Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques
Abstract
:1. Introduction
2. Results
2.1. Hippocampus and Cerebrospinal Fluid Viral Load
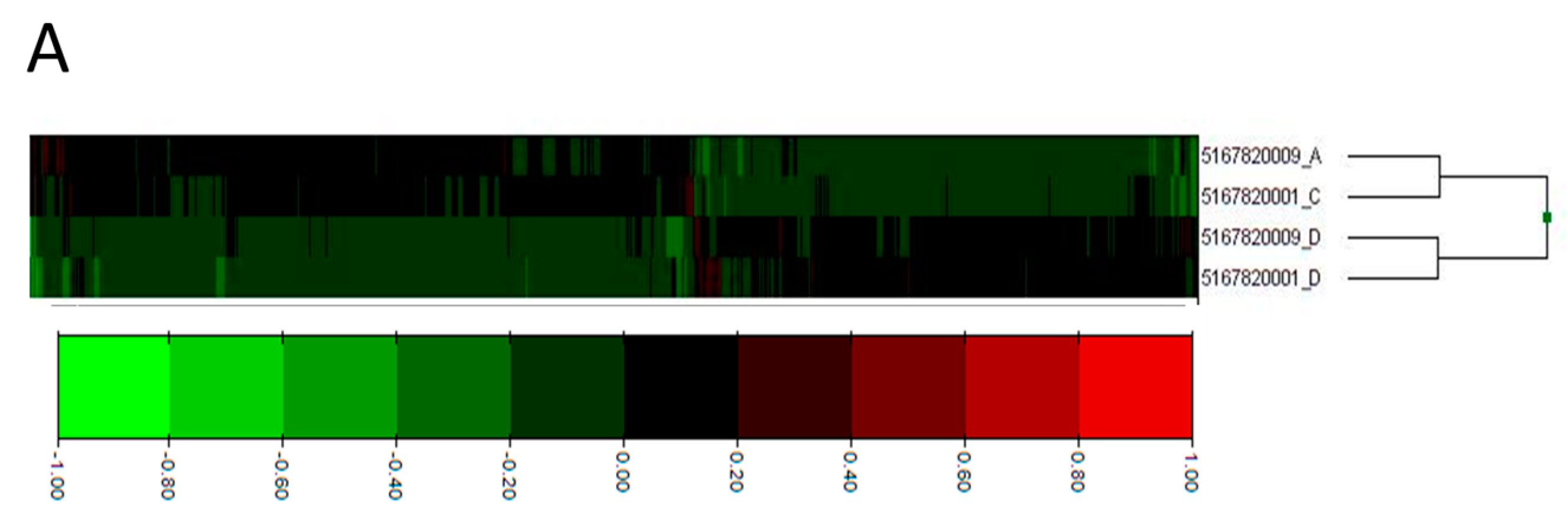
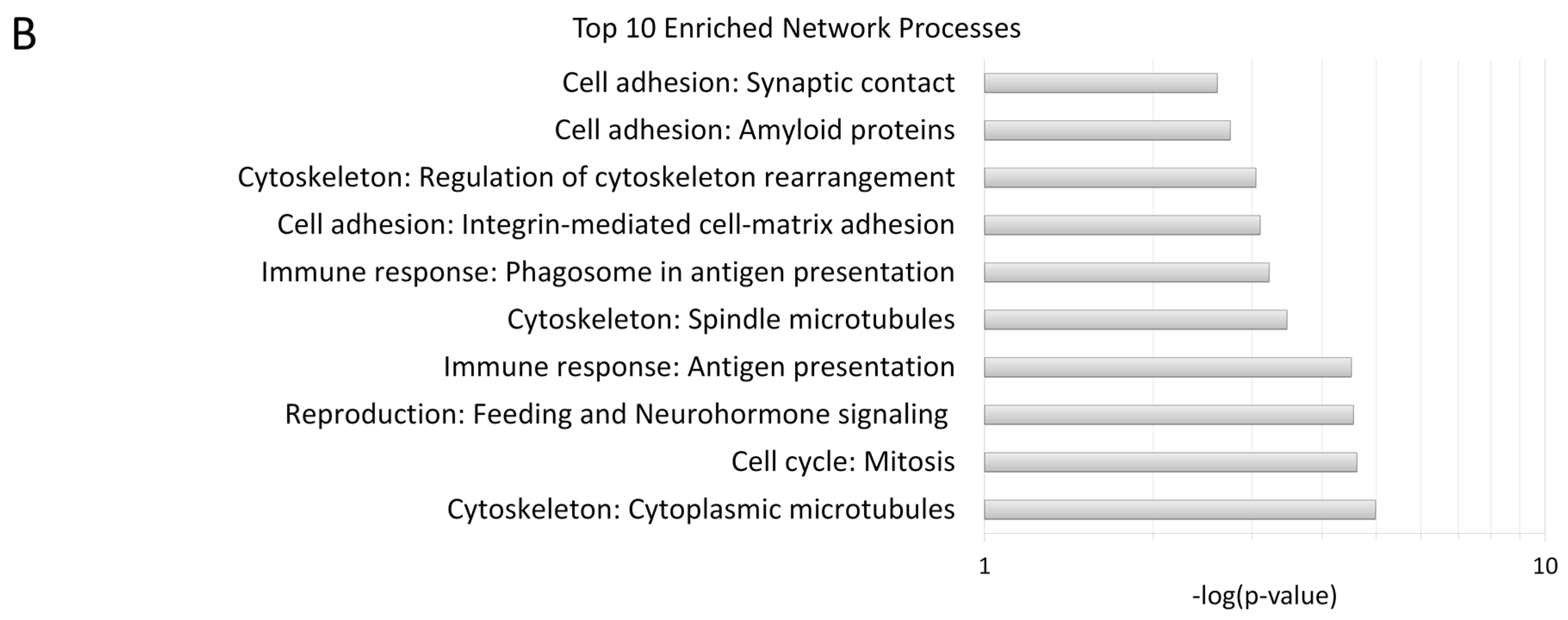
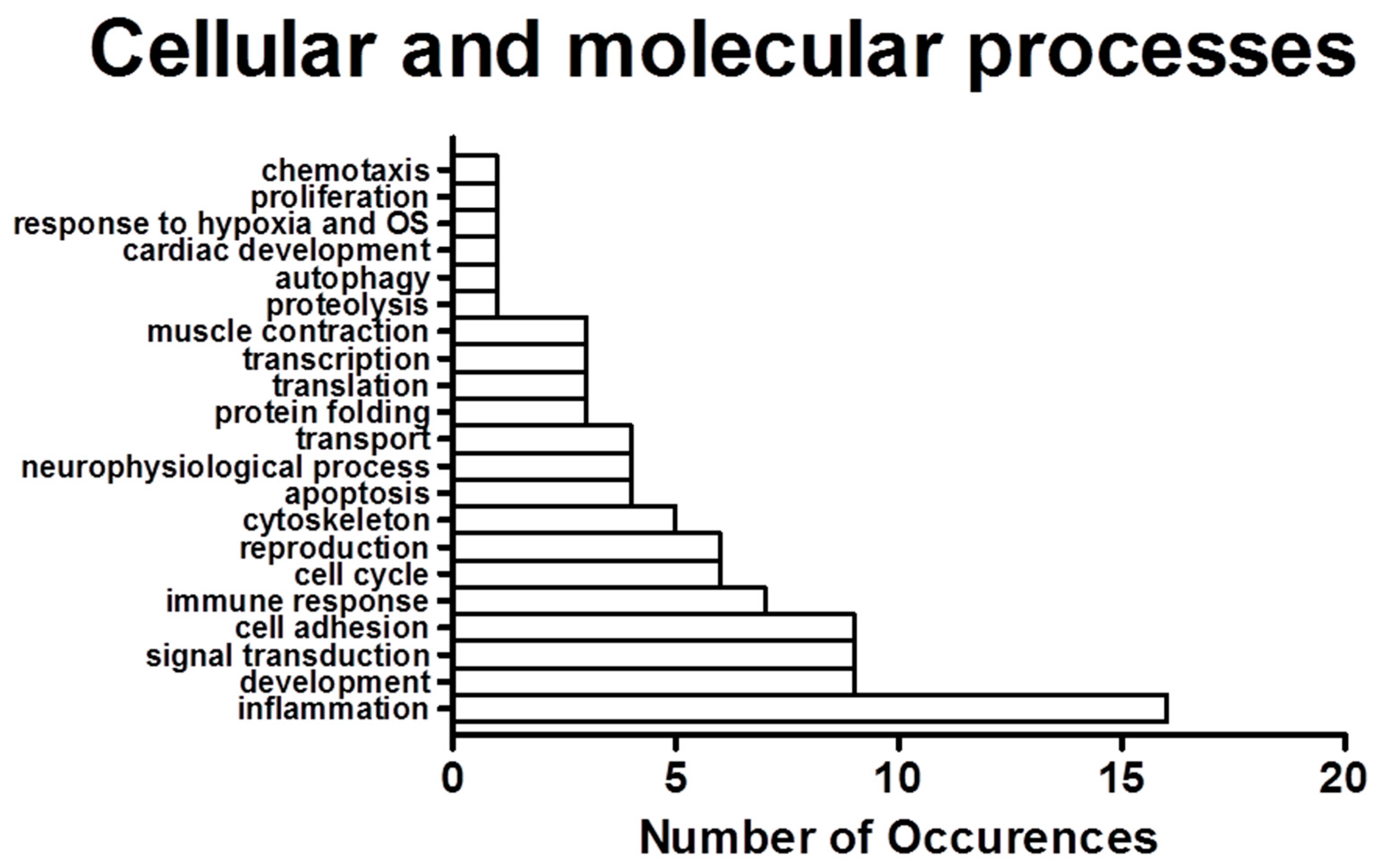
2.2. Process Networks Enriched with Differentially Expressed Genes
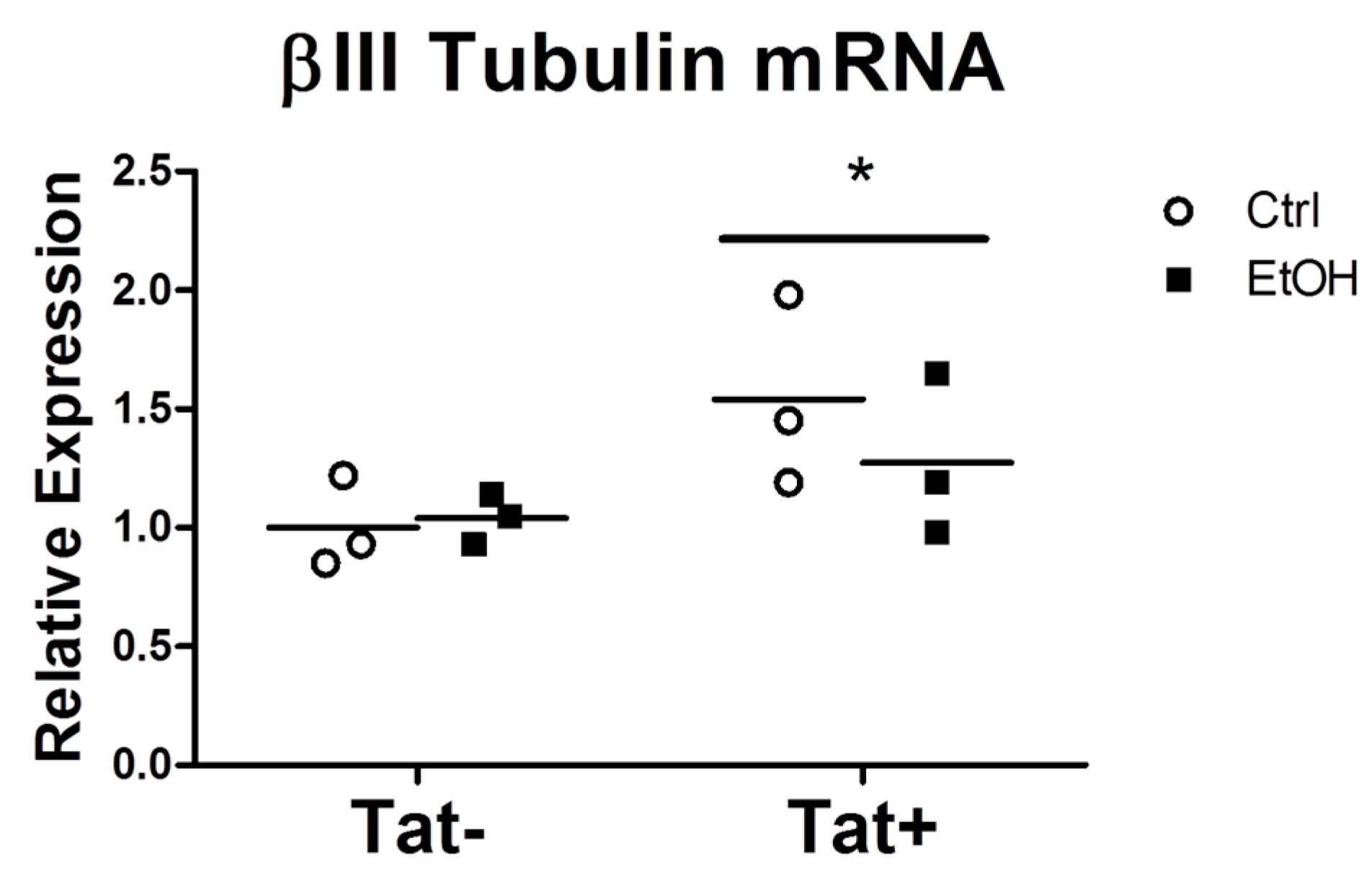
2.3. EtOH Treatment Alters NPC Differentiation in the Presence or Absence of HIV Tat
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Hippocampus and RNA Isolation
4.3. Cerebrospinal Fluid (CSF) Collection
4.4. Viral Load Quantification
4.5. Microarray Data Analysis
4.6. NPC Isolation and Culture
4.7. NPC RNA Isolation and qPCR
4.8. NPC Immunocytochemistry
4.9. Data Analysis
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Heaton, R.K.; Franklin, D.R.; Ellis, R.J.; McCutchan, J.A.; Letendre, S.L.; Leblanc, S.; Corkran, S.H.; Duarte, N.A.; Clifford, D.B.; Woods, S.P.; et al. HIV-associated neurocognitive disorders before and during the era of combination antiretroviral therapy: Differences in rates, nature, and predictors. J. Neurovirol. 2011, 17, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Grant, B.F.; Goldstein, R.B.; Saha, T.D.; Chou, S.P.; Jung, J.; Zhang, H.; Pickering, R.P.; Ruan, W.J.; Smith, S.M.; Huang, B.; et al. Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry 2015, 72, 757–766. [Google Scholar] [CrossRef] [PubMed]
- Joska, J.A.; Fincham, D.S.; Stein, D.J.; Paul, R.H.; Seedat, S. Clinical correlates of HIV-associated neurocognitive disorders in South Africa. AIDS Behav. 2010, 14, 371–378. [Google Scholar] [CrossRef] [PubMed]
- Gongvatana, A.; Morgan, E.E.; Iudicello, J.E.; Letendre, S.L.; Grant, I.; Woods, S.P.; The HIV Neurobehavioral Research Program (HNRP) Group. A history of alcohol dependence augments HIV-associated neurocognitive deficits in persons aged 60 and older. J Neurovirol. 2014, 20, 505–513. [Google Scholar] [CrossRef] [PubMed]
- Molina, P.E.; Bagby, G.J.; Nelson, S. Biomedical consequences of alcohol use disorders in the HIV-infected host. Curr. HIV Res. 2014, 12, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Woods, S.P.; Moore, D.J.; Weber, E.; Grant, I. Cognitive neuropsychology of HIV-associated neurocognitive disorders. Neuropsychol. Rev. 2009, 19, 152–168. [Google Scholar] [CrossRef] [PubMed]
