Protein Quality Control in the Nucleus
Abstract
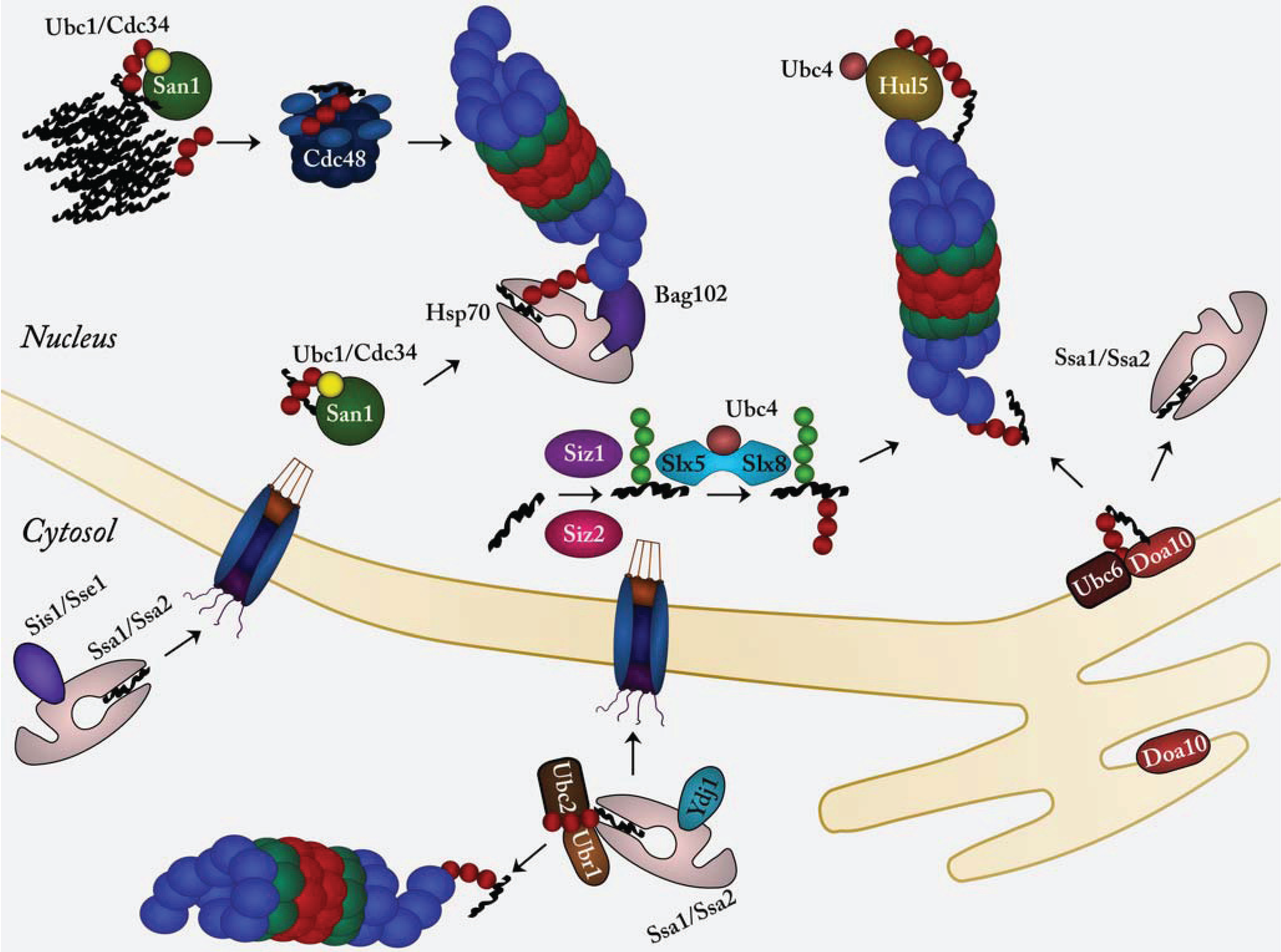
:1. Introduction
| Budding Yeast | Human |
|---|---|
| San1 | − |
| Ubr1 | UBR1/N-recognin 1 |
| Ubr2 | UBR2/N-recognin 2 |
| Doa10 | TEB4 |
| Slx5-Slx8 | RNF4 |
| Hul5 | UBE3 |
| − | Arkadia/RNF111 |
| − | E6-AP |
| − | UHRF2 |
| − | CHIP |
2. Nuclear Protein Quality Control
2.1. The E3 Ubiquitin-Protein Ligase CHIP Targets Chaperone Clients for Degradation
2.2. San1, an Intrinsically Disordered E3 Ubiquitin-Protein Ligase Specific for Misfolded Proteins
2.3. Hul5, a Proteasome-Associated E3 Ubiquitin-Protein Ligase
2.4. Doa10, an ER/Nuclear Envelope Bound E3 Involved in Nuclear Protein Quality Control
2.5. SUMO-Targeted E3 Ubiquitin-Protein Ligases in Nuclear Protein Quality Control
2.6. Ubr1 in Nuclear Protein Quality Control
2.7. Misfolded Cytosolic Proteins Are Transported to the Nucleus for Degradation
3. Concluding Remarks
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hartl, F.U.; Hayer-Hartl, M. Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 2009, 16, 574–581. [Google Scholar]
- Powers, E.T.; Morimoto, R.I.; Dillin, A.; Kelly, J.W.; Balch, W.E. Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem. 2009, 78, 959–991. [Google Scholar]
- Kettern, N.; Dreiseidler, M.; Tawo, R.; Hohfeld, J. Chaperone-assisted degradation: Multiple paths to destruction. Biol. Chem. 2010, 391, 481–489. [Google Scholar]
- Esser, C.; Alberti, S.; Hohfeld, J. Cooperation of molecular chaperones with the ubiquitin/proteasome system. Biochim. Biophys. Acta 2004, 1695, 171–188. [Google Scholar]
- Kriegenburg, F.; Poulsen, E.G.; Koch, A.; Kruger, E.; Hartmann-Petersen, R. Redox control of the ubiquitin-proteasome system: From molecular mechanisms to functional significance. Antioxid. Redox Signal. 2011, 15, 2265–2299. [Google Scholar] [CrossRef]
- Rubinsztein, D.C. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006, 443, 780–786. [Google Scholar] [CrossRef]
- Vembar, S.S.; Brodsky, J.L. One step at a time: Endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008, 9, 944–957. [Google Scholar] [CrossRef]
- Meacham, G.C.; Patterson, C.; Zhang, W.; Younger, J.M.; Cyr, D.M. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat. Cell Biol. 2001, 3, 100–105. [Google Scholar] [CrossRef]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef]
- Van Wijk, S.J.; Timmers, H.T. The family of ubiquitin-conjugating enzymes (E2s): Deciding between life and death of proteins. FASEB J. 2010, 24, 981–993. [Google Scholar] [CrossRef]
- Deshaies, R.J.; Joazeiro, C.A. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009, 78, 399–434. [Google Scholar] [CrossRef]
- Rosenbaum, J.C.; Fredrickson, E.K.; Oeser, M.L.; Garrett-Engele, C.M.; Locke, M.N.; Richardson, L.A.; Nelson, Z.W.; Hetrick, E.D.; Milac, T.I.; Gottschling, D.E.; et al. Disorder targets misorder in nuclear quality control degradation: A disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol. Cell 2011, 41, 93–106. [Google Scholar] [CrossRef]
