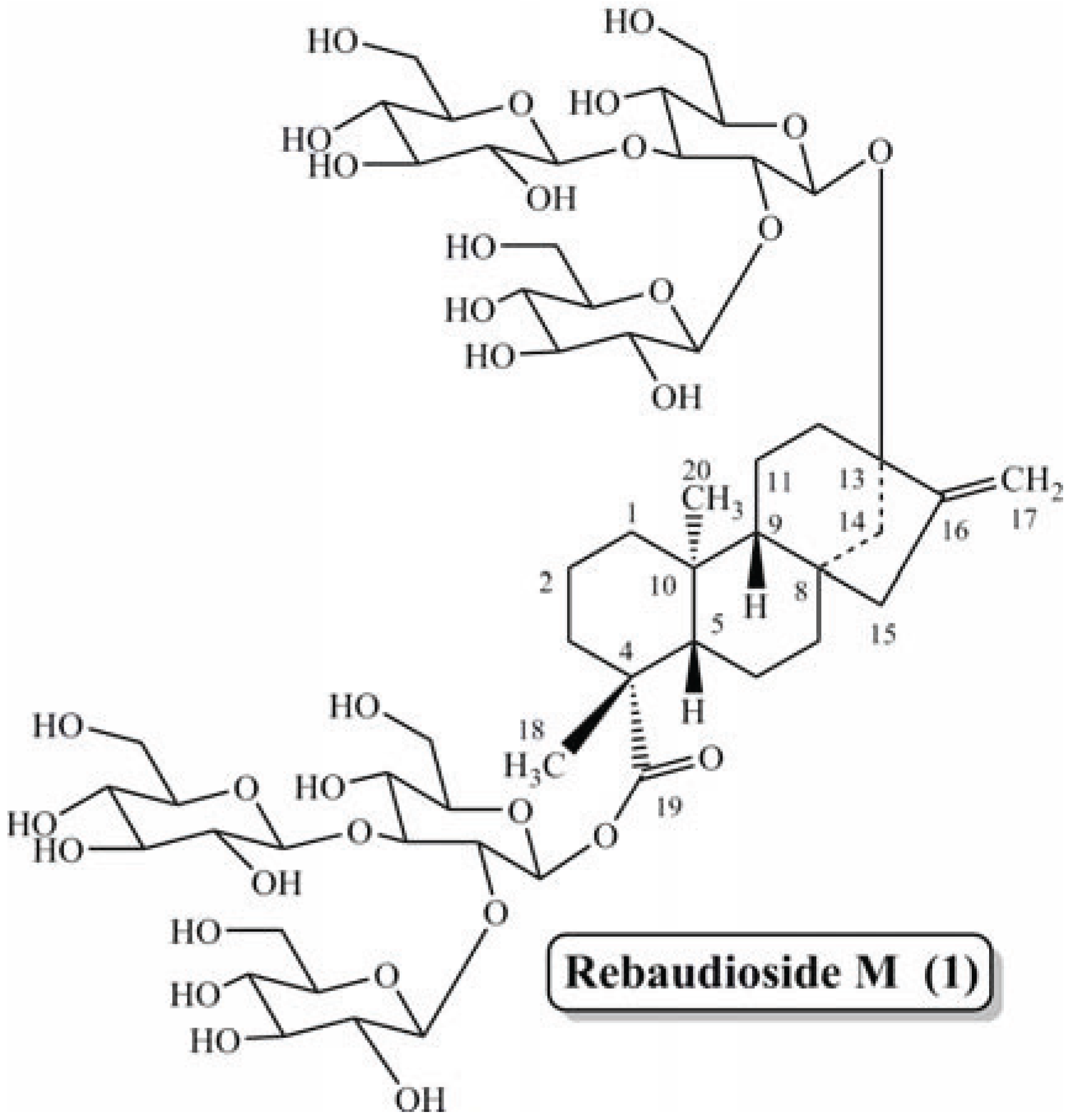
Isolation and Characterization of a Novel Rebaudioside M Isomer from a Bioconversion Reaction of Rebaudioside A and NMR Comparison Studies of Rebaudioside M Isolated from Stevia rebaudiana Bertoni and Stevia rebaudiana Morita
Abstract
:1. Introduction

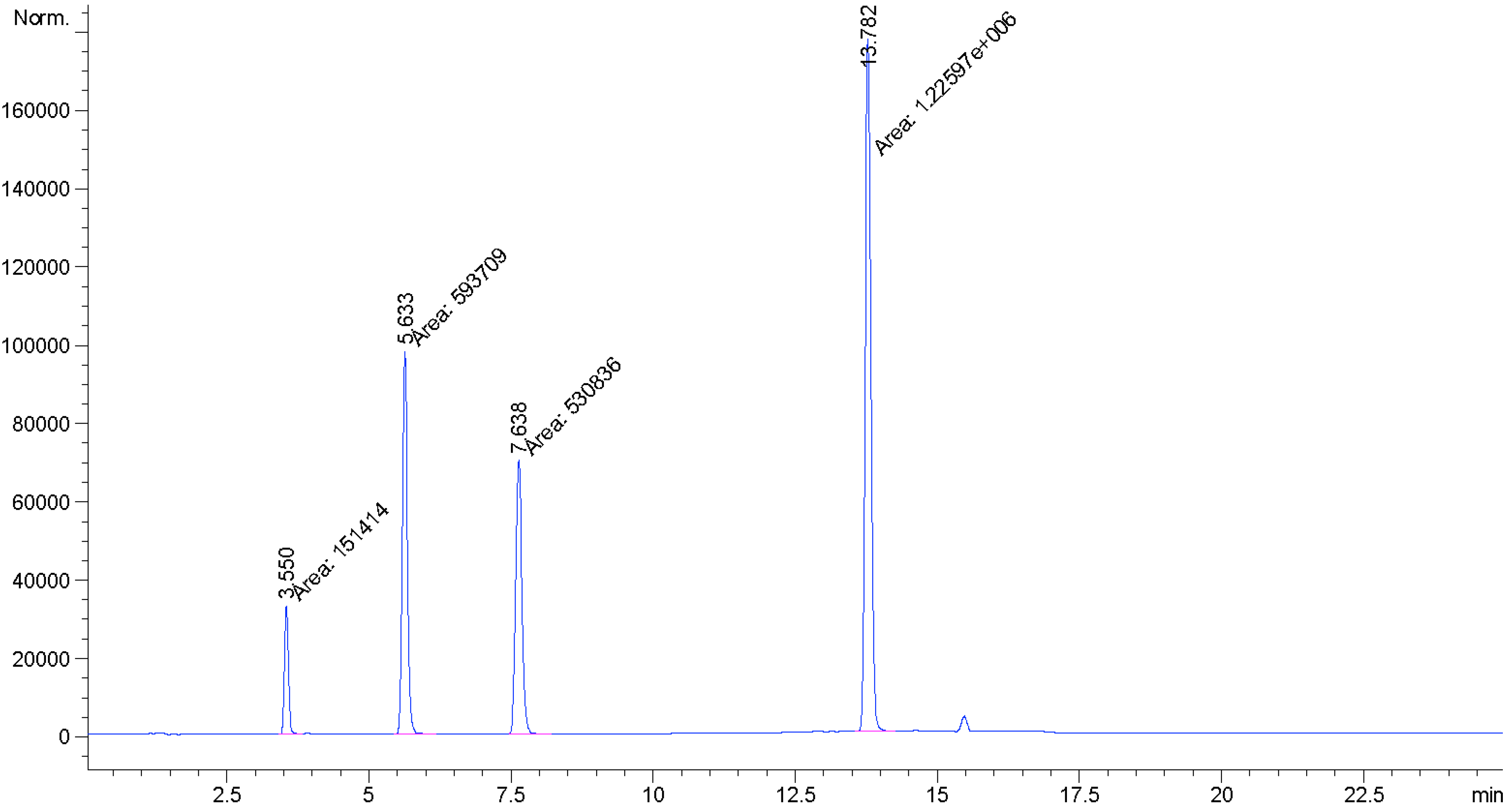
| Compound | Retention Time | MW | Peak Area |
|---|---|---|---|
| Rebaudioside M2 (2) | 3.550 | 1291 | 151,414 |
| Rebaudioside D (3) | 5.633 | 1129 | 593,709 |
| Unknown I | 7.638 | 1129 | 530,836 |
| Rebaudioside A (4) | 13.782 | 967 | 1,225,970 |
2. Results and Discussion
2.1. Rebaudioside M2 (2)
| Sugar | Position | 1H-NMR | 13C-NMR |
|---|---|---|---|
| 1 | 0.93 m,1.93 m | 41.9 | |
| 2 | 1.49 m, 1.86 m | 21.8 | |
| 3 | 1.16 m, 2.28 d (13.4) | 39.8 | |
| 4 | - | 43.7 | |
| 5 | 1.24 d (12.1) | 59.2 | |
| 6 | 1.73 m, 1.94 m | 24.4 | |
| 7 | 1.49 m, 1.56 m | 44.2 | |
| 8 | - | 46.9 | |
| 9 | 1.09 d (7.7) | 55.5 | |
| 10 | - | 42.4 | |
| 11 | 1.66 m, 1.70 m | 22.6 | |
| 12 | 1.60 m, 2.00 m | 39.9 | |
| 13 | - | 90.9 | |
| 14 | 1.53 d (12.6), 2.21 d (13.6) | 46.9 | |
| 15 | 2.15 d (17.2), 2.18 d (18.1) | 49.4 | |
| 16 | - | 164.0 | |
| 17 | 4.98 s, 5.16 s | 107.0 | |
| 18 | 1.29 s | 31.0 | |
| 19 | - | 181.5 | |
| 20 | 0.92 s | 19.1 | |
| I | 1' | 5.65 d (7.6) | 95.5 |
| 2' | 3.96 m | 80.5 | |
| 3' | 3.89 m | 79.0 | |
| 4' | 3.71 m | 71.5 | |
| 5' | 3.73 m | 79.0 | |
| 6' | 4.00 m, 4.15 d (11.7) | 70.9 | |
| II | 1'' | 4.85 d (7.8) | 98.4 |
| 2'' | 3.75 m | 81.7 | |
| 3'' | 3.98 m | 88.0 | |
| 4'' | 3.54 m | 71.3 | |
| 5'' | 3.96 m | 80.5 | |
| 6'' | 3.45 m, 3.77 m | 63.6 | |
| III | 1''' | 4.92 d (7.9) | 104.9 |
| 2''' | 3.32 m | 76.3 | |