- Heaton, R.K.; Clifford, D.B.; Franklin, D.R., Jr.; Woods, S.P.; Ake, C.; Vaida, F.; Ellis, R.J.; Letendre, S.L.; Marcotte, T.D.; Atkinson, J.H.; et al. HIV-associated neurocognitive disorders persist in the era of potent antiretroviral therapy: CHARTER Study. Neurology 2010, 75, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Winsauer, P.J.; Moerschbaecher, J.M.; Brauner, I.N.; Purcell, J.E.; Lancaster, J.R., Jr.; Bagby, G.J.; Nelson, S. Alcohol unmasks simian immunodeficiency virus-induced cognitive impairments in rhesus monkeys. Alcohol Clin. Exp. Res. 2002, 26, 1846–1857. [Google Scholar] [CrossRef] [PubMed]
- Bagby, G.J.; Zhang, P.; Purcell, J.E.; Didier, P.J.; Nelson, S. Chronic binge ethanol consumption accelerates progression of simian immunodeficiency virus disease. Alcohol Clin. Exp. Res. 2006, 30, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Molina, P.E.; Lang, C.H.; McNurlan, M.; Bagby, G.J.; Nelson, S. Chronic alcohol accentuates simian acquired immunodeficiency syndrome-associated wasting. Alcohol Clin. Exp. Res. 2008, 32, 138–147. [Google Scholar] [CrossRef] [PubMed]
- LeCapitaine, N.J.; Wang, Z.Q.; Dufour, J.P.; Potter, B.J.; Bagby, G.J.; Nelson, S.; Cefalu, W.T.; Molina, P.E. Disrupted anabolic and catabolic processes may contribute to alcohol-accentuated SAIDS-associated wasting. J. Infect. Dis. 2011, 204, 1246–1255. [Google Scholar] [CrossRef] [PubMed]
- Dodd, T.; Simon, L.; LeCapitaine, N.J.; Zabaleta, J.; Mussell, J.; Berner, P.; Ford, S.; Dufour, J.; Bagby, G.J.; Nelson, S.; et al. Chronic binge alcohol administration accentuates expression of pro-fibrotic and inflammatory genes in the skeletal muscle of simian immunodeficiency virus-infected macaques. Alcohol Clin. Exp. Res. 2014, 38, 2697–2706. [Google Scholar] [CrossRef] [PubMed]
- Chanraud, S.; Martelli, C.; Delain, F.; Kostogianni, N.; Douaud, G.; Aubin, H.J.; Reynaud, M.; Martinot, J.L. Brain morphometry and cognitive performance in detoxified alcohol-dependents with preserved psychosocial functioning. Neuropsychopharmacology 2007, 32, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Gongvatana, A.; Correia, S.; Dunsiger, S.; Gauthier, L.; Devlin, K.N.; Ross, S.; Navia, B.; Tashima, K.T.; DeLaMonte, S.; Cohen, R.A. Plasma cytokine levels are related to brain volumes in HIV-infected individuals. J. Neuroimmune Pharmacol. 2014, 9, 740–750. [Google Scholar] [CrossRef] [PubMed]
- Ketchum, M.J.; Weyand, T.G.; Weed, P.F.; Winsauer, P.J. Learning by subtraction: Hippocampal activity and effects of ethanol during the acquisition and performance of response sequences. Hippocampus 2016, 26, 601–622. [Google Scholar] [CrossRef] [PubMed]