- Kriegenburg, F.; Ellgaard, L.; Hartmann-Petersen, R. Molecular chaperones in targeting misfolded proteins for ubiquitin-dependent degradation. FEBS J. 2012, 279, 532–542. [Google Scholar] [CrossRef]
- Arndt, V.; Rogon, C.; Hohfeld, J. To be, or not to be—Molecular chaperones in protein degradation. Cell. Mol. Life Sci. 2007, 64, 2525–2541. [Google Scholar] [CrossRef]
- Qian, S.B.; McDonough, H.; Boellmann, F.; Cyr, D.M.; Patterson, C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 2006, 440, 551–555. [Google Scholar] [CrossRef]
- Hartmann-Petersen, R.; Seeger, M.; Gordon, C. Transferring substrates to the 26S proteasome. Trends Biochem. Sci. 2003, 28, 26–31. [Google Scholar] [CrossRef]
- Stolz, A.; Hilt, W.; Buchberger, A.; Wolf, D.H. Cdc48: A power machine in protein degradation. Trends Biochem. Sci. 2011, 36, 515–523. [Google Scholar] [CrossRef]
- Finley, D. Misfolded proteins driven to destruction by Hul5. Nat. Cell Biol. 2011, 13, 1290–1292. [Google Scholar] [CrossRef]
- Crosas, B.; Hanna, J.; Kirkpatrick, D.S.; Zhang, D.P.; Tone, Y.; Hathaway, N.A.; Buecker, C.; Leggett, D.S.; Schmidt, M.; King, R.W.; et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 2006, 127, 1401–1413. [Google Scholar] [CrossRef]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef]
- Hanna, J.; Finley, D. A proteasome for all occasions. FEBS Lett. 2007, 581, 2854–2861. [Google Scholar] [CrossRef]
- Mazumdar, T.; Gorgun, F.M.; Sha, Y.; Tyryshkin, A.; Zeng, S.; Hartmann-Petersen, R.; Jorgensen, J.P.; Hendil, K.B.; Eissa, N.T. Regulation of NF-κB activity and inducible nitric oxide synthase by regulatory particle non-ATPase subunit 13 (Rpn13). Proc. Natl. Acad. Sci. USA 2010, 107, 13854–13859. [Google Scholar] [CrossRef]
- Verma, R.; Aravind, L.; Oania, R.; McDonald, W.H.; Yates, J.R., III; Koonin, E.V.; Deshaies, R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 2002, 298, 611–615. [Google Scholar] [CrossRef]
- Mao, P.; Smerdon, M.J. Yeast deubiquitinase Ubp3 interacts with the 26S proteasome to facilitate Rad4 degradation. J. Biol. Chem. 2010, 285, 37542–37550. [Google Scholar] [CrossRef]
- Sontag, E.M.; Vonk, W.I.; Frydman, J. Sorting out the trash: The spatial nature of eukaryotic protein quality control. Curr. Opin. Cell Biol. 2014, 26, 139–146. [Google Scholar] [CrossRef]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. Biochim. Biophys. Acta 2014, 1843, 182–196. [Google Scholar] [CrossRef]
- Park, S.H.; Kukushkin, Y.; Gupta, R.; Chen, T.; Konagai, A.; Hipp, M.S.; Hayer-Hartl, M.; Hartl, F.U. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell 2013, 154, 134–145. [Google Scholar] [CrossRef]
- Brooks, P.; Fuertes, G.; Murray, R.Z.; Bose, S.; Knecht, E.; Rechsteiner, M.C.; Hendil, K.B.; Tanaka, K.; Dyson, J.; Rivett, J. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 2000, 346, 155–161. [Google Scholar] [CrossRef]
- Wilkinson, C.R.; Wallace, M.; Morphew, M.; Perry, P.; Allshire, R.; Javerzat, J.P.; McIntosh, J.R.; Gordon, C. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998, 17, 6465–6476. [Google Scholar] [CrossRef]
