, 7007–7012. [Google Scholar] [CrossRef] [PubMed]
- Moeller, K.; Nguyen, G.S.; Hollmann, F.; Hanefeld, U. Expression and characterization of the nitrile reductase QueF from E. coli. Enzyme Microb. Technol. 2013, 52, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Chikwana, V.M.; Stec, B.; Lee, B.W.; de Crecy-Lagard, V.; Iwata-Reuyl, D.; Swairjo, M.A. Structural basis of biological nitrile reduction. J. Biol. Chem. 2012, 287, 30560–30570. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.J.M.; Yang, L.; Ramos, M.J.; Fernandes, P.A.; Liang, Z.-X.; Hirao, H. Insight into enzymatic nitrile reduction: QM/MM study of the catalytic mechanism of QueF nitrile reductase. ACS Catal. 2015, 5, 3740–3751. [Google Scholar] [CrossRef]
- Jung, J.; Czabany, T.; Wilding, B.; Klempier, N.; Nidetzky, B. Kinetic analysis and probing with substrate analogues of the reaction pathway of the nitrile reductase QueF from Escherichia coli. J. Biol. Chem. 2016, 291, 25411–25426. [Google Scholar] [CrossRef] [PubMed]
- Colloc’h, N.; Poupon, A.; Mornon, J.-P. Sequence and structural features of the T-fold, an original tunneling building unit. Proteins 2000, 39, 142–154. [Google Scholar] [CrossRef]
- Chi, B.K.; Roberts, A.A.; Huyen, T.T.; Basell, K.; Becher, D.; Albrecht, D.; Hamilton, C.J.; Antelmann, H. S-bacillithiolation protects conserved and essential proteins against hypochlorite stress in Firmicutes bacteria. Antioxid. Redox Signal. 2013, 18, 1273–1295. [Google Scholar] [CrossRef] [PubMed]
- Arifuzzaman, M.; Maeda, M.; Itoh, A.; Nishikata, K.; Takita, C.; Saito, R.; Ara, T.; Nakahigashi, K.; Huang, H.C.; Hirai, A.; et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006, 16, 686–691. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, B.; Ho, L.; Hogg, P.J. Allosteric disulfide bonds. Biochemistry 2006, 45, 7429–7433. [Google Scholar] [CrossRef] [PubMed]
- Hogg, P.J. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 2003, 28, 210–214. [Google Scholar] [CrossRef]