- Mankowski, J.L.; Queen, S.E.; Clements, J.E.; Zink, M.C. Cerebrospinal fluid markers that predict SIV CNS disease. J. Neuroimmunol. 2004, 157, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Zink, M.C.; Coleman, G.D.; Mankowski, J.L.; Adams, R.J.; Tarwater, P.M.; Fox, K.; Clements, J.E. Increased macrophage chemoattractant protein-1 in cerebrospinal fluid precedes and predicts simian immunodeficiency virus encephalitis. J. Infect. Dis. 2001, 184, 1015–1021. [Google Scholar] [CrossRef] [PubMed]
- Zink, M.C.; Suryanarayana, K.; Mankowski, J.L.; Shen, A.; Piatak, M., Jr.; Spelman, J.P.; Carter, D.L.; Adams, R.J.; Lifson, J.D.; Clements, J.E. High viral load in the cerebrospinal fluid and brain correlates with severity of simian immunodeficiency virus encephalitis. J. Virol. 1999, 73, 10480–10488. [Google Scholar] [PubMed]
- Crews, F.T.; Sarkar, D.K.; Qin, L.; Zou, J.; Boyadjieva, N.; Vetreno, R.P. Neuroimmune Function and the Consequences of Alcohol Exposure. Alcohol Res. 2015, 37, 331–351. [Google Scholar] [PubMed]
- Zhao, C.; Den, W.; Gage, F.H. Mechanisms and functional implications of adult neurogenesis. Cell 2008, 132, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Marin-Burgin, A.; Schinder, A.F. Requirement of adult-born neurons for hippocampus-dependent learning. Behav. Brain. Res. 2012, 227, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Iosif, R.E.; Ekdahl, C.T.; Ahlenius, H.; Pronk, C.J.; Bonde, S.; Kokaia, Z.; Jacobsen, S.E.; Lindvall, O. Tumor necrosis factor receptor 1 is a negative regulator of progenitor proliferation in adult hippocampal neurogenesis. J. Neurosci. 2006, 26, 9703–9712. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Palmer, T.D. Differential roles of TNFR1 and TNFR2 signaling in adult hippocampal neurogenesis. Brain Behav. Immun. 2013, 30, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.M.; Nolan, Y.M. Neuroinflammation negatively affects adult hippocampal neurogenesis and cognition: can exercise compensate? Neurosci. Biobehav. Rev. 2016, 61, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Winocur, G.; Wojtowicz, J.M.; Sekeres, M.; Snyder, J.S.; Wang, S. Inhibition of neurogenesis interferes with hippocampus-dependent memory function. Hippocampus 2006, 16, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Dupret, D.; Revest, J.M.; Koehl, M.; Ichas, F.; De Giorgi, F.; Costet, P.; Abrous, D.N.; Piazza, P.V. Spatial relational memory requires hippocampal adult neurogenesis. PLoS ONE 2008, 3, e1959. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.S.; Choe, J.S.; Clifford, M.A.; Jeurling, S.I.; Hurley, P.; Brown, A.; Kamhi, J.F.; Cameron, H.A. Adult-born hippocampal neurons are more numerous, faster maturing, and more involved in behavior in rats than in mice. J. Neurosci. 2009, 29, 14484–14495. [Google Scholar] [CrossRef] [PubMed]