| 3''' | 3.51 m | 78.8 | |
| 4''' | 3.26 t (9.5) | 73.3 | |
| 5''' | 3.44 m | 78.8 | |
| 6''' | 3.75 m, 3.94 m | 64.4 | |
| IV | 1'''' | 4.84 * d (7.8) | 105.0 * |
| 2'''' | 3.41 m | 76.1 | |
| 3'''' | 3.46 m | 78.8 | |
| 4'''' | 3.45 m | 72.5 | |
| 5'''' | 3.75 m | 81.7 | |
| 6'''' | 3.55 m, 3.78 m | 65.8 | |
| V | 1''''' | 4.83 * d (8.0) | 105.3 * |
| 2''''' | 3.32 m | 78.5 | |
| 3''''' | 3.51 m | 78.7 | |
| 4''''' | 3.38 m | 72.9 | |
| 5''''' | 3.55 m | 78.8 | |
| 6''''' | 3.76 m, 3.97 m | 63.6 | |
| VI | 1'''''' | 4.50 d (7.9) | 105.7 |
| 2'''''' | 3.33 m | 78.1 | |
| 3'''''' | 3.49 m | 78.6 | |
| 4'''''' | 3.45 m | 72.3 | |
| 5'''''' | 3.48 m | 78.8 | |
| 6'''''' | 3.92 m, 3.94 m | 64.1 |
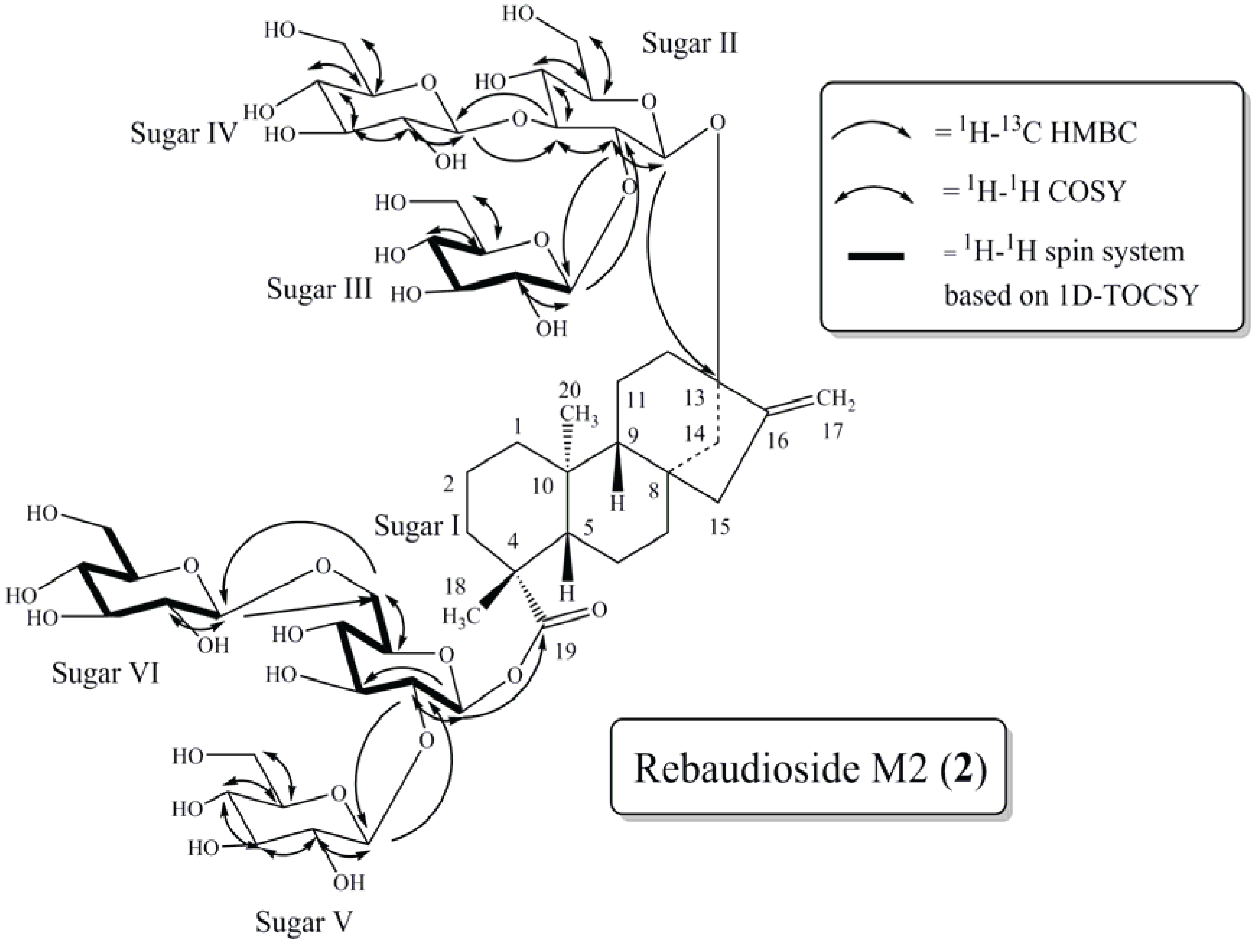
2.2. NMR Study of Rebaudioside M, Rebaudioside D and SG17
| Position No. | Exp. 1 13C Assignment for >95% Reb M (1) in Pyr-d5 + TMS | Exp. 2 13C Assignment for >95% Reb D (3) in Pyr-d5 + TMS | Exp. 3 13C Assignment for (1) 82% + (3) (18%) in Pyr-d5 + TMS | Exp. 4 13C Assignment for (3) 82% + (1) 18% in Pyr-d5 + TMS | Exp. 5 13C Assignment for 80% Reb M (1) in Pyr-d5 + TMS | 13C Assignment [24] in Pyr-d5 + TMS for SG17 | ||
|---|---|---|---|---|---|---|---|---|
| Reb M | Reb D | Reb D | Reb M | |||||
| 1 | 40.3 | 40.6 | 40.3 | 40.6 | 40.6 | 40.3 | 40.3 | 40.5 |
| 2 | 19.6 | 20.0 | 19.6 | 20.0 | 20.0 | 19.6 | 19.6 | 19.8 |
| 3 | 38.4 | 37.8 | 38.4 | 37.8 | 37.8 | 38.4 | 38.4 | 38.3 |
| 4 | 44.2 | 44.3 | 44.2 | 44.3 | 44.3 | 44.3 | 44.2 | 44.3 |
| 5 | 57.3 | 57.4 | 57.3 | 57.4 | 57.4 | 57.3 | 57.3 | 57.3 |
| 6 | 23.4 | 22.2 * | 23.4 | 22.2 | 22.2 | 23.4 | 23.4 | 22.2 * |
| 7 | 42.5 | 42.2 | 42.5 | 42.2 | 42.2 | 42.5 | 42.5 | 41.1 * |