- Cook, K.M.; Hogg, P.J. Post-translational control of protein function by disulfide bond cleavage. Antioxid. Redox Signal. 2013, 18, 1987–2015. [Google Scholar] [CrossRef] [PubMed]
- Berndt, C.; Lillig, C.H.; Holmgren, A. Thiol-based mechanisms of the thioredoxin and glutaredoxin systems: Implications for diseases in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2007, 292, H1227–H1236. [Google Scholar] [CrossRef] [PubMed]
- Swairjo, M.A.; Reddy, R.P.; Lee, B.; Van Lanen, S.G.; Brown, A.; De Crécy Lagard, V.; Iwata_Reuyl, D.; Schimmel, P. Crystallization and preliminary X-ray characterization of the nitrile reducatse QueF: A queuosine biosynthesis enzyme. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2005, 61, 945–948. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar] [PubMed]
- Storoni, L.C.; McCoy, A.J.; Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60, 432–438. [Google Scholar] [CrossRef] [PubMed]
- Murshudov, G.N.; Vagin, A.A.; Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 1997, D53, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, D60, 2126–2132. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Milne, I.; Lindner, D.; Bayer, M.; Husmeier, D.; McGuire, G.; Marshall, D.F.; Wright, F. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 2009, 25, 126–127. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
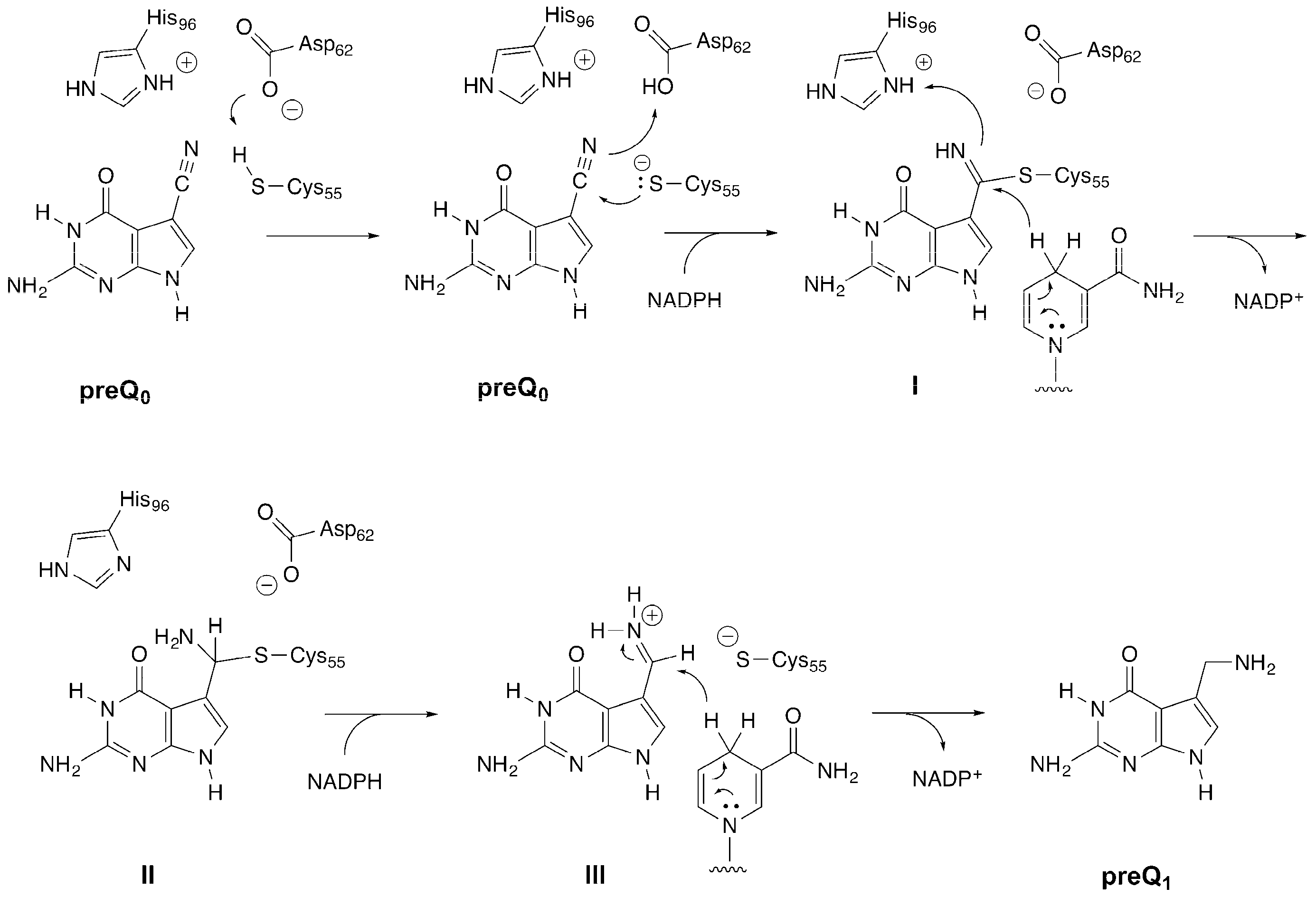
| Enzyme | Relative Activity 1 (%) |
|---|---|
| Wild-type QueF | 100 ± 4 |
| Cys99Ala | 80 ± 4 |
| Cys99Ser | 74 ± 3 |
| Glu97Gln | 1.9 ± 0.1 |
| Data Collection: | Value |
|---|---|
| Space group | P3221 |
| Unit cell (Å) | 87.31, 87.31, 196.73 |
| Wavelength (Å) | 1.12709 |
| Resolution range (Å) | 50–2.5 (2.54–2.50) 1 |
| Completeness (%) | 98.0 (92.5) |
| Redundancy | 5.0 (3.0) |
| Rmerge, Rpim (%) 2 | 0.087, 0.060 (0.630, 0.627) |
| <I/σ(I)> | 13.10 (1.17) |
| Refinement: | |
| Number of reflections | |
| Working/free | 28,753/1466 (1937/109) |
| Number of atoms | |
| Total | 6417 |
| Water/Mg2+ | 285/7 |
| PEG | 37 |
| R-cryst 3/R-free 4 (%) | 0.189/0.257 (0.303/0.409) |
| Rmsd bond lengths (Å) | 0.019 |
| Rmsd bond angles (°) | 2.007 |