- Pla, A.; Pascual, M.; Renau-Piqueras, J.; Guerri, C. TLR4 mediates the impairment of ubiquitin-proteasome and autophagy-lysosome pathways induced by ethanol treatment in brain. Cell Death Dis. 2014, 5, e1066. [Google Scholar] [CrossRef] [PubMed]
- Angeles, A.; Fung, G.; Luo, H. Immune and non-immune functions of the immunoproteasome. Front Biosci. (Landmark Ed.) 2012, 17, 1904–1916. [Google Scholar] [CrossRef] [PubMed]
- Tavazzi, E.; Morrison, D.; Sullivan, P.; Morgello, S.; Fischer, T. Brain inflammation is a common feature of HIV-infected patients without HIV encephalitis or productive brain infection. Curr. HIV Res. 2014, 12, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Elmer, B.M.; McAllister, A.K. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci. 2012, 35, 660–670. [Google Scholar] [CrossRef] [PubMed]
- Glynn, M.W.; Elmer, B.M.; Garay, P.A.; Liu, X.B.; Needleman, L.A.; El-Sabeawy, F.; McAllister, A.K. MHCI negatively regulates synapse density during the establishment of cortical connections. Nat. Neurosci. 2011, 14, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Bilousova, T.; Dang, H.; Xu, W.; Gustafson, S.; Jin, Y.; Wickramasinghe, L.; Won, T.; Bobarnac, G.; Middleton, B.; Tian, J.; et al. Major histocompatibility complex class I molecules modulate embryonic neuritogenesis and neuronal polarization. J. Neuroimmunol. 2012, 247, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Goddard, C.A.; Butts, D.A.; Shatz, C.J. Regulation of CNS synapses by neuronal MHC class I. Proc. Natl. Acad. Sci. USA 2007, 104, 6828–6833. [Google Scholar] [CrossRef] [PubMed]
- Arikkath, J.; Reichardt, L.F. Cadherins and catenins at synapses: Roles in synaptogenesis and synaptic plasticity. Trends Neurosci. 2008, 31, 487–494. [Google Scholar] [CrossRef] [PubMed]
- Fitting, S.; Ignatowska-Jankowska, B.M.; Bull, C.; Skoff, R.P.; Lichtman, A.H.; Wise, L.E.; Fox, M.A.; Su, J.; Medina, A.E.; Krahe, T.E.; et al. Synaptic dysfunction in the hippocampus accompanies learning and memory deficits in human immunodeficiency virus type-1 Tat transgenic mice. Biol. Psychiatry 2013, 73, 443–453. [Google Scholar] [CrossRef] [PubMed]