| 8 | 41.2 | 41.8 | 41.1 | 41.8 | 41.8 | 41.1 | 41.2 | 42.1 * |
| 9 | 54.2 | 53.9 * | 54.2 | 53.9 | 53.9 | 54.2 | 54.2 | 53.9 * |
| 10 | 39.7 | 39.7 | 39.7 | 39.7 | 39.7 | 39.7 | 39.7 | 39.7 |
| 11 | 20.2 | 20.5 * | 20.2 | 20.5 | 20.5 | 20.2 | 20.2 | 20.5 * |
| 12 | 38.4 | 37.8 * | 38.4 | 37.8 | 37.8 | 38.4 | 38.4 | 37.8 * |
| 13 | 87.6 | 86.7 * | 87.6 | 86.7 | 86.7 | 87.5 | 87.6 | 86.6 * |
| 14 | 43.3 | 44.1 * | 43.3 | 44.1 | 44.1 | 43.3 | 43.3 | 44.2 * |
| 15 | 46.5 | 47.7 * | 46.5 | 47.7 | 47.7 | 46.4 | 46.5 | 47.6 * |
| 16 | 153.2 | 154.0 * | 153.3 | 154.0 | 154.0 | 153.3 | 153.2 | 154.0 * |
| 17 | 104.9 | 104.8 | 104.9 | 104.8 | 104.8 | 104.9 | 104.9 | 104.0 |
| 18 | 28.2 | 29.2 * | 28.2 | 29.2 | 29.2 | 28.2 | 28.2 | 29.3 * |
| 19 | 176.9 | 175.8 * | 176.9 | 175.8 | 175.8 | 176.8 | 176.9 | 175.7 * |
| 20 | 16.7 | 16.8 | 16.7 | 16.8 | 16.8 | 16.7 | 16.7 | 16.8 |
| C-1' | 94.9 | 93.6 * | 94.9 | 93.6 | 93.6 | 94.9 | 94.9 | 93.6 * |
| C-1'' | 96.2 | 97.8 * | 96.2 | 97.8 | 97.8 | 96.2 | 96.2 | 97.8 * |
| C-1''' | 104.7 | 104.5 | 104.8 | 104.5 | 104.5 | 104.8 | 104.7 | 104.6 |
| C-1'''' | 103.9 | 104.7 * | 103.9 | 104.5 | 104.7 | 103.9 | 103.9 | 104.7 * |
| C-1''''' | 104.1 | 105.7 | 104.1 | 105.7 | 105.7 | 104.2 | 104.1 | 104.1 |
| C-1'''''' | 104.1 | NA | 104.1 | NA | NA | 104.1 | 104.1 | 105.1 |
| Position No. | Exp. 1 1H Assignment for >95% Rebaudioside M (1) in Pyr-d5 + TMS | Exp. 2 1H Assignment for >95% Rebaudioside D (3) in Pyr-d5 + TMS | 1H Assignment [24] in Pyr-d5 + TMS for SG17 |
|---|---|---|---|
| 1 | 0.77 t (11.5) | 0.75 t (11.4) | NR |
| 1.78 m | 1.72 m | ||
| 2 | 1.39 m | 1.44 m | NR |
| 2.27 m | 2.16 m | ||
| 3 | 1.04 m | 1.10 m | NR |