| Wilson B-factor (Å2) | 50.2 |
| Average B-factor | |
| Protein | 45.5 |
| Metals | 79.7 |
| Water | 46.76 |
| Ramachandran Plot (%) | |
| Favored | 94.0 |
| Allowed | 4.5 5 |
| Subunit | χ1 (°) | χ2 (°) | χ3 (°) | Bond Length (Å) | χ2′ (°) | χ1′ (°) | Disulfide Strain Energy (kJ/mol) |
|---|---|---|---|---|---|---|---|
| A | −59.85 | −126.76 | −105.12 | 2.03 | 176.01 | −66.90 | 15.023 |
| B | −56.90 | −123.68 | −89.05 | 2.03 | 171.20 | −81.83 | 16.120 |
| C | −59.70 | −116.71 | −102.91 | 2.05 | 168.10 | −63.57 | 14.772 |
| D | −60.93 | −127.32 | −79.75 | 2.07 | 173.65 | −89.46 | 18.584 |
| E | −55.16 | −120.73 | −94.72 | 2.04 | 172.66 | −72.34 | 13.938 |
| Unimodular QueF | Bimodular QueF | |
|---|---|---|
| Total sequences | 2074 | 1375 |
| % with disulfide forming cysteine (Cys99 in unimodular QueF, Cys236 in bimodular QueF) | 61% | 100% |
| % with any potentially disulfide forming cysteine (Cys99 or Cys53 in unimodular QueF) | 83% | NA |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mohammad, A.; Bon Ramos, A.; Lee, B.W.K.; Cohen, S.W.; Kiani, M.K.; Iwata-Reuyl, D.; Stec, B.; Swairjo, M.A. Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide. Biomolecules 2017, 7, 30. https://doi.org/10.3390/biom7010030
Mohammad A, Bon Ramos A, Lee BWK, Cohen SW, Kiani MK, Iwata-Reuyl D, Stec B, Swairjo MA. Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide. Biomolecules. 2017; 7(1):30. https://doi.org/10.3390/biom7010030
Chicago/Turabian StyleMohammad, Adeba, Adriana Bon Ramos, Bobby W. K. Lee, Spencer W. Cohen, Maryam K. Kiani, Dirk Iwata-Reuyl, Boguslaw Stec, and Manal A. Swairjo. 2017. "Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide" Biomolecules 7, no. 1: 30. https://doi.org/10.3390/biom7010030
APA StyleMohammad, A., Bon Ramos, A., Lee, B. W. K., Cohen, S. W., Kiani, M. K., Iwata-Reuyl, D., Stec, B., & Swairjo, M. A. (2017). Protection of the Queuosine Biosynthesis Enzyme QueF from Irreversible Oxidation by a Conserved Intramolecular Disulfide. Biomolecules, 7(1), 30. https://doi.org/10.3390/biom7010030