- Roberts, E.S.; Masliah, E.; Fox, H.S. CD163 identifies a unique population of ramified microglia in HIV encephalitis (HIVE). J. Neuropathol. Exp. Neurol. 2004, 63, 1255–1264. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Crews, F.T. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp. Neurol. 2008, 210, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Ekdahl, C.T.; Claasen, J.H.; Bonde, S.; Kokaia, Z.; Lindvall, O. Inflammation is detrimental for neurogenesis in adult brain. Proc. Natl. Acad. Sci. USA 2003, 100, 13632–13637. [Google Scholar] [CrossRef] [PubMed]
- Vereker, E.; O'Donnell, E.; Lynch, M.A. The inhibitory effect of interleukin-1beta on long-term potentiation is coupled with increased activity of stress-activated protein kinases. J. Neurosci. 2000, 20, 6811–6819. [Google Scholar] [PubMed]
- Masliah, E.; Roberts, E.S.; Langford, D.; Everall, I.; Crews, L.; Adame, A.; Rockenstein, E.; Fox, H.S. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J. Neuroimmunol. 2004, 157, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Gelman, B.B.; Chen, T.; Lisinicchia, J.G.; Soukup, V.M.; Carmical, J.R.; Starkey, J.M.; Masliah, E.; Commins, D.L.; Brandt, D.; Grant, I.; et al. The National NeuroAIDS Tissue Consortium brain gene array: two types of HIV-associated neurocognitive impairment. PLoS ONE 2012, 7, e46178. [Google Scholar] [CrossRef] [PubMed]
- Borjabad, A.; Morgello, S.; Chao, W.; Kim, S.Y.; Brooks, A.I.; Murray, J.; Potash, M.J.; Volsky, D.J. Significant effects of antiretroviral therapy on global gene expression in brain tissues of patients with HIV-1-associated neurocognitive disorders. PLoS Pathog. 2011, 7, e1002213. [Google Scholar] [CrossRef] [PubMed]
- Mishra, M.; Taneja, M.; Malik, S.; Khalique, H.; Seth, P. Human immunodeficiency virus type 1 Tat modulates proliferation and differentiation of human neural precursor cells: Implication in NeuroAIDS. J. Neurovirol. 2010, 16, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Rashid, M.A.; Kim, H.Y. N-Docosahexaenoylethanolamine ameliorates ethanol-induced impairment of neural stem cell neurogenic differentiation. Neuropharmacology 2015, 102, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Chao, J.; Yang, L.; Yao, H.; Buch, S. Platelet-derived growth factor-BB restores HIV Tat-mediated impairment of neurogenesis: role of GSK-3beta/beta-catenin. J. Neuroimmune Pharmacol. 2014, 9, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Moore, D.J.; Masliah, E.; Rippeth, J.D.; Gonzalez, R.; Carey, C.L.; Cherner, M.; Ellis, R.J.; Achim, C.L.; Marcotte, T.D.; Heaton, R.K.; et al. Cortical and subcortical neurodegeneration is associated with HIV neurocognitive impairment. AIDS 2006, 20, 879–887. [Google Scholar] [CrossRef] [PubMed]