- Mishra, A.; Godavarthi, S.K.; Maheshwari, M.; Goswami, A.; Jana, N.R. The ubiquitin ligase E6-AP is induced and recruited to aggresomes in response to proteasome inhibition and may be involved in the ubiquitination of Hsp70-bound misfolded proteins. J. Biol. Chem. 2009, 284, 10537–10545. [Google Scholar]
- Westhoff, B.; Chapple, J.P.; van der Spuy, J.; Hohfeld, J.; Cheetham, M.E. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr. Biol. 2005, 15, 1058–1064. [Google Scholar]
- Demand, J.; Alberti, S.; Patterson, C.; Hohfeld, J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001, 11, 1569–1577. [Google Scholar]
- Alberti, S.; Demand, J.; Esser, C.; Emmerich, N.; Schild, H.; Hohfeld, J. Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 2002, 277, 45920–45927. [Google Scholar]
- Kriegenburg, F.; Jakopec, V.; Poulsen, E.G.; Nielsen, S.V.; Roguev, A.; Krogan, N.; Gordon, C.; Fleig, U.; Hartmann-Petersen, R. A chaperone-assisted degradation pathway targets kinetochore proteins to ensure genome stability. PLoS Genet. 2014, 10, e1004140. [Google Scholar]
- Seeger, M.; Hartmann-Petersen, R.; Wilkinson, C.R.; Wallace, M.; Samejima, I.; Taylor, M.S.; Gordon, C. Interaction of the anaphase-promoting complex/cyclosome and proteasome protein complexes with multiubiquitin chain-binding proteins. J. Biol. Chem. 2003, 278, 16791–16796. [Google Scholar] [CrossRef]
- Connell, P.; Ballinger, C.A.; Jiang, J.; Wu, Y.; Thompson, L.J.; Hohfeld, J.; Patterson, C. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 2001, 3, 93–96. [Google Scholar] [CrossRef]
- Dai, Q.; Qian, S.B.; Li, H.H.; McDonough, H.; Borchers, C.; Huang, D.; Takayama, S.; Younger, J.M.; Ren, H.Y.; Cyr, D.M.; et al. Regulation of the cytoplasmic quality control protein degradation pathway by BAG2. J. Biol. Chem. 2005, 280, 38673–38681. [Google Scholar] [CrossRef]
- Arndt, V.; Daniel, C.; Nastainczyk, W.; Alberti, S.; Hohfeld, J. BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP. Mol. Biol. Cell 2005, 16, 5891–5900. [Google Scholar] [CrossRef]
- Saito, Y.; Takeda, J.; Okada, M.; Kobayashi, J.; Kato, A.; Hirota, K.; Taoka, M.; Matsumoto, T.; Komatsu, K.; Isobe, T. The proteasome factor Bag101 binds to Rad22 and suppresses homologous recombination. Sci. Rep. 2013. [Google Scholar] [CrossRef]
- Xu, W.; Marcu, M.; Yuan, X.; Mimnaugh, E.; Patterson, C.; Neckers, L. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA 2002, 99, 12847–12852. [Google Scholar] [CrossRef]
- Gotz, R.; Kramer, B.W.; Camarero, G.; Rapp, U.R. BAG-1 haplo-insufficiency impairs lung tumorigenesis. BMC Cancer. 2004. [Google Scholar] [CrossRef]
- Gardner, R.G.; Nelson, Z.W.; Gottschling, D.E. Degradation-mediated protein quality control in the nucleus. Cell 2005, 120, 803–815. [Google Scholar] [CrossRef]
- Matsuo, Y.; Kishimoto, H.; Tanae, K.; Kitamura, K.; Katayama, S.; Kawamukai, M. Nuclear protein quality is regulated by the ubiquitin-proteasome system through the activity of Ubc4 and San1 in fission yeast. J. Biol. Chem. 2011, 286, 13775–13790. [Google Scholar]