| 2.32 d (13.7) | 2.72 d (13.0) | ||
| 4 | - | - | NR |
| 5 | 1.08 d (13.4) | 1.00 d (13.0) | NR |
| 6 | 2.25 m | 1.90 m | NR |
| 2.42 q (12.9) | 2.18 m | ||
| 7 | 1.44 m | 1.29 m | NR |
| 1.82 m | 1.42 m | ||
| 8 | - | - | NR |
| 9 | 0.93 d (7.8) | 0.90 d (6.9) | NR |
| 10 | - | - | NR |
| 11 | 1.67 m | 1.67 m | NR |
| 1.78 m | 1.70 m | ||
| 12 | 1.88 m | 1.91 m | NR |
| 2.74 m | 2.24 m | ||
| 13 | - | - | NR |
| 14 | 2.04 m | 1.79 d (12.3) | NR |
| 2.75 m | 2.53 d (10.9) | ||
| 15 | 1.91 d (17.7) | 2.02 m | NR |
| 2.06 m | 2.06 m | ||
| 16 | - | - | NR |
| 17 | 4.92 s | 5.05 s | NR |
| 5.71 s | 5.68 s | ||
| 18 | 1.35 s | 1.42 s | NR |
| 19 | - | - | NR |
| 20 | 1.39 s | 1.17 s | NR |
| H-1' | 6.40 d (8.3) | 6.32 d (7.6) * | 6.33 d (7.4) * |
| H-1'' | 5.47 d (7.9) | 5.10 d (7.5) * | 5.07 d (7.8) * |
| H-1''' | 5.50 d (7.5) | 5.58 d (7.8) | 5.41 d (7.8) |
| H-1'''' | 5.47 d (7.9) | 5.40 d (7.9) | 5.59 d (7.5) |
| H-1''''' | 5.82 d (7.4) | 5.48 d (7.7) * | 5.48 d (7.7) * |
| H-1'''''' | 5.33 d (8.0) | NA | 5.33 d (7.7) |
3. Experimental
3.1. General Experimental Procedures for Rebaudioside M2 (2)
3.1.1. Isolation and Purification
3.1.2. Mass Spectrometry
3.1.3. Nuclear Magnetic Resonance
3.2. Material Sources
3.3. Bioconversion Reaction
3.4. General Experimental Procedures for Rebaudioside D (3)
3.4.1. Isolation and Purification
3.4.2. Mass Spectrometry
3.4.3. Nuclear Magnetic Resonance
4. Conclusions
Supplementary Materials
Supplementary File 1Acknowledgements
Author Contributions
Conflicts of Interest
References
- Ahmed, J.; Preissner, S.; Dunkel, M.; Worth, C.L.; Eckert, A.; Preissner, R. SuperSweet—A resource on natural and artificial sweetening agents. Nucleic Acid Res. 2011, 39, D377–D382. [Google Scholar]