- Nath, A.; Conant, K.; Chen, P.; Scott, C.; Major, E.O. Transient exposure to HIV-1 Tat protein results in cytokine production in macrophages and astrocytes. A hit and run phenomenon. J. Biol. Chem. 1999, 274, 17098–17102. [Google Scholar] [CrossRef] [PubMed]
- Buscemi, L.; Ramonet, D.; Geiger, J.D. Human immunodeficiency virus type-1 protein Tat induces tumor necrosis factor-alpha-mediated neurotoxicity. Neurobiol. Dis. 2007, 26, 661–670. [Google Scholar] [CrossRef] [PubMed]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. [Google Scholar]
- Bagby, G.J.; Stoltz, D.A.; Zhang, P.; Kolls, J.K.; Brown, J.; Bohm, R.P., Jr.; Rockar, R.; Purcell, J.; Murphey-Corb, M.; Nelson, S. The effect of chronic binge ethanol consumption on the primary stage of SIV infection in rhesus macaques. Alcohol Clin. Exp. Res. 2003, 27, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Molina, P.E.; McNurlan, M.; Rathmacher, J.; Lang, C.H.; Zambell, K.L.; Purcell, J.; Bohm, R.P.; Zhang, P.; Bagby, G.J.; Nelson, S. Chronic alcohol accentuates nutritional, metabolic, and immune alterations during asymptomatic simian immunodeficiency virus infection. Alcohol Clin. Exp. Res. 2006, 30, 2065–2078. [Google Scholar] [CrossRef] [PubMed]
- Purcell, D.W.; Parsons, J.T.; Halkitis, P.N.; Mizuno, Y.; Woods, W.J. Substance use and sexual transmission risk behavior of HIV-positive men who have sex with men. J. Subst. Abuse 2001, 13, 185–200. [Google Scholar] [CrossRef]
- Pandrea, I.; Happel, K.I.; Amedee, A.M.; Bagby, G.J.; Nelson, S. Alcohol's role in HIV transmission and disease progression. Alcohol Res. Health 2010, 33, 203–218. [Google Scholar] [PubMed]
- Cook, R.L.; McGinnis, K.A.; Kraemer, K.L.; Gordon, A.J.; Conigliaro, J.; Maisto, S.A.; Samet, J.H.; Crystal, S.; Rimland, D.; Bryant, K.J.; et al. Intoxication before intercourse and risky sexual behavior in male veterans with and without human immunodeficiency virus infection. Med. Care 2006, 44, S31–S36. [Google Scholar] [CrossRef] [PubMed]
- Stein, M.D.; Hanna, L.; Natarajan, R.; Clarke, J.; Marisi, M.; Sobota, M.; Rich, J. Alcohol use patterns predict high-risk HIV behaviors among active injection drug users. J. Subst. Abuse Treat 2000, 18, 359–363. [Google Scholar] [CrossRef]
- Friedman, L.S. Complications associated with blood alcohol concentration following injury. Alcohol 2014, 48, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Center for Behavioral Health Statistics and Quality. Behavioral Health Trends in the United States: Results from the 2014 National Survey on Drug Use and Health; HHS Publication: Washington, DC, USA, 2015.