- Bhattacharyya, J.; Das, K.P. Molecular chaperone-like properties of an unfolded protein, alpha(s)-casein. J. Biol. Chem. 1999, 274, 15505–15509. [Google Scholar] [CrossRef]
- Stromer, T.; Fischer, E.; Richter, K.; Haslbeck, M.; Buchner, J. Analysis of the regulation of the molecular chaperone Hsp26 by temperature-induced dissociation: The N-terminal domail is important for oligomer assembly and the binding of unfolding proteins. J. Biol. Chem. 2004, 279, 11222–11228. [Google Scholar] [CrossRef]
- Jaya, N.; Garcia, V.; Vierling, E. Substrate binding site flexibility of the small heat shock protein molecular chaperones. Proc. Natl. Acad. Sci. USA 2009, 106, 15604–15609. [Google Scholar] [CrossRef]
- Fredrickson, E.K.; Clowes Candadai, S.V.; Tam, C.H.; Gardner, R.G. Means of self-preservation: How an intrinsically disordered ubiquitin-protein ligase averts self-destruction. Mol. Biol. Cell 2013, 24, 1041–1052. [Google Scholar] [CrossRef]
- Gallagher, P.S.; Clowes Candadai, S.V.; Gardner, R.G. Requirement for Cdc48/p97 in nuclear protein quality control degradation varies with the substrate and correlates with substrate insolubility. J. Cell Sci. 2014, 127, 1980–1991. [Google Scholar] [CrossRef]
- Iwata, A.; Nagashima, Y.; Matsumoto, L.; Suzuki, T.; Yamanaka, T.; Date, H.; Deoka, K.; Nukina, N.; Tsuji, S. Intranuclear degradation of polyglutamine aggregates by the ubiquitin-proteasome system. J. Biol. Chem. 2009, 284, 9796–9803. [Google Scholar] [CrossRef]
- Leggett, D.S.; Hanna, J.; Borodovsky, A.; Crosas, B.; Schmidt, M.; Baker, R.T.; Walz, T.; Ploegh, H.; Finley, D. Multiple associated proteins regulate proteasome structure and function. Mol. Cell 2002, 10, 495–507. [Google Scholar] [CrossRef]
- Aviram, S.; Kornitzer, D. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol. Cell. Biol. 2010, 30, 985–994. [Google Scholar]
- Fang, N.N.; Ng, A.H.; Measday, V.; Mayor, T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat. Cell Biol. 2011, 13, 1344–1352. [Google Scholar] [CrossRef]
- Torres, E.M.; Dephoure, N.; Panneerselvam, A.; Tucker, C.M.; Whittaker, C.A.; Gygi, S.P.; Dunham, M.J.; Amon, A. Identification of aneuploidy-tolerating mutations. Cell 2010, 143, 71–83. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Chu, B.W.; Kovary, K.M.; Guillaume, J.; Chen, L.C.; Teruel, M.N.; Wandless, T.J. The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J. Biol. Chem. 2013, 288, 34575–34587. [Google Scholar] [CrossRef]
- Denic, V.; Quan, E.M.; Weissman, J.S. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell 2006, 126, 349–359. [Google Scholar] [CrossRef]
- Carvalho, P.; Goder, V.; Rapoport, T.A. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell 2006, 126, 361–373. [Google Scholar] [CrossRef]
- Swanson, R.; Locher, M.; Hochstrasser, M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001, 15, 2660–2674. [Google Scholar] [CrossRef]
- Ravid, T.; Kreft, S.G.; Hochstrasser, M. Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 2006, 25, 533–543. [Google Scholar] [CrossRef]
- Deng, M.; Hochstrasser, M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature 2006, 443, 827–831. [Google Scholar] [CrossRef]