- Priya, K.; Gupta, V.R.M.; Srikanth, K. Natural sweeteners: A complete review. J. Pharm. Res. 2011, 4, 2034–2039. [Google Scholar]
- Weerasinghe, D.P.; DuBois, G.E. Sweetener and sweetness modulators: Requirements for commercial viability. In Sweetness and Sweeteners: Biology, Chemistry, and Psychophysics; Weerasinghe, D.P., DuBois, G.E., Eds.; Oxford University Press: Danvers, MA, USA, 2008; pp. 444–462. [Google Scholar]
- DuBois, G.E.; Prakash, I. Non-caloric sweeteners, sweetness modulators, and sweetener enhancers. Annu. Rev. Food Sci. Technol. 2012, 3, 353–380. [Google Scholar] [CrossRef]
- Prakash, I.; DuBois, G.E.; Clos, J.F.; Wilkens, K.L.; Fosdick, L.E. Development of rebiana, a natural, non-caloric sweetener. Food Chem. Toxicol. 2008, 46, S75–S82. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. A new diterpene glycoside from Stevia rebaudiana. Molecules 2011, 15, 2937–2943. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Prakash, I. Structures of the novel diterpene glycosides from Stevia rebaudiana. Carbohydr. Res. 2011, 34, 1057–1060. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Rhea, J.; Milanowski, D.; Mocek, U.; Prakash, I. Two minor diterpenoid glycosides from the leaves of Stevia rebaudiana. Nat. Prod. Commun. 2011, 6, 175–178. [Google Scholar]
- Chaturvedula, V.S.P.; Clos, J.F.; Rhea, J.; Milanowski, D.; Mocek, U.; DuBois, G.E.; Prakash, I. Minor diterpene glycosides from the leaves of Stevia rebaudiana. Phytochem. Lett. 2011, 4, 209–212. [Google Scholar] [CrossRef]