- Paxinos, G.; Huang, X.F.; Petrides, M.; Toga, A.W. The Rhesus Monkey Brain in Stereotaxic Coordinates, 2nd ed.; Elsevier Academic Press: San Diego, CA, USA, 2009; p. 163. [Google Scholar]
- Molina, P.E.; Amedee, A.M.; LeCapitaine, N.J.; Zabaleta, J.; Mohan, M.; Winsauer, P.J.; Vande Stouwe, C.; McGoey, R.R.; Auten, M.W.; LaMotte, L.; et al. Modulation of gut-specific mechanisms by chronic delta(9)-tetrahydrocannabinol administration in male rhesus macaques infected with simian immunodeficiency virus: A systems biology analysis. AIDS Res. Hum. Retrovir. 2014, 30, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Clark-Langone, K.M.; Wu, J.Y.; Sangli, C.; Chen, A.; Snable, J.L.; Nguyen, A.; Hackett, J.R.; Baker, J.; Yothers, G.; Kim, C.; et al. Biomarker discovery for colon cancer using a 761 gene RT-PCR assay. BMC Genom. 2007, 8, 279. [Google Scholar] [CrossRef] [PubMed]
- Workman, C.; Jensen, L.J.; Jarmer, H.; Berka, R.; Gautier, L.; Nielser, H.B.; Saxild, H.H.; Nielsen, C.; Brunak, S.; Knudsen, S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002, 3, research0048.1–research0048.16. [Google Scholar] [CrossRef] [PubMed]
- Li, M.D.; Cao, J.; Wang, S.; Wang, J.; Sarkar, S.; Vigorito, M.; Ma, J.Z.; Chang, S.L. Transcriptome sequencing of gene expression in the brain of the HIV-1 transgenic rat. PLoS ONE 2013, 8, e59582. [Google Scholar] [CrossRef] [PubMed]
- Nesil, T.; Cao, J.; Yang, Z.; Chang, S.L.; Li, M.D. Nicotine attenuates the effect of HIV-1 proteins on the neural circuits of working and contextual memories. Mol. Brain 2015, 8, 43. [Google Scholar] [CrossRef] [PubMed]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: The human gene integrator. Database (Oxf.) 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Aprea, S.; Del Valle, L.; Mameli, G.; Sawaya, B.E.; Khalili, K.; Peruzzi, F. Tubulin-mediated binding of human immunodeficiency virus-1 Tat to the cytoskeleton causes proteasomal-dependent degradation of microtubule-associated protein 2 and neuronal damage. J. Neurosci. 2006, 26, 4054–4062. [Google Scholar] [CrossRef] [PubMed]
- Vemuri, M.C.; Chetty, C.S. Alcohol impairs astrogliogenesis by stem cells in rodent neurospheres. Neurochem. Int. 2005, 47, 129–135. [Google Scholar] [CrossRef] [PubMed]


| SIV DNA and RNA in Central Nervous System Compartments | ||||
|---|---|---|---|---|
| Viral Load in Hippocampal Tissue | CSF Viral Load | Plasma Viral Load | ||
| Animal ID | DNA (SIV Copies/1 × 106 Cells) | RNA (SIV Copies/μg RPS13) | RNA (SIV Copies/mL) | RNA (SIV Copies/mL, Log Transformed) |
| EK21 (SUC/SIV+) | <25 | <25 | <100 | 6.41 |
| EJ59 (SUC/SIV+) | 510 | <25 | 5705 | 5.97 |
| EN44 (CBA/SIV+) | 140 | <25 | 7560 | 5.59 |
| EN72 (CBA/SIV+) | 660 | 2267 | 112,000 | 5.79 |
| Inflammation and Immune Response Associated Genes | ||
|---|---|---|
| Gene | Description | FC |
| TXNIP | Mediator of oxidative stress, over-expression induces G0/G1 cell cycle arrest, required for maturation of NK cells | 2.43 |
| CCL8 | Chemotactic factor for monocytes, lymphocytes, eosinophils, and basophils | 1.91 |
| PROS1 | Anticoagulant plasma protein, stimulates fibrinolysis | 1.82 |
| HLA-A | Presentation of antigens to the immune system, major histocompatibility class (MHC) I | 1.65 |
| C1QC | Associates with C1r and C1s to yield the first component of the complement system | 1.64 |
| CFB | Complement Factor B, cleaved into Ba and Bb which further activate the complement cascade and monocyte response, Ba inhibits proliferation of pre-activated B-lymphocytes | 1.60 |
| HLA-F | Presentation of antigens to the immune system, MHC class I | 1.58 |
| HLA-G | Presentation of antigens to the immune system, MHC class I | 1.57 |
| HLA-DRA | MHC class II-type, expressed by B lymphocytes, dendritic cells, and macrophages; presentation of antigens to the immune system | 1.54 |
| HLA-H | Presentation of antigens to the immune system, MHC class I | 1.52 |
| MSN | Moesin, cross-linker between plasma membrane and actin cytoskeleton, localized to filopodia | 1.52 |
| APP | Functions in neurite outgrowth, neuronal adhesion, and axogenesis. Cleavage products of APP activate caspases and form amyloid plaques found in Alzheimer’s. | 1.50 |
| TFRC | Transferrin receptor, cellular uptake of iron, necessary for nervous system development | 1.50 |
| ITPR1 | Intracellular inositol 1,4,5-triphosphate receptor, mediates release of Ca2+ from endoplasmic reticulum. | 1.46 |
| MTOR | Mediates cellular responses to DNA damage and nutrient deprivation | 1.45 |
| JAK1 | Kinase in interferon-α, -β, and -γ signal transduction pathways | 1.45 |
| EDG1 | Role in cell migration | 1.45 |
| HLA-E | Binds peptide-derived signal sequence of human leukocyte antigen (HLA) subtypes | 1.45 |