- Furth, N.; Gertman, O.; Shiber, A.; Alfassy, O.S.; Cohen, I.; Rosenberg, M.M.; Doron, N.K.; Friedler, A.; Ravid, T. Exposure of bipartite hydrophobic signal triggers nuclear quality control of Ndc10 at the endoplasmic reticulum/nuclear envelope. Mol. Biol. Cell 2011, 22, 4726–4739. [Google Scholar] [CrossRef]
- Shiber, A.; Breuer, W.; Brandeis, M.; Ravid, T. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol. Biol. Cell 2013, 24, 2076–2087. [Google Scholar] [CrossRef]
- Zavacki, A.M.; Arrojo E Drigo, R.; Freitas, B.C.; Chung, M.; Harney, J.W.; Egri, P.; Wittmann, G.; Fekete, C.; Gereben, B.; Bianco, A.C. The E3 ubiquitin ligase TEB4 mediates degradation of type 2 iodothyronine deiodinase. Mol. Cell. Biol. 2009, 29, 5339–5347. [Google Scholar] [CrossRef]
- Sriramachandran, A.M.; Dohmen, R.J. SUMO-targeted ubiquitin ligases. Biochim. Biophys. Acta 2014, 1843, 75–85. [Google Scholar] [CrossRef]
- Tatham, M.H.; Matic, I.; Mann, M.; Hay, R.T. Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci. Signal. 2011. [Google Scholar] [CrossRef]
- Yang, L.; Mullen, J.R.; Brill, S.J. Purification of the yeast Slx5-Slx8 protein complex and characterization of its DNA-binding activity. Nucleic Acids Res. 2006, 34, 5541–5551. [Google Scholar] [CrossRef]
- Kosoy, A.; Calonge, T.M.; Outwin, E.A.; O’Connell, M.J. Fission yeast Rnf4 homologs are required for DNA repair. J. Biol. Chem. 2007, 282, 20388–20394. [Google Scholar] [CrossRef]
- Uzunova, K.; Gottsche, K.; Miteva, M.; Weisshaar, S.R.; Glanemann, C.; Schnellhardt, M.; Niessen, M.; Scheel, H.; Hofmann, K.; Johnson, E.S.; et al. Ubiquitin-dependent proteolytic control of SUMO conjugates. J. Biol. Chem. 2007, 282, 34167–34175. [Google Scholar] [CrossRef]
- Xie, Y.; Kerscher, O.; Kroetz, M.B.; McConchie, H.F.; Sung, P.; Hochstrasser, M. The yeast Hex3·Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J. Biol. Chem. 2007, 282, 34176–34184. [Google Scholar]
- Wang, Z.; Jones, G.M.; Prelich, G. Genetic analysis connects Slx5 and Slx8 to the SUMO pathway in Saccharomyces cerevisiae. Genetics 2006, 172, 1499–1509. [Google Scholar] [CrossRef]
- Wang, Z.; Prelich, G. Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol. Cell. Biol. 2009, 29, 1694–1706. [Google Scholar] [CrossRef]
- Plechanovova, A.; Jaffray, E.G.; McMahon, S.A.; Johnson, K.A.; Navratilova, I.; Naismith, J.H.; Hay, R.T. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat. Struct. Mol. Biol. 2011, 18, 1052–1059. [Google Scholar] [CrossRef]
- De Thé, H.; le Bras, M.; Lallemand-Breitenbach, V. The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 2012, 198, 11–21. [Google Scholar] [CrossRef]
- Bernardi, R.; Pandolfi, P.P. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007, 8, 1006–1016. [Google Scholar] [CrossRef]
- Tatham, M.H.; Geoffroy, M.C.; Shen, L.; Plechanovova, A.; Hattersley, N.; Jaffray, E.G.; Palvimo, J.J.; Hay, R.T. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 2008, 10, 538–546. [Google Scholar] [CrossRef]