- Chaturvedula, V.S.P.; Mani, U.; Prakash, I. Structures of the novel α-glucosyl linked diterpene glycosides from Stevia rebaudiana. Carbohydr. Res. 2011, 346, 2034–2038. [Google Scholar] [CrossRef]
- Prakash, I.; Campbell, M.; San Miguel, R.I.; Chaturvedula, V.S.P. Synthesis and sensory evaluation of ent-Kaurane diterpene glycosides. Molecules 2012, 17, 8908–8916. [Google Scholar] [CrossRef]
- Prakash, I.; Campbell, M.; San Miguel, R.I.; Chaturvedula, V.S.P. Catalytic hydrogenation of the sweet principles of Stevia rebaudiana, rebaudiosde B, rebaudioside C and rebaudioside D and sensory evaluation of their reduced derivatives. Int. J. Mol. Sci. 2013, 13, 15126–15136. [Google Scholar]
- Brandle, J.E.; Starratt, A.N.; Gijzen, M. Stevia rebaudiana: Its agricultural, biological, and chemical properties. Can. J. Plant Sci. 1998, 78, 527–536. [Google Scholar] [CrossRef]
- Lewis, W.H. Early uses of Stevia rebaudiana (Asteraceae) leaves as a sweetener in Paraguay. Econ. Bot. 1992, 46, 336–337. [Google Scholar] [CrossRef]
- Madan, S.; Ahmad, S.; Singh, G.N.; Kohli, K.; Kumar, Y.; Singh, R.; Garg, M. Stevia rebaudiana (Bert.) Bertoni—A review. Indian J. Nat. Prod. Resour. 2010, 1, 267–286. [Google Scholar]
- Kinghorn, A.A.D.; Soejarto, D.D.; Nanayakkara, N.P.D.; Compadre, C.M.; Makapugay, H.C.; Hovanec-Brown, J.M.; Medon, P.J.; Kamath, S.K. A phytochemical screening procedure for sweet ent-Kaurene glycosides in the genus Stevia. J. Nat. Prod. 1984, 47, 439–444. [Google Scholar] [CrossRef]
- Staratt, A.N.; Kirby, C.W.; Pocs, R.; Brandle, J.E. Rebaudioside F, a diterepene glycoside from Stevia rebaudiana. Photochemistry 2002, 59, 367–370. [Google Scholar] [CrossRef]