| AP1G1 | Adaptin subunit, promotes formation of clathrin-coated vesicles for delivery from Golgi to lysosomes | 1.44 |
| PIM1 | Proto-oncogene involved in cell survival, kinase activity suppresses pro-apoptotic proteins | 1.44 |
| TNFRSF1A | Tumor necrosis factor (TNF) receptor, activates nuclear factor kappa-light-chain-enhancer of activated B cells (NFkB), apoptosis, and inflammation | 1.40 |
| ELK1 | Stimulates transcription induced by mitogen-activated protein kinase (MAPK) signaling cascade | 1.39 |
| PSMA6 | Peptidase that is a component of the 20S subunit of the proteasome | 1.39 |
| ODC1 | Important enzyme for polyamine synthesis, converts ornithine to putrescine | 1.38 |
| NME1-NME2 | Naturally occurring read-through region between NME1-NME2 genes | 1.37 |
| HLA-DPA1 | MHC class II-type, expressed by B lymphocytes, dendritic cells, and macrophages; presentation of antigens to the immune system | 1.37 |
| GNG10 | G protein subunit gamma | 1.35 |
| GNG2 | G protein subunit gamma | 1.35 |
| VWF | Maintains hemostasis, promotes adhesion of platelets to sites of vascular injury | 1.34 |
| PSMD10 | Regulates 26S proteasome, p53-independent apoptosis and NFkB | 1.34 |
| PPP2CB | Serine/threonine phosphatase, implicated in negative control of cell growth and division | 1.34 |
| PSMD6 | Subunit of 26S proteasome which co-localizes with DNA damage, degrades ubiquiniated proteins | 1.33 |
| SEC61A1 | Crucial role in insertion of membrane and secretory proteins in to endoplasmic reticulum | 1.33 |
| CEBPA | Coordinates proliferation and differentiation of myeloid progenitors | 1.32 |
| PIK3CB | Catalytic sub-unit of Phosphoinositide 3-kinase (PI3K), activation of signaling cascades involved in cell growth, survival, and proliferation | 1.32 |
| AP3B1 | Subunit of adaptor protein complex, target cargos into vesicles for delivery into neurites and nerve terminals | 1.31 |
| HLA-DMA | Prepares MHCII peptide binding site for antigen binding | 1.30 |
| AVP | Antidiuretic action in kidney, vasoconstriction | 1.30 |
| MAPK11 | p38 MAPK, mediates activation of cellular response to pro-inflammatory cytokines | 0.75 |
| ACTN1 | F-actin cross-linking protein, anchors actin to intracellular structures | 0.75 |
| ELMO1 | Involved in cytoskeletal rearrangements needed of phagocytosis of apoptotic cells | 0.74 |
| PSMD13 | Subunit of 19S regulator of the 26S proteasome. | 0.74 |
| TRAP1 | TNF-receptor associated protein, maintains mitochondrial function | 0.69 |
| LPL | Lipoprotein lipase, hydrolysis of triglycerides in circulating chylomicrons and very low density lipoprotein (VLDL) | 0.65 |
| CYCS | Cytochrome C, election carrier in mitochondrial election transport chain, initiates apoptosis when released from mitochondria | 0.64 |
| TIAM1 | Connects extracellular signals to cytoskeletal activities, activates Ras-related C3 botulinum toxin substrate 1 (RAC1), Cell division control protein 42 homolog (CDC42) | 0.64 |
| ISG15 | Important for innate immune response against virus, inhibits HIV viral budding. Secreted interferon-stimulated gene 15 (ISG15) induces natural killer cell proliferation, neutrophil chemotaxis, induces interferon-gamma (IFNγ). | 0.63 |
| CEBPB | Transcription factor that regulates immune and inflammatory responses | 0.59 |
| RYR1 | Ryanodine receptor, mediates release of calcium from intracellular stores | 0.57 |
| HSPA1B | Member of heat shock protein 70 family, stabilizes proteins against aggregation and mediates folding of new proteins | 0.53 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maxi, J.K.; Dean, M.; Zabaleta, J.; Reiss, K.; Bagby, G.J.; Nelson, S.; Winsauer, P.J.; Peruzzi, F.; Molina, P.E. Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques. Biomolecules 2016, 6, 43. https://doi.org/10.3390/biom6040043
Maxi JK, Dean M, Zabaleta J, Reiss K, Bagby GJ, Nelson S, Winsauer PJ, Peruzzi F, Molina PE. Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques. Biomolecules. 2016; 6(4):43. https://doi.org/10.3390/biom6040043
Chicago/Turabian StyleMaxi, John K., Matt Dean, Jovanny Zabaleta, Krzysztof Reiss, Gregory J. Bagby, Steve Nelson, Peter J. Winsauer, Francesca Peruzzi, and Patricia E. Molina. 2016. "Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques" Biomolecules 6, no. 4: 43. https://doi.org/10.3390/biom6040043
APA StyleMaxi, J. K., Dean, M., Zabaleta, J., Reiss, K., Bagby, G. J., Nelson, S., Winsauer, P. J., Peruzzi, F., & Molina, P. E. (2016). Chronic Binge Alcohol Administration Dysregulates Hippocampal Genes Involved in Immunity and Neurogenesis in Simian Immunodeficiency Virus-Infected Macaques. Biomolecules, 6(4), 43. https://doi.org/10.3390/biom6040043