- Lallemand-Breitenbach, V.; Jeanne, M.; Benhenda, S.; Nasr, R.; Lei, M.; Peres, L.; Zhou, J.; Zhu, J.; Raught, B.; de Thé, H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 2008, 10, 547–555. [Google Scholar]
- Zhang, X.W.; Yan, X.J.; Zhou, Z.R.; Yang, F.F.; Wu, Z.Y.; Sun, H.B.; Liang, W.X.; Song, A.X.; Lallemand-Breitenbach, V.; Jeanne, M.; et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science 2010, 328, 240–243. [Google Scholar]
- Jeanne, M.; Lallemand-Breitenbach, V.; Ferhi, O.; Koken, M.; le Bras, M.; Duffort, S.; Peres, L.; Berthier, C.; Soilihi, H.; Raught, B.; et al. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell 2010, 18, 88–98. [Google Scholar]
- Erker, Y.; Neyret-Kahn, H.; Seeler, J.S.; Dejean, A.; Atfi, A.; Levy, L. Arkadia, a novel SUMO-targeted ubiquitin ligase involved in PML degradation. Mol. Cell. Biol. 2013, 33, 2163–2177. [Google Scholar] [CrossRef]
- Bartel, B.; Wunning, I.; Varshavsky, A. The recognition component of the N-end rule pathway. EMBO J. 1990, 9, 3179–3189. [Google Scholar]
- Eisele, F.; Wolf, D.H. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 2008, 582, 4143–4146. [Google Scholar] [CrossRef]
- Heck, J.W.; Cheung, S.K.; Hampton, R.Y. Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc. Natl. Acad. Sci. USA 2010, 107, 1106–1111. [Google Scholar] [CrossRef]
- Prasad, R.; Kawaguchi, S.; Ng, D.T. A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell 2010, 21, 2117–2127. [Google Scholar] [CrossRef]
- Summers, D.W.; Wolfe, K.J.; Ren, H.Y.; Cyr, D.M. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One 2013, 8, e52099. [Google Scholar] [CrossRef]
- Guerriero, C.J.; Weiberth, K.F.; Brodsky, J.L. Hsp70 targets a cytoplasmic quality control substrate to the San1p ubiquitin ligase. J. Biol. Chem. 2013, 288, 18506–18520. [Google Scholar] [CrossRef]
- Kitamura, K.; Nakase, M.; Tohda, H.; Takegawa, K. The Ubiquitin ligase Ubr11 is essential for oligopeptide utilization in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2012, 11, 302–310. [Google Scholar] [CrossRef]
- Nillegoda, N.B.; Theodoraki, M.A.; Mandal, A.K.; Mayo, K.J.; Ren, H.Y.; Sultana, R.; Wu, K.; Johnson, J.; Cyr, D.M.; Caplan, A.J. Ubr1 and Ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 2010, 21, 2102–2116. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nielsen, S.V.; Poulsen, E.G.; Rebula, C.A.; Hartmann-Petersen, R. Protein Quality Control in the Nucleus. Biomolecules 2014, 4, 646-661. https://doi.org/10.3390/biom4030646
Nielsen SV, Poulsen EG, Rebula CA, Hartmann-Petersen R. Protein Quality Control in the Nucleus. Biomolecules. 2014; 4(3):646-661. https://doi.org/10.3390/biom4030646
Chicago/Turabian StyleNielsen, Sofie V., Esben G. Poulsen, Caio A. Rebula, and Rasmus Hartmann-Petersen. 2014. "Protein Quality Control in the Nucleus" Biomolecules 4, no. 3: 646-661. https://doi.org/10.3390/biom4030646
APA StyleNielsen, S. V., Poulsen, E. G., Rebula, C. A., & Hartmann-Petersen, R. (2014). Protein Quality Control in the Nucleus. Biomolecules, 4(3), 646-661. https://doi.org/10.3390/biom4030646