- Sakamoto, I.; Yamasaki, K.; Tanaka, O. Application of 13C-NMR spectroscopy to chemistry of natural glycosides: Rebaudioside-C, a new sweet diterpene glycosides of Stevia rebaudiana. Chem. Pharm. Bull. 1977, 25, 844–846. [Google Scholar] [CrossRef]
- Sakamoto, I.; Yamasaki, K.; Tanaka, O. Application of 13C-NMR spectroscopy to chemistry of plant glycosides: Rebaudioside-D and -E, new sweet diterpene-glucosides of Stevia rebaudiana Bertoni. Chem. Pharm. Bull. 1977, 25, 3437–3439. [Google Scholar] [CrossRef]
- Steinmetz, W.E.; Lin, A. NMR studies of the confirmation of the natural sweetener rebaudioside A. Carbohydr. Res. 2009, 344, 2533–2538. [Google Scholar] [CrossRef]
- Prakash, I.; Chaturvedula, V.S.P.; Markosyan, A. Isolation, characterization and sensory evaluation of a Hexa β-d-glucopyranosyl diterpene from Stevia rebaudiana. Nat. Prod. Commun. 2013, 8, 1523–1526. [Google Scholar]
- Prakash, I.; Markosyan, A.; Chaturvedula, V.S.P.; Campbell, M.; San Miguel, R.I.; Purkayastha, S.; Johnson, M. Methods for purifying steviol glycosides. PCT Patent Application WO 2013/096420, 27 June 2013. [Google Scholar]
- Agency Response Letter GRAS Notice No. GRN 000473. Purified Steviol Glycosides with Rebaudioside X (also Known as Rebaudioside M) as the Principal Component. Available online: http://www.fda.gov/Food/IngredientsPackagingLabeling/GRAS/NoticeInventory/ucm382202 (accessed on 1 June 2013).
- Ohta, M.; Sasa, S.; Inoue, A.; Tamai, T.; Fujita, I.; Morita, K.; Matsuura, F. Characterization of novel steviol glycosides from leaves of Stevia rebaudiana Morita. J. Appl. Glycosci. 2010, 57, 199–209. [Google Scholar] [CrossRef]
- Morita, T.; Isao, F.; Fumito, M.; Masaya, O. New Steviol Glycoside. U.S. Patent Application No. 2011/0183056, 28 July 2011. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Prakash, I.; Bunders, C.; Devkota, K.P.; Charan, R.D.; Ramirez, C.; Priedemann, C.; Markosyan, A. Isolation and Characterization of a Novel Rebaudioside M Isomer from a Bioconversion Reaction of Rebaudioside A and NMR Comparison Studies of Rebaudioside M Isolated from Stevia rebaudiana Bertoni and Stevia rebaudiana Morita. Biomolecules 2014, 4, 374-389. https://doi.org/10.3390/biom4020374
Prakash I, Bunders C, Devkota KP, Charan RD, Ramirez C, Priedemann C, Markosyan A. Isolation and Characterization of a Novel Rebaudioside M Isomer from a Bioconversion Reaction of Rebaudioside A and NMR Comparison Studies of Rebaudioside M Isolated from Stevia rebaudiana Bertoni and Stevia rebaudiana Morita. Biomolecules. 2014; 4(2):374-389. https://doi.org/10.3390/biom4020374
Chicago/Turabian StylePrakash, Indra, Cynthia Bunders, Krishna P. Devkota, Romila D. Charan, Catherine Ramirez, Christopher Priedemann, and Avetik Markosyan. 2014. "Isolation and Characterization of a Novel Rebaudioside M Isomer from a Bioconversion Reaction of Rebaudioside A and NMR Comparison Studies of Rebaudioside M Isolated from Stevia rebaudiana Bertoni and Stevia rebaudiana Morita" Biomolecules 4, no. 2: 374-389. https://doi.org/10.3390/biom4020374
APA StylePrakash, I., Bunders, C., Devkota, K. P., Charan, R. D., Ramirez, C., Priedemann, C., & Markosyan, A. (2014). Isolation and Characterization of a Novel Rebaudioside M Isomer from a Bioconversion Reaction of Rebaudioside A and NMR Comparison Studies of Rebaudioside M Isolated from Stevia rebaudiana Bertoni and Stevia rebaudiana Morita. Biomolecules, 4(2), 374-389. https://doi.org/10.3390/biom4020374




